Abstract
In the budding yeast Saccharomyces cerevisiae, chromosome end protection is provided by a heterotrimeric complex composed of Cdc13 in association with the RPA-like proteins Stn1 and Ten1. We report here that the high affinity and specificity of the S. cerevisiae Cdc13 DNA binding domain for single-stranded telomeric DNA are not widely shared by other fungal Cdc13 proteins, suggesting that restriction of this complex to telomeres may be limited to the Saccharomyces clade. We propose that the evolutionarily conserved task of Stn1 and Ten1 (and their associated large subunit) is a genome-wide role in DNA replication rather than a telomere-dedicated activity.
The single-stranded extension of the G-rich strand of chromosome ends is a highly conserved feature of eukaryotic telomere structure, as is the need to protect these natural DNA ends. Experiments in a number of organisms have demonstrated that unprotected telomeres are subjected to a variety of DNA processing insults, with resulting catastrophic consequences for genome integrity.1
In budding yeast, telomere integrity relies on a heterotrimeric complex composed of Cdc13, Stn1, and Ten1,2 which is targeted to chromosome ends through the exceptionally high affinity and specificity that Cdc13 displays for G-rich telomeric single-stranded DNA (ssDNA).3,4 Defects in any of these three proteins lead to the extensive loss of the telomeric C strand and cell cycle arrest,5–7 showing that this complex protects telomeres from unregulated nucleolytic resection. Recent evidence indicates that these three proteins function as a telomere-dedicated RPA-like complex,8–10 which we have called the t-RPA complex. While the canonical RPA complex binds to double-strand breaks and subsequently blocks cell cycle progression to coordinate DNA repair, the t-RPA complex is proposed to protect yeast telomeres from such events and thereby ensure cell cycle progression.
A potentially similar heterotrimeric complex has also been identified in human cells as an activity called AAF (α-accessory factor) that stimulates the DNA polymerase α—primase complex in vitro.11,12 More recently, this complex has been proposed to perform a telomere capping function in parallel with the well-characterized shelter-in complex.13,14 However, the yeast and human complexes exhibit one key difference, which is that the human complex lacks telomere-specific DNA binding activity,14 in sharp contrast to the specificity displayed by the Saccharomyces cerevisiae Cdc13 protein.
Because S. cerevisiae has played such a key role as a model organism for telomere biology, we asked whether the sequence-specific DNA binding behavior of Cdc13 is a conserved property. Specifically, we have analyzed Cdc13 proteins from the subphylum Saccharomycotina (budding yeast), which comprises the Saccharomyces, Kluveromyces, and Candida clades and represents an evolutionary distance equivalent to that from humans to tunicates.15 Alignment of Cdc13 proteins from the Saccharomyces and Kluveromyces clades revealed the presence of five distinct domains (Figure 1A).16 In contrast, Cdc13 homologues from the Candida clade are composed of only the DNA binding domain (DBD) and the C-terminal domain (Figure 1A).10,17 The absence of the three N-terminal domains is not due to incorrect predictions of gene boundaries, as examination of upstream regions did not reveal the existence of reading frames that had been missed during automated gene annotation. Furthermore, analysis of syntenic relationships provided evidence of substantial genomic rearrangements upstream of the CDC13 genes in the Candida clade.17
Figure 1.
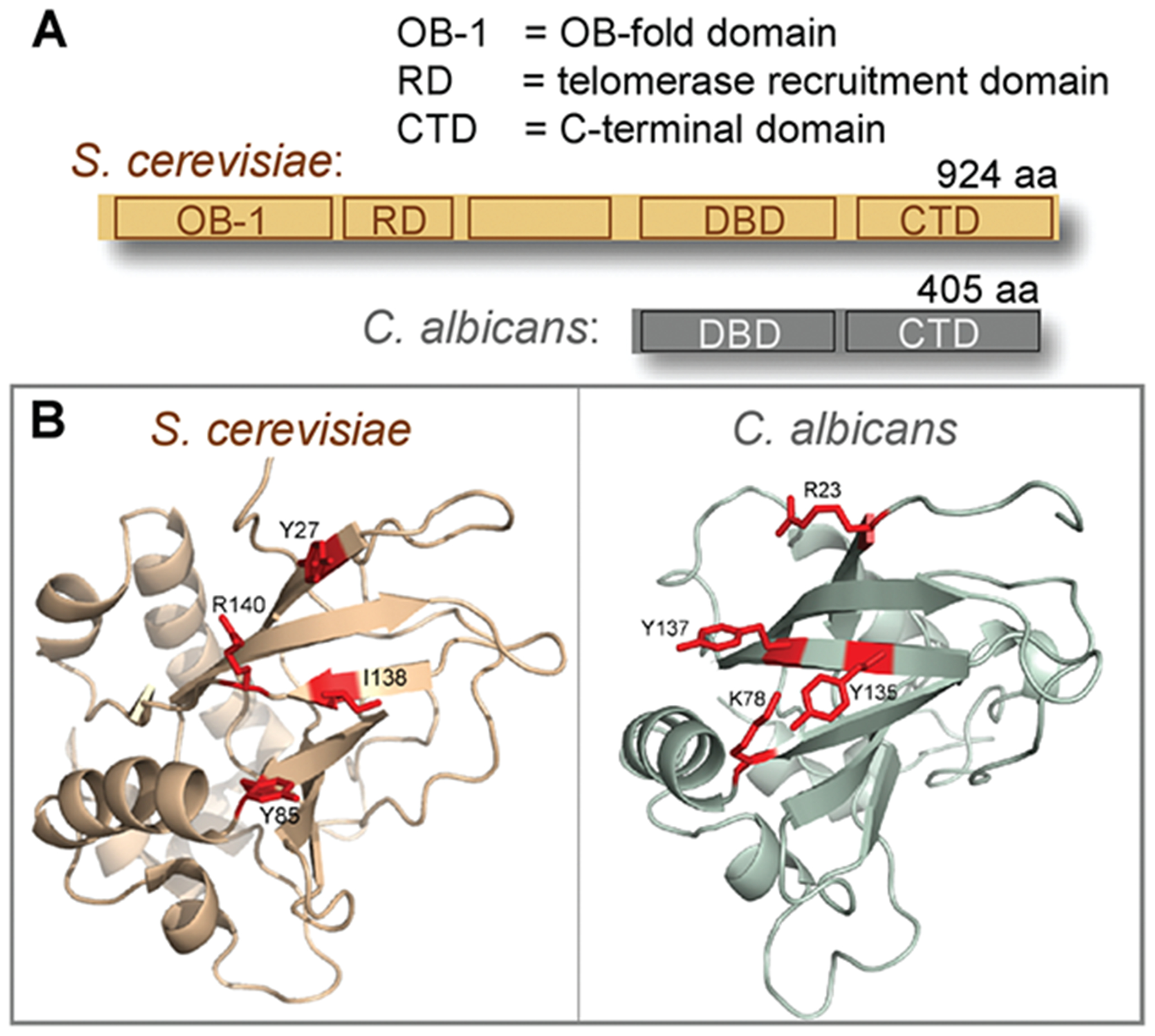
(A) Schematic comparison of the domain structure of Cdc13 and (B) the structure of the DBD, from S. cerevisiae and C. albicans, with residues that comprise the hot spot” for binding affinity colored red.
In addition to loss of the N-terminal half of the protein, changes in the Cdc13 DBD were also observed. Examination of the Cdc13 DBDs from the Saccharomyces and Kluveromyces clades revealed a significant degree of similarity both at the sequence level and in the predicted secondary and tertiary structures. In contrast, the same domain from the Candida clade had undergone substantial sequence divergence, both within the clade and when compared to the rest of the Saccharomycotina subphylum. Nevertheless, structure predictions indicated with a high degree of confidence that the Candida clade DBDs adopted an OB fold that was structurally similar to that of the S. cerevisiae Cdc13 DBD18 (Figure 1B and Supporting Information). Comparison of the predicted C. albicans structure with the S. cerevisiae DBD structure revealed a key difference, however. In the S. cerevisiae Cdc13 DBD, a “hot spot” for binding affinity is formed by a cluster of four DNA contact residues,19 which are conserved among members of the Saccharomyces clade. In contrast, this hot spot cluster was not observed in the predicted C. albicans DBD (Figure 1B). The absence of this hot spot was not restricted to the C. albicans DBD, as a high frequency of substitutions occurred at these four amino acid positions among all members of the Candida clade.
These changes at the predicted ssDNA–protein interface suggested that the properties of interaction with ssDNA would be substantially different. To examine this, a panel of Cdc13 DBDs were expressed in E. coli and initially examined for solubility; three DBDs (Candida glabrata from the Saccharomyces clade and Candida parapsilosis and C. albicans both from the Candida clade) were chosen for further analysis (see Figure S1 of the Supporting Information for a phylogenetic tree of Cdc13 sequences). Figure 2 and Table 1 illustrate the binding properties of the C. albicans Cdc13 DBD with a telomeric 26-mer, compared to nontelomeric ssDNA oligomers of comparable length. Surprisingly, the C. albicans DBD bound telomeric ssDNA oligomers with an affinity that was reduced by ~1000-fold, when compared to the affinity of the binding of the S. cerevisiae DBD to comparable oligomers.4 Furthermore, the affinity of the C. albicans DBD for a nontelomeric substrate relative to a telomeric oligomer was reduced only ~5-fold, in contrast to S. cerevisiae Cdc13, which discriminates telomeric ssDNA by at least 1000-fold.3 The level of binding to a 13-mer telomeric ssDNA was reduced relative to that of the 26-mer substrate (Table 1), which further differentiates the C. albicans Cdc13 DBD from the S. cerevisiae DBD.4
Figure 2.
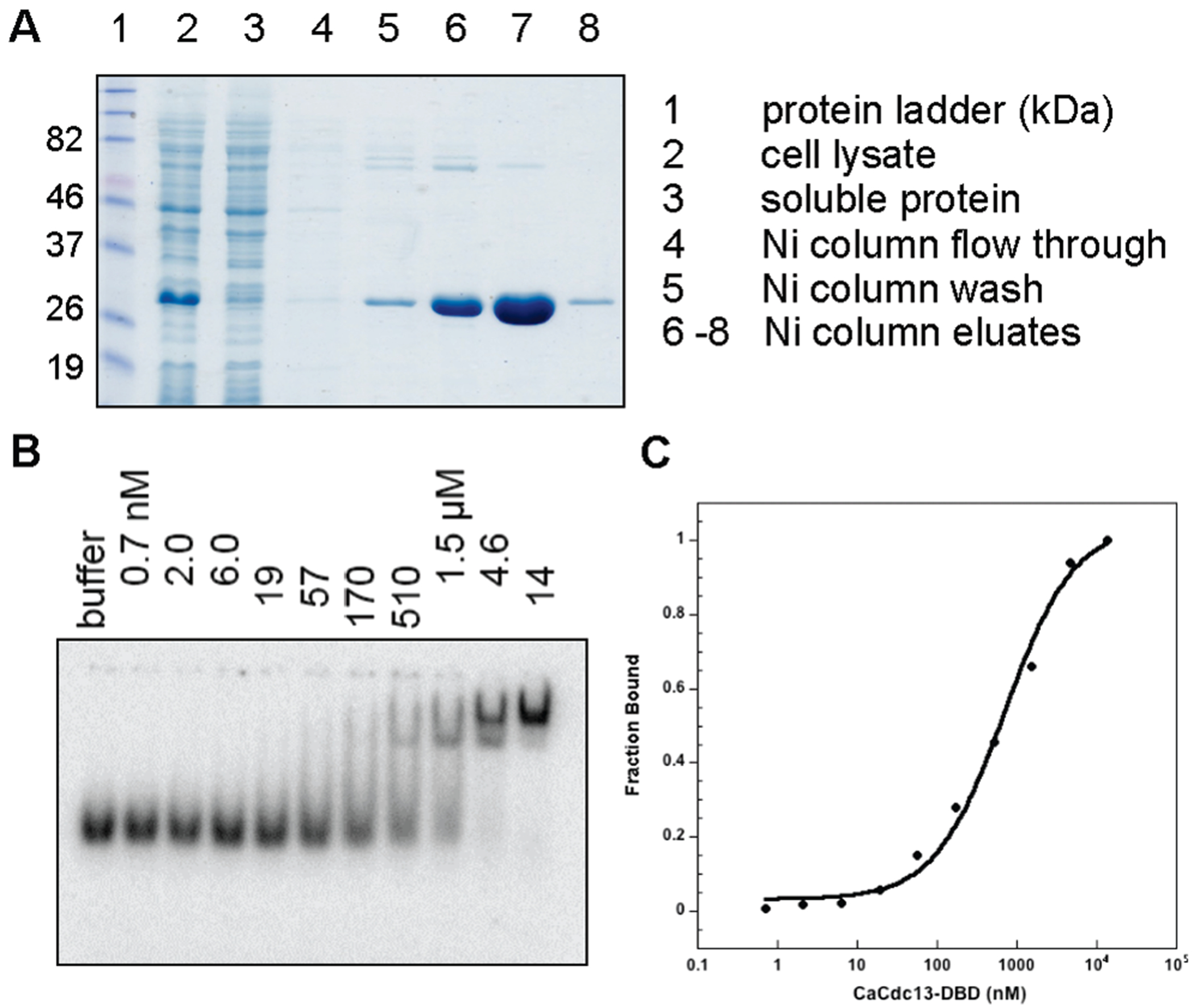
Expression, purification, and representative binding data of the C. albicans DBD with a 26-mer telomeric oligomer (Ca T1; see Table S1 of the Supporting Information). Affinity purification (A) and electrophoretic mobility shift assays (B) were performed as described in the Supporting Information, and the data were fit to a standard two-state binding model (C).
Table 1.
KD Values for the Wild-Type C. albicans Cdc13 DBD, or the DBD Bearing the Indicated Mutations, Bound to Single-Stranded Oligonucleotides
| C. albicans DBD | oligonucleotidea | KD (μM) | x-fold change |
|---|---|---|---|
| wild type | Ca T1 (26-mer) | 0.65 ± 0.10 | – |
| Ca T2 (13-mer) | 1.8 ± 0.27 | 2.8 | |
| R1 (26-mer) | 2.0 ± 0.58 | 3.1 | |
| R2 (46-mer) | 1.90 ± 0.24 | 3.2 | |
| Ca T3 (46-mer) | 0.59 ± 0.09 | 0.9 | |
| R23A | Ca T3 (46-mer) | 2.70 ± 0.33 | 4.6 |
| K78A | Ca T3 (46-mer) | 1.90 ± 0.15 | 3.2 |
| Y135A | Ca T3 (46-mer) | 3.20 ± 0.33 | 5.4 |
| Y137A | Ca T3 (46-mer) | 1.20 ± 0.17 | 2.0 |
Oligonucleotide sequences are listed in Table S1 of the Supporting Information.
The lack of strong discrimination for telomeric substrates was not specific for the C. albicans Cdc13 DBD, as the C. parapsilosis DBD also exhibited a similar binding affinity for a telomeric 26-mer (KD = 0.21 ± 0.026 μM), which was enhanced only 7-fold relative to those of nontelomeric substrates.
As an additional point of comparison with the S. cerevisiae Cdc13 DBD, alanine missense mutations were introduced into the four residues indicated in Figure 1 of the C. albicans Cdc13 DBD. Alanine replacements in the four comparable residues of the S. cerevisiae Cdc13 DBD comprising the hot spot result in a 500—700-fold reduction in binding affinity, as shown previously.19 In striking contrast, the four mutant C. albicans Cdc13 DBDs exhibited at most a 5-fold impact on binding affinity, relative to that of the wild-type DBD (Table 1).
These observations indicate that the C. albicans and C. parapsilosis Cdc13 DBDs exhibit biochemical properties that are quite distinct from those of the S. cerevisiae Cdc13 DBD, which correlates with the changes predicted for the ssDNA—protein interface for the C. albicans and C. parapsilosis DBDs (Figure 1 and data not shown). In addition to the loss of the affinity hot spot, the unusual β2—β3 loop that plays a central role in ssDNA recognition by the S. cerevisiae Cdc13 DBD18 was shorter in the C. albicans and C. parapsilosis Cdc13 DBD models and also lacked the high content of aromatic residues that contributed to binding by the S. cerevisiae DBD. These two changes may be sufficient to account for the dramatic reduction in the level of binding to telomeric ssDNA substrates by the C. albicans and C. parapsilosis DBDs.
Assessment of the predicted ssDNA—protein interface for other Cdc13 members of the Saccharomycotina subphylum suggests that an attenuated preference for telomeric substrates may be a general feature, as Cdc13 proteins from several members of the Kluveromyces clade exhibited amino acid replacements at both the hot spot for binding affinity and the β2—β3 loop. We therefore examined the DNA binding properties of the Cdc13 DBD from C. glabrata, which diverged from the common ancestor of Saccharomyces sensu stricto species after whole-genome duplication occurred and is an outlier of the Saccharomyces clade.20 The C. glabrata DBD bound a ssDNA 26-mer oligomer composed of G-rich telomeric DNA with a KD of 0.079 ± 0.02 μM, which was several orders of magnitude weaker than the affinity of the S. cerevisiae DBD for its telomeric substrate. Furthermore, the C. glabrata DBD favored a telomeric substrate by only 40—80-fold in binding affinity, relative to nontelomeric substrates. This suggests that a transition from nonspecific ssDNA binding to an increased affinity for telomeric ssDNA took place in the common ancestor of C. glabrata and the rest of the Saccharomyces clade, with increased specificity evolving along the branch leading to S. cerevisiae.
These observations raise the provocative question of whether telomere-specific localization of Cdc13, and its associated proteins Stn1 and Ten1, is restricted to members of the Saccharomyces clade. Specifically, we propose that only Cdc13 from S. cerevisiae (and presumably closely related species) possesses the sequence-specific DNA binding properties that are necessary to target this complex to a specific region of the genome. This proposal is also consistent with the absence of the N-terminal domain of Cdc13 within the Candida clade, which would prevent inappropriate recruitment of telomerase to nontelomeric regions of the genome. The corollary of this proposal is that the evolutionarily conserved task of Stn1 and Ten1 (and their associated large subunit) is a genome-wide role in DNA replication, rather than a direct role in telomere maintenance or chromosome end protection. Consistent with this hypothesis, the human Stn1 protein was first discovered as a subunit of a stimulatory factor for DNA polymerase α/primase.11,12 The exclusive localization of this complex to telomeres in S. cerevisiae may reflect niche specialization, with the acquisition of specificity for telomeric ssDNA driven by evolutionary pressures within the Saccharomyces clade.
Supplementary Material
Funding Sources
Supported by the Lebensfeld Foundation (to V.L.) and by National Science Foundation Grant 0617956 (to D.S.W.).
Footnotes
Supporting Information. Supplementary Table S1, Figure S1, and detailed experimental methods. This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES
- (1).de Lange T (2009) How telomeres solve the end-protection problem. Science 326, 948–952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (2).Lustig AJ (2001) Cdc13 subcomplexes regulate multiple telomere functions. Nat. Struct. Mol. Biol 8, 297–299. [DOI] [PubMed] [Google Scholar]
- (3).Nugent CI, Hughes TR, Lue NF, and Lundblad V (1996) Cdc13p: A single-strand telomeric DNA-binding protein with a dual role in yeast telomere maintenance. Science 274, 249–252. [DOI] [PubMed] [Google Scholar]
- (4).Anderson EM, Halsey WA, and Wuttke DS (2002) Delineation of the high-affinity single-stranded telomeric DNA-binding domain of Saccharomyces cerevisiae Cdc13. Nucleic Acids Res. 30, 4305–4313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (5).Garvik B, Carson M, and Hartwell L (1995) Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol. Cell. Biol 15, 6128–6138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Grandin N, Reed SI, and Charbonneau M (1997) Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 11, 512–527. [DOI] [PubMed] [Google Scholar]
- (7).Grandin N, Damon C, and Charbonneau M (2001) Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 20, 1173–1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Gao H, Cervantes RB, Mandell EK, Otero JH, and Lundblad V (2007) RPA-like proteins mediate yeast telomere function. Nat. Struct. Mol. Biol 14, 208–214. [DOI] [PubMed] [Google Scholar]
- (9).Gelinas AD, Paschini M, Reyes FE, Héroux A, Batey RT, Lundblad V, and Wuttke DS (2009) Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc. Natl. Acad. Sci U.S.A 106, 19298–19303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, Lue NF, and Lei M (2009) Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 23, 2900–2914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Goulian M, Heard CJ, and Grimm SL (1990) Purification and properties of an accessory protein for DNA polymerase α/primase. J. Biol. Chem 265, 13221–13230. [PubMed] [Google Scholar]
- (12).Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, and Pilz RB (2009) A DNA polymerase α/primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J. Biol. Chem 284, 5807–5818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, Leehy K, Heacock M, Price CM, and Shippen DE (2009) Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol. Cell 36, 207–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, Saito M, and Ishikawa F (2009) RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol. Cell 36, 193–206. [DOI] [PubMed] [Google Scholar]
- (15).Dujon B (2006) Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 22, 375–387. [DOI] [PubMed] [Google Scholar]
- (16).Sun J, Yang Y, Wan K, Mao N, Yu T-Y, Lin Y-C, DeZwaan DC, Freeman BC, Lin J-J, Lue NF, and Lei M (2011) Structural bases of dimerization of yeast telomere protein Cdc13 and its interaction with the catalytic subunit of DNA polymerase α. Cell Res. 21, 258–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Mandell EK (2009) Structure function analysis of telomeric proteins. Thesis Dissertation, Baylor College of Medicine, Houston. [Google Scholar]
- (18).Milton-Fry RM, Anderson EM, Hughes TR, Lundblad V, and Wuttke DS (2002) Conserved elements for single-stranded telomeric DNA recognition. Science 296, 145–147. [DOI] [PubMed] [Google Scholar]
- (19).Anderson EM, Halsey WA, and Wuttke DS (2003) Site-directed mutagenesis reveals the thermodynamic requirements for single-stranded DNA recognition by the telomere-binding protein Cdc13. Biochemistry 42, 3751–3758. [DOI] [PubMed] [Google Scholar]
- (20).Scannell DR, Byrne KP, Gordon JL, Wong S, and Wolfe KH (2006) Multiple rounds of speciation associated with reciprocal gene loss in polyploid yeasts. Nature 440, 341–345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


