Abstract
The human cytomegalovirus-encoded US2 glycoprotein targets endoplasmic reticulum-resident major histocompatibility complex (MHC) class I heavy chains for rapid degradation by the proteasome. We demonstrate that the endoplasmic reticulum-lumenal domain of US2 allows tight interaction with class I molecules encoded by the HLA-A locus. Recombinant soluble US2 binds properly folded, peptide-containing recombinant HLA-A2 molecules in a peptide sequence-independent manner, consistent with US2's ability to broadly downregulate class I molecules. The physicochemical properties of the US2/MHC class I complex suggest a 1:1 stoichiometry. These results demonstrate that US2 does not require additional cellular proteins to specifically interact with soluble class I molecules. Binding of US2 does not significantly alter the conformation of class I molecules, as a soluble T-cell receptor can simultaneously recognize class I molecules associated with US2. The lumenal domain of US2 can differentiate between the products of distinct class I loci, as US2 binds several HLA-A locus products while being unable to bind recombinant HLA-B7, HLA-B27, HLA-Cw4, or HLA-E. We did not observe interaction between soluble US2 and either recombinant HLA-DR1 or recombinant HLA-DM. The substrate specificity of US2 may help explain the presence in human cytomegalovirus of multiple strategies for downregulation of MHC class I molecules.
The human cytomegalovirus (HCMV) is a ubiquitous betaherpesvirus that causes persistent infection following primary exposure. Infectious HCMV is secreted for extended periods from numerous sites upon primary infection, and in common with other known human herpesviruses, HCMV establishes latency (4). HCMV can reactivate from latent infection of myeloid lineage cells years after acute infection, even in a fully primed immunocompetent host (28). This lifestyle requires HCMV to disarm several components of the immune response, in particular the major histocompatibility complex (MHC) class I antigen presentation pathway.
MHC class I molecules are heterotrimeric complexes composed of a polymorphic heavy chain, the invariant β2-microglobulin light chain (β2m), and an antigenic peptide (34). Proteasomal cleavage of cellular and microbial cytosolic proteins yields peptides, some of which are delivered to the endoplasmic reticulum (ER) by the transporter associated with antigen processing and presentation (37). Within the ER, chaperones facilitate peptide loading of empty class I molecules (6). Upon receipt of peptide, class I molecules travel to the cell surface to present the cargo to CD8+ T cells (13).
Although CD8+ cytotoxic T lymphocytes respond to peptides derived from several HCMV immediate-early virus gene products, it has been difficult to find cytotoxic T lymphocytes specific for the more abundant and diverse set of proteins expressed later in the virus life cycle (3, 26). The concerted action of a series of glycoproteins encoded by HCMV unique short (US) genes disrupts surface expression of class I molecules and likely contributes to this absence of CD8+ T-cell recognition (17). Indeed, HCMV prevents the production and display of antigenic peptides by several seemingly independent mechanisms (32). The US3 glycoprotein, whose ER-lumenal domain shares approximately 20% sequence identity with US2, retains class I molecules in the ER during the immediate-early period of virus infection (1, 18). The US3 ER-lumenal and transmembrane domains are each required for association with class I molecules (20). During the early period of virus infection, the ER-resident glycoproteins US2 and US11 independently bind to newly synthesized MHC class I molecules in the ER and redirect the class I heavy chains to the cytosol, via the Sec61 translocon. Within the cytosol, the class I heavy chains are rapidly deglycosylated and degraded by the proteasome (35, 36). The dislocation reaction is sensitive to changes in redox conditions, and the cytoplasmic tail of the class I heavy chain is required for degradation (30, 33). However, the molecular details of how either US2 or US11 targets class I for destruction remain obscure.
A growing number of viruses are known to encode factors that downregulate the surface expression of class I molecules posttranslationally, including human immunodeficiency virus (HIV), Kaposi's sarcoma-associated herpesvirus (KSHV), adenovirus, murine cytomegalovirus, and murine gammaherpesvirus 68 (29, 32). Several of these virus gene products selectively target HLA-A and HLA-B locus products, including US2 and US11, KSHV K5, and HIV Nef. The locus specificity of the KSHV K5 product derives largely from interactions within the membrane (15), while HIV Nef relies on polymorphisms in the cytoplasmic tail of class I molecules to selectively interfere with class I presentation (5).
While HCMV interferes with transcription of MHC class II molecules (24), it has further been suggested that US2 might target MHC class II HLA-DR and HLA-DM for degradation (31). HLA-DM facilitates peptide loading of class II molecules, which present peptides derived from extracellular proteins to CD4+ T cells (16). Notwithstanding their similar folding patterns, the considerable sequence disparity between class I molecules and the DR or DM MHC class II complexes raises the question of how US2 can engage this rather divergent set of glycoproteins.
US2 is a 199-amino-acid membrane protein, with an ER-lumenal portion, a predicted single transmembrane domain, and a short cytoplasmic tail. The absence of a cleavable signal sequence at the N terminus is unusual for a glycoprotein that otherwise resembles a typical type I membrane protein. The regions of US2 that are responsible for class I degradation are not characterized, and we have so far failed to detect significant homologies between US2 and non-HCMV-encoded polypeptides. To better understand US2 function, we have reconstituted the interaction between recombinant forms of US2 and MHC class I. We show that the US2 ER-lumenal domain specifically interacts with class I molecules and enables a delineation of its class I binding preferences.
MATERIALS AND METHODS
Bacterial strains and constructs.
US2 amino-terminal and carboxy-terminal deletion mutants were constructed by PCR with Pfu DNA polymerase (Stratagene) using a US2 cDNA template. The 5′ oligonucleotide primer GGGAATTCCATATGGGTCCCTTGATCCGCCTGCC and the 3′ primer CCGCTCGAGTTAATCCACTCGCAGTTCGGGGACGC were used to amplify US215–140. PCR products were doubly digested using NdeI and XhoI (New England Biolabs) and ligated into the pET27 expression vector (Novagen). Full-length and additional deletion mutant constructs of US2 were also cloned into pET27 following PCR from a cDNA template.
The expression construct for soluble HLA-A2 class I heavy chain (amino acids G1 to E275) was constructed by PCR with Pfu DNA polymerase using a cDNA template. The 5′ oligonucleotide primer TGGGCTCTCACTCCATGAGGTATTTC and the 3′ primer CCGCTCGAGTTACTCCCATCTCAGGGTGAGGGGCT were used to amplify the heavy chain. The XhoI-digested PCR products were ligated into pET27, which had been cut with NdeI, blunted with Klenow DNA polymerase (New England Biolabs), and digested with XhoI. The HLA-E class I heavy chain (amino acids G1 to E275) was similarly amplified and cloned into pET27 using the 5′ primer GGAATTCCATATGGGCTCCCACTCCTTGAAGTATTTCC and the 3′ primer CCGCTCGAGTCACTTCCATCTCAGGGTGACGGGCTC. All constructs obtained from PCR amplification were sequenced to verify the absence of mutations.
BL21(DE3)plysS strains carrying expression constructs for either HLA-A68, HLA-B7, or HLA-B27 heavy chains or β2m were obtained from the laboratory of Don C. Wiley, Harvard University, Cambridge, Mass. Inclusion bodies of US2 constructs, HLA-A2, and HLA-E were produced in BL21(DE3).
Protein expression and inclusion body purification.
Single bacterial colonies containing either US2, HLA-A2, or HLA-E heavy chain constructs were grown at 37°C in Luria-Bertani medium containing 30 μg of kanamycin sulfate/ml. One hundred micrograms of ampicillin per milliliter was added to cultures expressing HLA-A68, HLA-B7, HLA-B27, HLA-B51, or HLA-Cw4 heavy chains or β2-microglobulin. Cultures were induced at an optical density at 600 nm of 0.6 with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Inclusion bodies were purified as described previously (9). Inclusion bodies were dissolved in 8 M urea–25 mM 2-(N-morpholino)ethanesulfonic acid (MES; pH 6.0)–10 mM EDTA–1 mM dithiothreitol (DTT) (20 mM DTT was added to US2 inclusion body resuspensions). HLA-A2–ELAGIGILTV and HLA-A2–ALGIGILTV peptide complexes were generous gifts from Olivier Michielin, Strasbourg, France. HLA-DM and HLA-DR1 were generous gifts from Don Wiley.
Renaturation by dilution.
A 7.15-mg quantity of US2 denatured in 8 M urea–25 mM MES (pH 6.5)–10 mM sodium EDTA–20 mM DTT was added to 7.5 ml of injection buffer (3 M guanidine HCl, 10 mM sodium acetate, pH 4.2). The US2 mixture was then injected through a 27-gauge needle into 500 ml of rapidly stirred 10°C refolding buffer (100 mM Tris HCl [pH 8.3], 2 mM EDTA, 400 mM l-arginine HCl, 5 mM reduced glutathione, 0.5 mM oxidized glutathione) at a final protein concentration of 1 mM. Phenylmethylsulfonyl fluoride (0.1 mM) in isopropanol was added immediately prior to addition of US2. Refolding reaction mixtures were incubated at >10°C. US2 (7.15 mg/liter) was added twice more at 6- to 12-h intervals to the refolding mixture, and the refolding reaction mixture was incubated for an additional 24 h.
Class I molecules were refolded as described previously (7, 9). HLA-A2 was refolded with either the human T-cell leukemia virus (HTLV) Tax peptide LLFGYPVYV, the HIV reverse transcriptase (polymerase [Pol]) peptide ILKEPVHGV, or the hepatitis B virus peptide FLPSDFFPSV; HLA-A*6801 was refolded with the influenza A virus matrix protein epitope KTGGPIYKR; HLA-B*0702 was refolded with nonamer peptide APRTVALTA; HLA-B27 was refolded with peptide GRIDKPILK; HLA-Cw4 was refolded using QYDDAVYKL; and HLA-E was refolded with the HLA-B8 leader epitope VMAPRTVLL.
Protein purification.
US2 refolding reaction mixtures were concentrated with a pressurized stirred cell (Amicon) across membranes with a 10,000-molecular-weight cutoff and further concentrated to a volume of 200 to 500 μl using a Centricon 10 concentrator (Amicon). The resultant protein was separated by fast protein liquid chromatography on Superdex 200 or Superdex 75 gel filtration columns (Pharmacia) in 10 mM Tris (pH 8.0)–150 mM NaCl. Typical refolding yields for US215–140 ranged from 2 to 5%, although the yield could be increased somewhat by refolding US2 in the presence of purified HLA-A2. Soluble US215–140 aggregates at room temperature in the absence of class I molecules and was kept at 4 to 10°C at all times.
Class I refolding reaction mixtures were dialyzed twice for 12 h against 10 mM Tris (pH 8.0)–1 mM EDTA (molecular weight cutoff = 6,000 to 8,000). The dialyzed protein was concentrated with a DEAE cellulose (DE-52) anion-exchange column packed with 20 g of refold per liter. The partially purified class I molecules were further concentrated by a Centricon 10 concentrator and separated from the remaining contaminants by Superdex 200 fast protein liquid chromatography gel filtration in 10 mM Tris (pH 8.0)–150 mM NaCl.
Native gel band shift assay.
Samples were incubated in a native gel loading buffer (final concentrations, 250 mM Tris [pH 8.8], 10% glycerol) to a final volume of 40 μl. Gels of 1.5 mm in thickness and containing either 8 or 12% polyacrylamide, without a stacking gel, were run at 4°C using 25 mM Tris–190 mM glycine running buffer. Proteins were visualized with Coomassie brilliant blue R-250.
Superdex gel filtration elution assay.
HLA-A2 (300 μg), US215–140 (190 μg), or HLA-A2 and US2 (300 and 190 μg, respectively) were incubated for 15 min and loaded onto an analytical Superdex 75 gel filtration column in a 100-μl volume. HLA-B7 (360 μg), US215–140 (580 μg), or HLA-B7 and US215–140 (360 and 580 μg, respectively) were incubated for 15 min and loaded onto Superdex 75 in a 200-μl volume. HLA-B27 (360 μg), US215–140 (580 μg), or HLA-B27 and US215–140 (360 and 580 μg, respectively) were likewise incubated for 15 min and loaded onto Superdex 75 in a 200-μl volume. All columns were run in 10 mM Tris (pH 8.0)–150 mM NaCl at 4°C.
RESULTS
Production of soluble US2.
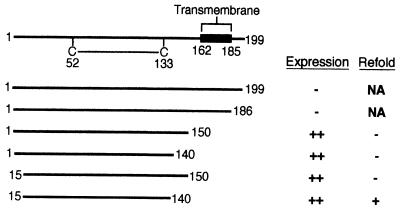
US2 expression constructs that included the predicted transmembrane coding region (residues 162 to 185) failed to yield significant levels of protein in Escherichia coli (Fig. 1). Since preliminary studies suggested that US2 mutants comprising residues 1 to 150 retain the ability to bind to class I in vivo, efforts to produce recombinant US2 therefore focused on the ER-lumenal segment. The US2 amino terminus is significantly less hydrophobic than are canonical signal sequences (22) and fails to be cleaved upon insertion into the ER (unpublished data). Initial attempts to produce soluble US2 were therefore carried out with mutants possessing a native amino terminus (Fig. 1). E. coli transformants containing expression constructs for US21–150 and US21–140 produced high levels of inclusion body protein. However, attempts to refold the urea-denatured inclusion bodies resulted in insoluble protein aggregates, which consistently eluted in the void volume upon size exclusion chromatography.
FIG. 1.
Regions of US2 used for expression in E. coli. A schematic diagram of the US2 primary structure is shown at the top, with the single US2 disulfide bond and predicted transmembrane domain indicated. The series of US2 deletion mutants constructed for expression in E. coli are displayed beneath. The extent to which the polypeptides can be isolated from E. coli and refolded to yield soluble material is indicated to the right of each US2 mutant. NA, not applicable.
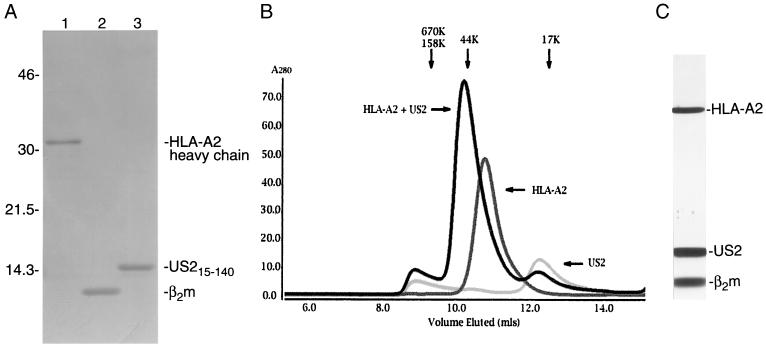
Additional US2 expression constructs utilize a methionine at residue 15 to initiate translation (Fig. 1). Both US215–150 and US215–140 expression plasmids yielded inclusion bodies at >100 mg of bacteria per liter. Only refolding of US215–140 yielded soluble monodisperse protein, as shown by size exclusion chromatography. The elution profile suggests that US215–140 exists as a monomer in solution (Fig. 2).
FIG. 2.
The US2 ER-lumenal domain is sufficient for association with class I molecules. (A) Reducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis of A21–275 (lane 1), β2m1–99 (lane 2), and US215–140 (lane 3) inclusion body material (10 μg each). The proteins were visualized by using Coomassie brilliant blue R-250 (Sigma). Numbers at left are molecular masses in kilodaltons. (B) Superimposed Superdex 75 gel filtration chromatograms of soluble US215–140, HLA-A2/Tax, and HLA-A2/Tax plus US215–140. The magnitude of the shift between the peak of the HLA-A2/Tax/US215/140 complex and free HLA-A2/Tax indicates 1:1 complex stoichiometry. The elution positions of molecular weight standards are shown above the chromatogram. (C) The HLA-A2/Tax/US215–140 complex associates with sufficient affinity to be further purified by Mono Q anion-exchange chromatography. Protein from the peak fraction was separated by nonreducing sodium dodecyl sulfate-polyacrylamide gel electrophoresis and visualized by silver staining.
US215–140 associates with class I molecules in vitro
To examine whether soluble US215–140 retains the ability to interact with soluble MHC class I, the HLA-A2 heavy chain lacking its transmembrane and cytoplasmic tail segments (G1 to E275) and β2m (Met1 to Met99) were refolded in the presence of the HTLV-1 Tax peptide LLFGYPVYV. Incubation of soluble US215–140 with purified HLA-A2 results in the formation of a complex of a Stokes' radius distinct from either HLA-A2 or US215–140 alone (Fig. 2B). The magnitude of the shift in elution position of the peak suggests a 1:1 stoichiometry of the US2/HLA-A2 complex (Fig. 2B). Preliminary analytical ultracentrifugation studies were also most consistent with a 1:1 stoichiometry (unpublished data). Further increasing the concentration of soluble US2 did not appear to alter the stoichiometry of the complex.
The strength of interaction between US215–140 and soluble HLA-A2/Tax is exemplified by the ability to remain associated during sequential gel filtration and ion-exchange chromatography (Fig. 2C). Thus, the US2 ER-lumenal domain alone is sufficient to mediate tight binding to HLA-A2 in the absence of other cellular or viral proteins. Although both US2 and MHC class I contain a single N-linked glycan in mammalian cells, glycosylation is not essential for interaction between the two proteins, as the bacterially produced subunits lack N-linked glycans.
US2 recognizes class I molecules independently of peptide sequence.
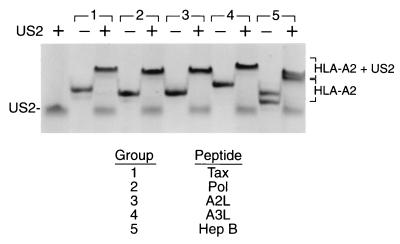
The ability of HLA-A2 to engage in complex formation with US2 is conveniently monitored by native gel electrophoresis. The binding characteristics and hence electrophoretic mobilities for several receptors that interact with class I molecules are influenced by the sequence of the MHC peptide cargo. We examined the ability of US2 to bind to HLA-A2 complexes containing different peptides of defined sequence, produced from recombinant subunits and synthetic peptides. As determined by native gel shift, US215–140 associates with five distinct HLA-A2–peptide complexes containing either HTLV Tax residues 11 to 19, HIV type 1 reverse transcriptase residues 309 to 317, hepatitis B virus nucleocapsid residues 18 to 27, melanoma peptide variant MART-1 27 to 35 (A2L), and variant MART-1 27 to 35 (A3L) (Fig. 3). The presence of US215–140 produced a complex of lower mobility with several additional HLA-A2–peptide complexes (unpublished data). Differences in electrophoretic mobilities among the US2/HLA-A2/peptide complexes result from peptide sequence variation (Fig. 3). Thus, US2 binds peptide-loaded class I molecules in a peptide sequence-independent fashion. Peptide remains associated with class I molecules upon US2 binding, as shown by incubation of the complex at 37°C, which allowed recovery of properly folded class I molecules (unpublished data). Empty class I molecules are not stable at 37°C and would have dissociated upon heating (2).
FIG. 3.
US215–140 binds to five distinct HLA-A2/peptide complexes. US215–140 (15 μg) gel shifts HLA-A2 complexes (7.5 μg) containing the following peptides: group 1, LLFGYPVYV; group 2, ILKEPVHGV; group 3, ELAGIGILTV; group 4, ALGIGILTV; group 5, FLPSDFFPSV.
MHC class I heavy chains require β2m and peptide to refold in vitro (9). We examined whether the presence of US215–140 facilitated the refolding of class I heavy chains in the absence of other class I components. Folding of class I heavy chains or association between free heavy chains and US2 was not evident in the presence of Tax peptide and refolded US215–140 (unpublished data). In the absence of peptide and ER chaperones, soluble class I heavy chains have a short half-life in solution, despite high concentrations of US215–140 and β2m. All in all, our data suggest that US2 prefers to interact with a properly folded class I molecule in a 1:1 stoichiometry.
US215–140 maintains locus-specific binding.
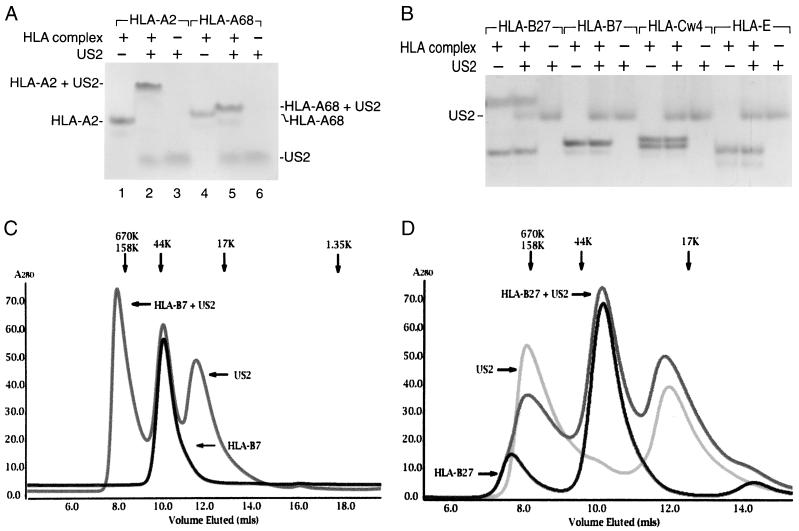
US2 demonstrates locus specificity in its ability to downregulate class I heavy chains in mammalian cells. While both HLA-A and HLA-B locus products are susceptible to attack in cells expressing US2, HLA-C, HLA-E, and HLA-G complexes resist US2-mediated degradation (8, 27). To examine the binding characteristics of US215–140, a series of class I complexes were refolded from bacterial inclusion bodies in the presence of the appropriate peptide ligands. Purified US215–140 was incubated with either HLA-A2, HLA-Aw68, HLA-B7, or HLA-B27. Association between class I molecules and US2 was assessed by native gel electrophoresis. US215–140 forms complexes with both HLA-A locus products (Fig. 4A), but an interaction with either HLA-B7 or HLA-B27 could not be detected at US2 concentrations that readily gel shift HLA-A2 (Fig. 4B). Further elevation of the US2 concentration by a factor of 10 did not result in either HLA-B7 or HLA-B27 gel shifts (unpublished data). Furthermore, neither HLA-Cw4 nor HLA-E showed any signs of interaction with soluble US2 under conditions where the HLA-A locus products readily bind (Fig. 4B). Elevating the US2 concentration by 10-fold over the minimal requirement to effectuate a quantitative gel shift for HLA-A2 did not produce a class I gel shift. US2 does not target H-2 Kb for degradation (21), and US215–140 likewise does not alter the mobility of soluble Kb in native gel electrophoresis (unpublished data).
FIG. 4.
The US2 ER-lumenal domain associates selectively with class I molecules. (A) US215–140 gel shifts HLA-A2 and HLA-Aw68. US215–140 (15 μg) was incubated with HLA-A2 and HLA-Aw68 complexes (10 μg each) as indicated, and proteins were separated by native gel electrophoresis. (B) US215–140 does not interact with similar affinity with molecules of other class I loci. US215–140 (15 μg) was incubated with HLA-B7, HLA-B27, HLA-Cw4, and HLA-E (10 μg each) as indicated and separated by native gel electrophoresis. (C) Superimposed Superdex 75 gel filtration chromatograms of HLA-B7 and HLA-B7 plus US215–140. (D) Superimposed Superdex 75 gel filtration chromatograms of US215–140, HLA-B27, and HLA-B27 plus US215–140. The presence of US215–140 does not alter the elution position of either HLA-B7 or HLA-B27. The elution positions of molecular weight standards are shown above the chromatograms in panels C and D.
Not only did we fail to detect such interaction by native gel electrophoresis, but also gel filtration chromatography failed to show evidence for an interaction between either HLA-B7 or HLA-B27 and US215–140 despite a fivefold molar excess of soluble US2 (Fig. 4C and D).
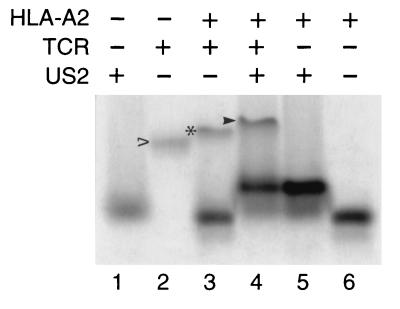
US215–140 and TCR can simultaneously interact with MHC class I.
It is not known whether the conformation of class I molecules changes upon association with US2, nor is it clear exactly where US2 interacts with MHC class I products. To examine this question in vitro, we asked whether US215–140 and T-cell receptor (TCR) can recognize the same class I complex. Purified A6 TCR, which is specific for the HLA-A2/Tax complex (10), was incubated with HLA-A2/Tax and US215–140. Complex assembly was monitored by band shift in native gel electrophoresis. A protein complex of distinct mobility is generated when HLA-A2, US215–140, and A6 TCR are incubated together (Fig. 5). The HLA-A2/US215–140/TCR complex formed regardless of the order of addition. Thus, the binding of US215–140 to class I molecules does not apparently induce major conformational changes in the class I complex in solution, as the A6 TCR maintains the ability to associate with US2/class I complexes. The formation of this novel complex further suggests that US2 and TCR interact with class I molecules at distinct and nonoverlapping binding sites.
FIG. 5.
US215–140 and a TCR can simultaneously interact with the same HLA-A2 molecules. US215–140 (15 μg), HLA-A2/Tax (17 μg), and soluble A6 TCR (6.6 μg) were incubated as indicated, and proteins were separated by native gel electrophoresis. TCR alone is shown by the empty arrowhead, HLA-A2/Tax/TCR complex is shown by the asterisk, and HLA-A2/Tax/US215–140/TCR complex is shown by the filled arrowhead.
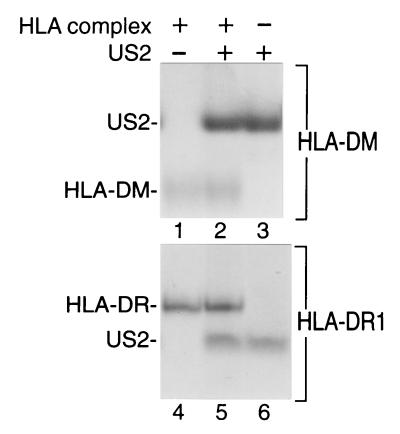
US215–140 does not interact with HLA-DR or HLA-DM.
Although US2 was originally shown to downregulate expression of class I products, US2 has been suggested to also target for destruction components of the MHC class II antigen presentation pathway, HLA-DRα and HLA-DMα. To determine whether US215–140 can associate with complexes in the class II pathway, recombinant soluble HLA-DR1 and HLA-DM were incubated with US215–140 and complex formation was monitored by native gel electrophoresis. As shown in Fig. 6, US215–140 does not alter the mobility of either DR1 or DM complexes in native gel electrophoresis. No gel shift was observed for DR1 or DM even in the presence of US2 concentrations 10-fold higher than the amount required to quantitatively bind HLA-A2 complexes. The mobility of a second DR allele was likewise not altered in the presence of US215–140. The preparation of DR used is known to produce a gel shift upon complexation with the TCR under conditions essentially identical to those used here (unpublished data). The batch of DR used in this study was shown to catalyze peptide exchange in MHC class II molecules.
FIG. 6.
US215–140 does not form high-affinity complexes with either HLA-DR or HLA-DM. US215–140 (15 μg) was incubated with HLA-DR (5 μg) or HLA-DM (10 μg) as indicated, and proteins were separated by native gel electrophoresis.
DISCUSSION
Many viruses that cause persistent infections have evolved strategies to interfere with MHC class I antigen presentation (32). This report provides the first evidence for an exclusive, direct interaction between a virus protein and class I MHC products in vitro.
The ability of US215–140 to form a stable complex with properly folded class I complexes is consistent with biochemical data from US2-expressing cells. US2 can be recovered by coimmunoprecipitation with a conformation-specific monoclonal antibody, W6/32, which recognizes properly folded class I complexes. W6/32-reactive material recovered from US2+ transfectants does not contain radiolabeled β2m (32). However, immunoblot analysis of W6/32 immunoprecipitates from US2+ cells reveals the presence of unlabeled β2m (unpublished data). Thus, initial complex formation, as assayed by an anti-class I antibody, involves preexisting (nonradioactive) β2m. In contrast, ER-resident glycosylated US2 cannot be recovered by polyclonal antibodies that recognize epitopes present in unfolded heavy chains (36). The distinct mobility of five US2/HLA-A2/peptide complexes (Fig. 3) and the ability of the A6 TCR to recognize the US2/HLA-A2 complex (Fig. 5) indicate that US2 can bind peptide-loaded class I molecules. It is unclear whether US2 interacts exclusively with peptide-loaded class I heavy chains or whether it can also recognize class I molecules prior to peptide loading. We conclude that US2 recognizes via its lumenal domain a fully assembled, peptide-loaded class I molecule, a conformation that apparently arises relatively late in biosynthesis. It is noteworthy that US2 nonetheless efficiently downregulates class I molecules, in spite of this late stage of recognition, just prior to exit of class I molecules from the ER.
The association between US215–140 and HLA-A2 is saturable. Upon native electrophoresis and size exclusion chromatography in the presence of excess US2, both free US2 and US2/HLA-A2 complexes are evident (Fig. 3 to 5 and unpublished data). US2 binding does not significantly alter the conformation of class I MHC molecules, as judged from the ability of soluble TCR to interact with the US2/class I complex. In contrast to many proteins that bind class I, including TCRs and several types of NK cell receptors, US2 recognizes class I molecules regardless of the sequence of the peptide cargo, consistent with the ability of US2 to bind class I molecules at a site distinct from that seen by the TCR. US215–140 interacts with apparently similar affinities with a series of HLA-A2 complexes that possess distinct peptides. While most immune system receptors for class I, including the TCR, several NK receptors, and CD8, are able to form homodimers or heterodimers, recombinant US2 is a monomer in solution and binds to class I molecules with a 1:1 stoichiometry. Studies with cell-permeable chemical cross-linkers have likewise failed to reveal dimerization of US2 in cellular transfectants (unpublished data).
In transfectants that express either US2 or US11, all HLA-A and HLA-B class I molecules appear susceptible to degradation, as judged from the reduction in their surface expression and from the accelerated degradation of the bulk of newly synthesized class I molecules (references 35 and 36 and unpublished observations). In other words, there is no evidence to suggest the selective resistance of the HLA-A or HLA-B locus products to the activity of US2 or US11. Surprisingly, our data show that the luminal domain of US2 is sufficient to allow stable complex formation with HLA-A2 and HLA-A68, whereas neither HLA-B7 nor HLA-B27 appears capable of doing so. If the luminal domain of US2 were the sole region of the molecule responsible for recognition of class I heavy chains, we would predict resistance of these HLA-B locus alleles to US2-mediated degradation, and this is not the case. Interestingly, the association between US3 and class I in vivo requires both the US3 ER-lumenal and transmembrane domains (20), and the recombinant US3 ER-lumenal domain does gel shift MHC class I in vitro (unpublished data). These observations suggest that perhaps the US2 and class I transmembrane segments may also interact to account for US2's ability to target HLA-B locus products for degradation in living cells.
The downregulation of class I molecules is a precarious act for viruses, as NK cells lyse cells that lose MHC class I expression (25). However, the known human NK receptors for class I focus primarily on the HLA-C and HLA-E locus products. By selectively downregulating HLA-A and HLA-B class I molecules, viruses can diminish antigen presentation to CD8+ T lymphocytes while limiting NK cell activation (27). The data presented in this paper suggest that the US2 ER-lumenal domain plays an important role in allowing US2 to distinguish between MHC class I locus products. In contrast, HIV Nef and KHSV K5 focus on the class I transmembrane and tail regions for distinguishing between class I locus products (5, 15). These segments contain an abundance of locus-specific residues (11). Most of the polymorphic residues in class I are concentrated in regions that contact peptide or TCR, and US2 does not compete with TCR for class I association. It is all the more unusual that US2 can use the polymorphisms in the class I lumenal domain to distinguish between class I molecules. Unlike US2 and US11, HCMV US3 does not demonstrate locus-specific interaction with class I (19).
It seems paradoxical that HCMV should encode two proteins, US2 and US11, with apparently redundant functions. As strong selection pressures constantly maintain compact virus genomes, it is a fair assumption that expression of both US2 and US11 benefits HCMV. Murine cytomegalovirus likewise encodes multiple proteins dedicated to class I downregulation, although without apparent homology to their HCMV paralogues and availing themselves of cell biologically distinct mechanisms to accomplish their goal (14). We have suggested that the presence of both US2 and US11 may ensure that HCMV will be able to counter the high degree of class I polymorphism (21). US11 associates with free class I heavy chains in coimmunoprecipitation experiments in cellular transfectants, which indicates recognition by US11 of a motif that arises earlier in class I assembly. Perhaps US2 and US11 safeguard against the escape of class I molecules from the ER by focusing on different stages of the class I biosynthetic pathway. Further structural and mechanistic analyses should provide an explanation for why HCMV employs both proteins.
US215–140 does not bind HLA-DR and HLA-DM with sufficient affinity to produce a shift on native gel electrophoresis. In contrast, it has previously been reported that US2 associates with HLA-DR and HLA-DM complexes and subsequently targets the DRα and DMα subunits for degradation (31). Several explanations could explain the discrepancy between these findings. It is possible that distinct regions of US2 mediate interaction with MHC class I and class II molecules. Should this be the case, we predict that the US2 transmembrane and/or cytoplasmic tail would contribute significantly to association with HLA-DM and HLA-DR complexes, as we suggest to be the case for the interaction of US2 with HLA-B locus products. Alternatively, it remains possible that another factor is required to stabilize the interaction between US2 and DR or DM. Finally, US2 may bind with higher affinity to class I molecules than to either DR or DM molecules. Consequently, it is plausible that US2 can be induced to associate with HLA-DR and HLA-DM when overexpressed in the ER.
The mechanism by which US2 targets class I heavy chains for dislocation remains unclear. We have hypothesized that US2 subverts a poorly understood homeostatic “quality control” process. The quality control machinery in the ER recognizes and removes misfolded proteins from the ER. We now suggest that association between the ER-luminal domains of US2 and class I initiates dislocation of class I heavy chains. However, interaction between US2 and class I does not appear to significantly alter the conformation of class I molecules. The details of the events that connect formation of a tight complex between US2 and class I in the ER and the appearance of class I heavy chains in the cytosol remain uncertain. Further, the stability of the US2/HLA-A2 complex and the absence of obvious conformational alterations raise important questions about the details of MHC I dislocation. Is the complex disassembled prior to discharge into the cytosol, and if so, which proteins assist in this unfolding? Alternatively, it is conceivable that the entire US2-class I complex is dislocated via the Sec61 channel, whose diameter is reported to range from 20 to 60 Å (12, 23). The data reported here conclusively demonstrate the lumenal domain's importance for binding and call attention to the proposed transmembrane segment and cytoplasmic tail as essential for the dislocation reaction.
ACKNOWLEDGMENTS
We thank Rachelle Gaudet, Joydeep Mitra, and members of the Ploegh lab for assistance.
This work was supported by a grant from the National Institutes of Health (5R37-AI33456). B.E.G. is a predoctoral fellow of the Howard Hughes Medical Institute. E.W.W. is supported by a National Cancer Institute Fellowship in Cancer Biology. D.T. is a Charles A. King Trust postdoctoral fellow. D.J.S. is supported by a Women's Reproductive Health Research Training Grant from the National Institutes of Health (K12-HD01255).
REFERENCES
- 1.Ahn K, Angulo A, Ghazal P, Peterson P A, Yang Y, Fruh K. Human cytomegalovirus inhibits antigen presentation by a sequential multistep process. Proc Natl Acad Sci USA. 1996;93:10990–10995. doi: 10.1073/pnas.93.20.10990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baas E J, van Santen H M, Kleijmeer M J, Geuze H J, Peters P J, Ploegh H L. Peptide-induced stabilization and intracellular localization of empty HLA class I complexes. J Exp Med. 1992;176:147–156. doi: 10.1084/jem.176.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Boppana S B, Britt W J. Recognition of human cytomegalovirus gene products by HCMV-specific cytotoxic T cells. Virology. 1996;222:293–296. doi: 10.1006/viro.1996.0424. [DOI] [PubMed] [Google Scholar]
- 4.Britt W J, Alford C A, editors. Cytomegalovirus. 3rd ed. Vol. 2. Philadelphia, Pa: Lippincott-Raven Publishers; 1996. [Google Scholar]
- 5.Cohen G B, Gandhi R T, Davis D M, Mandelboim O, Chen B K, Strominger J L, Baltimore D. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–671. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 6.Cresswell P, Bangia N, Dick T, Diedrich G. The nature of the MHC class I peptide loading complex. Immunol Rev. 1999;172:21–28. doi: 10.1111/j.1600-065x.1999.tb01353.x. [DOI] [PubMed] [Google Scholar]
- 7.Fan Q R, Garboczi D N, Winter C C, Wagtmann N, Long E O, Wiley D C. Direct binding of a soluble natural killer cell inhibitory receptor to a soluble human leukocyte antigen-Cw4 class I major histocompatibility complex molecule. Proc Natl Acad Sci USA. 1996;93:7178–7183. doi: 10.1073/pnas.93.14.7178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Furman H M, Ploegh H L, Schust D J. Can viruses help us to understand and classify the MHC class I molecules at the maternal-fetal interface? Hum Immunol. 2000;61:1169–1176. doi: 10.1016/s0198-8859(00)00203-2. [DOI] [PubMed] [Google Scholar]
- 9.Garboczi D N, Hung D T, Wiley D C. HLA-A2-peptide complexes: refolding and crystallization of molecules expressed in Escherichia coli and complexed with single antigenic peptides. Proc Natl Acad Sci USA. 1992;89:3429–3433. doi: 10.1073/pnas.89.8.3429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garboczi D N, Utz U, Ghosh P, Seth A, Kim J, VanTienhoven E A, Biddison W E, Wiley D C. Assembly, specific binding, and crystallization of a human TCR-alphabeta with an antigenic Tax peptide from human T lymphotropic virus type 1 and the class I MHC molecule HLA-A2. J Immunol. 1996;157:5403–5410. [PubMed] [Google Scholar]
- 11.Gussow D, Rein R S, Meijer I, de Hoog W, Seemann G H, Hochstenbach F M, Ploegh H L. Isolation, expression, and the primary structure of HLA-Cw1 and HLA-Cw2 genes: evolutionary aspects. Immunogenetics. 1987;25:313–322. doi: 10.1007/BF00404424. [DOI] [PubMed] [Google Scholar]
- 12.Hamman B D, Chen J C, Johnson E E, Johnson A E. The aqueous pore through the translocon has a diameter of 40–60 A during cotranslational protein translocation at the ER membrane. Cell. 1997;89:535–544. doi: 10.1016/s0092-8674(00)80235-4. [DOI] [PubMed] [Google Scholar]
- 13.Heemels T M, Ploegh H. Generation, translocation, and presentation of MHC class I-restricted peptides. Annu Rev Biochem. 1995;64:463–491. doi: 10.1146/annurev.bi.64.070195.002335. [DOI] [PubMed] [Google Scholar]
- 14.Hengel H, Reusch U, Gutermann A, Ziegler H, Jonjic S, Lucin P, Koszinowski U H. Cytomegaloviral control of MHC class I function in the mouse. Immunol Rev. 1999;168:167–176. doi: 10.1111/j.1600-065x.1999.tb01291.x. [DOI] [PubMed] [Google Scholar]
- 15.Ishido S, Wang C, Lee B S, Cohen G B, Jung J U. Downregulation of major histocompatibility complex class I molecules by Kaposi's sarcoma-associated herpesvirus K3 and K5 proteins. J Virol. 2000;74:5300–5309. doi: 10.1128/jvi.74.11.5300-5309.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jensen P E, Weber D A, Thayer W P, Chen X, Dao C T. HLA-DM and the MHC class II antigen presentation pathway. Immunol Res. 1999;20:195–205. doi: 10.1007/BF02790403. [DOI] [PubMed] [Google Scholar]
- 17.Jones T R, Hanson L K, Sun L, Slater J S, Stenberg R M, Campbell A E. Multiple independent loci within the human cytomegalovirus unique short region down-regulate expression of major histocompatibility complex class I heavy chains. J Virol. 1995;69:4830–4841. doi: 10.1128/jvi.69.8.4830-4841.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Jones T R, Wiertz E J, Sun L, Fish K N, Nelson J A, Ploegh H L. Human cytomegalovirus US3 impairs transport and maturation of major histocompatibility complex class I heavy chains. Proc Natl Acad Sci USA. 1996;93:11327–11333. doi: 10.1073/pnas.93.21.11327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jun Y, Kim E, Jin M, Sung H C, Han H, Geraghty D E, Ahn K. Human cytomegalovirus gene products US3 and US6 down-regulate trophoblast class I MHC molecules. J Immunol. 2000;164:805–811. doi: 10.4049/jimmunol.164.2.805. [DOI] [PubMed] [Google Scholar]
- 20.Lee S, Yoon J, Park B, Jun Y, Jin M, Sung H C, Kim I H, Kang S, Choi E J, Ahn B Y, Ahn K. Structural and functional dissection of human cytomegalovirus US3 in binding major histocompatibility complex class I molecules. J Virol. 2000;74:11262–11269. doi: 10.1128/jvi.74.23.11262-11269.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machold R P, Wiertz E J, Jones T R, Ploegh H L. The HCMV gene products US11 and US2 differ in their ability to attack allelic forms of murine major histocompatibility complex (MHC) class I heavy chains. J Exp Med. 1997;185:363–366. doi: 10.1084/jem.185.2.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martoglio B, Dobberstein B. Signal sequences: more than just greasy peptides. Trends Cell Biol. 1998;8:410–415. doi: 10.1016/s0962-8924(98)01360-9. [DOI] [PubMed] [Google Scholar]
- 23.Menetret J, Neuhof A, Morgan D G, Plath K, Radermacher M, Rapoport T A, Akey C W. The structure of ribosome-channel complexes engaged in protein translocation. Mol Cell. 2000;6:1219–1232. doi: 10.1016/s1097-2765(00)00118-0. [DOI] [PubMed] [Google Scholar]
- 24.Miller D M, Rahill B M, Boss J M, Lairmore M D, Durbin J E, Waldman J W, Sedmak D D. Human cytomegalovirus inhibits major histocompatibility complex class II expression by disruption of the Jak/Stat pathway. J Exp Med. 1998;187:675–683. doi: 10.1084/jem.187.5.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ravetch J V, Lanier L L. Immune inhibitory receptors. Science. 2000;290:84–89. doi: 10.1126/science.290.5489.84. [DOI] [PubMed] [Google Scholar]
- 26.Reddehase M J. The immunogenicity of human and murine cytomegaloviruses. Curr Opin Immunol. 2000;12:390–396. doi: 10.1016/s0952-7915(00)00106-0. [DOI] [PubMed] [Google Scholar]
- 27.Schust D J, Tortorella D, Seebach J, Phan C, Ploegh H L. Trophoblast class I major histocompatibility complex (MHC) products are resistant to rapid degradation imposed by the human cytomegalovirus (HCMV) gene products US2 and US11. J Exp Med. 1998;188:497–503. doi: 10.1084/jem.188.3.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Soderberg-Naucler C, Fish K N, Nelson J A. Reactivation of latent human cytomegalovirus by allogeneic stimulation of blood cells from healthy donors. Cell. 1997;91:119–126. doi: 10.1016/s0092-8674(01)80014-3. [DOI] [PubMed] [Google Scholar]
- 29.Stevenson P G, Efstathiou S, Doherty P C, Lehner P J. Inhibition of MHC class I-restricted antigen presentation by gamma 2-herpesviruses. Proc Natl Acad Sci USA. 2000;97:8455–8460. doi: 10.1073/pnas.150240097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Story C M, Furman M H, Ploegh H L. The cytosolic tail of class I MHC heavy chain is required for its dislocation by the human cytomegalovirus US2 and US11 gene products. Proc Natl Acad Sci USA. 1999;96:8516–8521. doi: 10.1073/pnas.96.15.8516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomazin R, Boname J, Hegde N R, Lewinsohn D M, Altschuler Y, Jones T R, Cresswell P, Nelson J A, Riddell S R, Johnson D C. Cytomegalovirus US2 destroys two components of the MHC class II pathway, preventing recognition by CD4+ T cells. Nat Med. 1999;5:1039–1043. doi: 10.1038/12478. [DOI] [PubMed] [Google Scholar]
- 32.Tortorella D, Gewurz B E, Furman M H, Schust D J, Ploegh H L. Viral subversion of the immune system. Annu Rev Immunol. 2000;18:861–926. doi: 10.1146/annurev.immunol.18.1.861. [DOI] [PubMed] [Google Scholar]
- 33.Tortorella D, Story C M, Huppa J B, Wiertz E J, Jones T R, Bacik I, Bennink J R, Yewdell J W, Ploegh H L. Dislocation of type I membrane proteins from the ER to the cytosol is sensitive to changes in redox potential. J Cell Biol. 1998;142:365–376. doi: 10.1083/jcb.142.2.365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Townsend A, Ohlen C, Bastin J, Ljunggren H G, Foster L, Karre K. Association of class I major histocompatibility heavy and light chains induced by viral peptides. Nature. 1989;340:443–448. doi: 10.1038/340443a0. [DOI] [PubMed] [Google Scholar]
- 35.Wiertz E J, Jones T R, Sun L, Bogyo M, Geuze H J, Ploegh H L. The human cytomegalovirus US11 gene product dislocates MHC class I heavy chains from the endoplasmic reticulum to the cytosol. Cell. 1996;84:769–779. doi: 10.1016/s0092-8674(00)81054-5. [DOI] [PubMed] [Google Scholar]
- 36.Wiertz E J, Tortorella D, Bogyo M, Yu J, Mothes W, Jones T R, Rapoport T A, Ploegh H L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 37.Yewdell J W, Bennink J R. Cut and trim: generating MHC class I peptide ligands. Curr Opin Immunol. 2001;13:13–18. doi: 10.1016/s0952-7915(00)00175-8. [DOI] [PubMed] [Google Scholar]