Abstract
The interplay between gut microbiota and host is crucial for maintaining host health. When this balance is broken, various diseases can arise, including colorectal cancer (CRC). However, the mechanism by which gut microbiota and host interactions mediate CRC development remains unclear. Here, we found that Gasdermin D (GSDMD), an inflammasome effector responsible for forming membrane pores to mediate cell pyroptosis, was upregulated in both human and mouse intestinal tumor samples. GSDMD deficiency significantly suppressed intestinal tumor development in Apcmin/+ mice, a spontaneous CRC mouse model. Apcmin/+Gsdmd−/− mice exhibited reduced IL-1β release in the intestine, and the administration of recombinant mouse IL-1β partially restored intestinal tumor development in Apcmin/+Gsdmd−/− mice. Moreover, 16s rRNA sequencing showed a substantial increase in Lactobacillus abundance in the feces of Apcmin/+Gsdmd−/− mice compared to Apcmin/+ mice. Concurrently, Kynurenine (Kyn), a metabolite derived from host tryptophan (Trp) metabolism, was significantly decreased in the feces of Apcmin/+Gsdmd−/− mice, as shown by metabolite analysis. Additionally, Kyn levels were inversely correlated with Lactobacillus abundance. Furthermore, the administration of exogenous Kyn also promoted intestinal tumor development in Apcmin/+Gsdmd−/− mice. Thus, GSDMD promotes spontaneous CRC development through increasing IL-1β release and Kyn production. Our data suggest an association between GSDMD, gut microbiota, the host Trp/Kyn pathway, and CRC development.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12964-024-01890-6.
Keywords: Colon cancer, GSDMD, Gut microbiota, Metabolite
Introduction
Colorectal cancer (CRC) is the third most common malignancy and one of the leading causes of death worldwide [1]. The adenomatous polyposis coli (Apc) gene functions as a tumor suppressor, and its loss leads to the development of intestinal tumor. The Apcmin/+ mice strain, which harbors an inactivating mutation in the Apc gene, is a widely used spontaneous mouse model for CRC [2, 3]. This model is clinically relevant because the intravillus adenomas in Apcmin/+ mice resemble those found in the duodenum of patients with familial adenomatous polyposis (FAP) [4].
CRC is closely linked to gut microbiota and inflammation [5]. The interaction between gut microbiota and the host is critical for maintaining intestinal homeostasis. When the balance is disrupted, aberrant inflammation and metabolic dysregulation may trigger diseases such as CRC. Lactobacillus is commonly observed to decrease in CRC cases and is considered a probiotic that may help prevent CRC development. However, the precise mechanisms by which Lactobacillus suppresses CRC development are not yet fully understood [5].
The inflammasome is an intracellular multiprotein complex responsible for the maturation of interleukin-1β (IL-1β) and IL-18, as well as the induction of cell pyroptosis [6]. While the primary function of the inflammasome is to protect the host from pathogenic infections, chronic activation leads to pathological inflammation, thereby exacerbating CRC progression. Several studies have demonstrated that the inflammasome pathway is critical in CRC development [7]. Major inflammasome components, such as NLRP3, NLRP1b, NLRC4, NLRP6, Pyrin, ASC, AIM2, Caspase 1, Caspase 11, IL-18, and IL-1β, play important roles in CRC development [1, 8]. On the other hand, some reports suggest that the inflammasome can suppress CRC development through IL-18 dependent and independent mechanisms [8]. IL-18 has been shown to suppress CRC development by promoting epithelial cells regeneration and inhibiting the expression of IL-22 binding protein (IL-22 bp) and IL-22 receptor (IL-22R). Conversely, IL-1β is widely recognized for its role in promoting CRC by enhancing tumor cell proliferation and shaping the tumor microenvironment [9–12].
GSDMD is a novel inflammasome effector which triggers the release of mature IL-1β and IL-18, and promotes cell pyroptosis [13, 14]. Upon activation by damage-associated molecular patterns (DAMPs), GSDMD is cleaved by Caspase 1/11 in mice or Caspase 1/4/5 in humans, yielding the active N-terminal domain (GSDMD-N) and the C-terminal domain (GSDMD-C). The GSDMD-N fragment translocates to the plasma membrane, mediating cell pyroptosis and the release of mature IL-1β and IL-18 [15]. In canonical inflammasome, Caspase 1 cleaves GSDMD to initiate cell pyroptosis, while in noncanonical inflammasome, lipopolysaccharide (LPS) directly activates Caspase 4/5/11 in cytoplasm, which in turn cleaves GSDMD to mediate pyroptosis [16]. GSDMD has been shown to enhance chemotherapy drug-induced cell death in colon cancer [17, 18]. However, its role in spontaneous colon cancer remains controversial. One study showed that GSDMD repressed inflammation-induced CRC progression [19], while another study found that a point mutation in leucine-rich repeat kinase 2 (LRRK2), G2019S, promoted inflammation-associated CRC through LRRK2-GSDMD-mediated gut inflammation [20].
Kynurenine (Kyn), a metabolite derived from tryptophan, is considered to be an oncometabolite in CRC [21]. Kyn levels are elevated in colon cancer tissues compared to healthy samples [22, 23]. Kyn facilitates CRC development through several mechanisms, including the promotion of regulatory T cell (Treg) differentiation [22] and CD8+ T cell exhaustion [24], increased proliferation of colon cancer cells, and the suppression of apoptosis [23, 25].
In the study, we found that GSDMD was upregulated in intestinal cancer and promoted IL-1β-dependent spontaneous intestinal cancer development. GSDMD deficiency led to an increased abundance of Lactobacillus and a decreased Kyn levels in the feces of spontaneous intestinal cancer mice. Kyn, a metabolite of tryptophan derived from the host rather than from bacteria, was shown to restore intestinal tumor development in Apcmin/+Gsdmd−/− mice. These findings suggest that GSDMD promotes CRC development through increasing IL-1β release and Kyn production. Additionally, our results indicate that GSDMD deficiency alters the gut microbiota composition, which is associated with changes in the host Trp/Kyn metabolic pathway during CRC development.
Materials and methods
Human colorectal cancer tissue microarray
A commercial tissue microarray (HColA160CS01) was purchased from Shanghai Outdo Biotech (Shanghai, China). Anti-GSDMD antibody (Proteintech) was used to evaluate GSDMD expression in intestinal cancer via immunohistochemistry (IHC). Microwave heating was used for antigen retrieval in citrate buffer (pH 6.0) for 5 min. The tissue microarray was incubated with the anti-GSDMD antibody at 4˚C overnight. Slides were analyzed independently by two pathologists. Staining intensity was graded from 0 (negative staining) to 3 (strong positive staining), with intermediate scores of 0.5, 1, and 2. Positive percentage scores ranged from 0 to 100%. The overall GSDMD expression score was calculated by multiplying the staining intensity by the positive percentage score.
Reagents
Anti-GSDMD antibody for immunoblot analysis was sourced from Abcam (Cambridge, UK), while the anti-GAPDH antibody was obtained from Cell Signaling Technology (Danvers, MA, US). ELISA kits for tryptophan (Trp) and kynurenine (Kyn) were purchased from Senbeijia Biotech (Nanjing, China).
Mice
Gsdmd−/− mice and Apcmin/+ mice on the C57BL/6 background were purchased from GemPharmatech (Nanjing, China). All mice were housed under specific pathogen-free conditions, and littermates from the same mouse line were bred as strict controls. Apcmin/+ and Apcmin/+Gsdmd−/− mice were sacrificed at 20 weeks of age for analysis. 8-week-old Apcmin/+Gsdmd−/− mice were injected intraperitoneally once per week with L-KYN (2922-83-0, Solarbio, Beijing, China) at 5 mg/kg body weight or with PBS (control mice) and sacrificed at 20 weeks for analysis. Macroscopic intestinal tumors were counted and measured with the caliper. Tumor load was calculated as the sum of all tumors’ diameters present in a given mouse. All animal experiments were performed in compliance with the guide for the care and use of laboratory animals and were approved by the institutional biomedical research ethics committee of Guangdong Medical University.
Recombinant mouse IL-1β administration
Recombinant mouse IL-1β (Novoprotein, Shanghai, China) or PBS as negative control was injected into eight-week-old Apcmin/+ and Apcmin/+Gsdmd−/− mice intraperitoneally at a dose of 0.5 µg per mouse (in 200 µl sterile PBS) twice weekly, and the mice were sacrificed at 20 weeks of age for analysis.
RT-PCR
The procedure for real-time PCR has been described previously [26]. Briefly, total RNA was extracted from intestinal tumors of Apcmin/+, Apcmin/+Gsdmd−/−, and IL-1β-treated Apcmin/+Gsdmd−/− mice, as well as from control intestinal tissues of WT and Gsdmd−/− mice using TRIzol® Reagent (Invitrogen, Shanghai, China). cDNA was synthesized with PrimeScript™ RT Master Mix (Takara Bio, Dalian, China). The levels of genes of the interest were quantified using TB Green®Premix Ex Taq™ (Tli RNaseH Plus) (Takara Bio). The expression levels of the genes were calculated by the 2−ΔΔCt method and normalized to β-actin. Amplification of cDNA was performed on a ViiA 7 Real-Time PCR system (Applied Biosystems, Foster City, CA, USA) with the sequences of oligonucleotide primers listed in Supplementary Table 1.
Immunoblot analysis
The protocol for immunoblot analysis has been previously reported [27]. In brief, the tissues were lysed for 30 min in ice-cold RIPA lysis buffer supplemented with 10 mM sodium fluoride (NaF), 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, and a complete protease inhibitor cocktail (Roche). Lysates were separated via SDS-PAGE and transferred to polyvinylidene fluoride (PVDF) membranes (Millipore, Shanghai, China). Membranes were blocked at room temperature for 1 h and incubated overnight at 4 °C with primary antibodies. After incubation with secondary antibodies at room temperature for 1 h, blots were visualized using enhanced chemiluminescence (ECL) reagents (Millipore).
Histology
Intestinal tissue was fixed in 4% paraformaldehyde (PFA) for 48 h and then embedded in paraffin wax. 5-mm sections were stained with hematoxylin-eosin (H&E). To assess cell proliferation, paraffin sections were stained using anti-Ki67 antibody (Servicebio, Wuhan, Hubei, China).
Enzyme-linked immunosorbent assay (ELISA)
To measure the protein level of IL-1β in intestinal tumors, intestinal tumors from Apcmin/+ and Apcmin/+Gsdmd−/− mice, as well as control intestinal tissues from WT and Gsdmd−/− mice were weighed and mechanically homogenized in PBS containing 1% NP-40 and a complete protease inhibitor cocktail (Roche). The protein level of IL-1β in intestine tissue homogenate was quantified using an IL-1β ELISA kit (R&D Systems, Minneapolis, MN) according to the manufacturer’s instructions.
To determine the levels of Kyn and Trp in the colon, the concentrations of these metabolites in colon tissues from Apcmin/+ and Apcmin/+Gsdmd−/− mice were quantified using corresponding ELISA kits (Senbeijia, Nanjing, China), in accordance with the manufacturer’s protocol.
To assess the levels of Kyn in serum and feces, eight-week-old Apcmin/+Gsdmd−/− mice received weekly injections of either Kyn or PBS. Two weeks later, Kyn levels in the serum and feces of these mice were measured using a Kyn ELISA kit (Senbeijia, Nanjing, China), following the manufacturer’s protocol.
Flow cytometry analysis
Freshly isolated tumor tissues were chopped into small pieces and digested with 0.01% (w/v) Liberase TH (Roche, 5401151001) and 100 U/ml DNase I (Roche, 10104159001) in RPMI 1640 for 20 min at 37 °C. Cells were filtered through a 40-µm cell strainer and washed with 5 ml wash buffer (1 × PBS with 2 mM EDTA and 0.5% BSA), followed by centrifugation at 200 g for 5 min. The cells were stained with fluorescence-labeled anti-mouse CD45 (Biolegend, 103132), anti-mouse CD4 (Biolegend, 100406), anti-mouse CD8 (Biolegend, 100712), anti-mouse Foxp3 antibodies (Biolegend, 118904) using the Foxp3/Transcription Factor Staining Buffer Set (ThermoFisher, 00-5523-00). Cells were washed, resuspended in PBS containing 1% BSA, and analyzed by BD FACS Aria II flow cytometer (Franklin Lakes, NJ, USA).
16S ribosomal RNA gene sequencing
Genomic DNA was extracted from fecal samples of 10-week-old Apcmin/+ and Apcmin/+Gsdmd−/− mice using the OMEGA Soil DNA Kit (M5635-02) (Omega Bio-Tek, Norcross, GA, USA), and stored at -20 °C for further analysis. DNA quality and quantity were assessed via agarose gel electrophoresis and NanoDrop NC2000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA). PCR amplification of the bacterial 16S rRNA genes V3-V4 region was performed using the forward primer 338 F (5’-ACTCCTACGGGAGGCAGCA-3’) and the reverse primer 806R (5’-GGACTACHVGGGTWTCTAAT-3’). Sample-specific 7-bp barcodes were incorporated into the primers for multiplex sequencing. The PCR products were purified with Vazyme VAHTSTM DNA Clean Beads (Vazyme, Nanjing, China) and quantified using the Quant-iT PicoGreen dsDNA Assay Kit (Invitrogen, Carlsbad, CA, USA). After the individual quantification step, the PCR amplicons were pooled in equal amounts, and pair-end 2*250 bp sequencing was performed using the Illumina NovaSeq platform with NovaSeq 6000 SP Reagent Kit (500 cycles). Data analyses were mainly performed using QIIME2 and R packages (v3.2.0). Alpha diversity indices, including Chao1 richness estimator and Simpson index, were calculated using the amplicon sequence variants (ASV) table in QIIME2. Principal coordinates analysis (PCoA) was performed by QIIME2 (2019.4). Venn diagram was generated to visualize the shared and unique ASVs among samples or groups using R package “VennDiagram”, based on the occurrence of ASVs across samples/groups regardless of their relative abundance (Zaura, Keijser et al. 2009). Taxa abundances at the ASV levels were statistically compared among samples or groups by MetagenomeSeq, and visualized as Manhattan plots (Zgadzaj, R et al. 2016). Linear discriminant analysis effect size (LEfSe) was performed to detect differentially abundant taxa across groups using the default parameters (Segata, Izard et al. 2011). Heatmaps were performed using R package “pheatmap”.
Preparation of fecal supernatant and analysis of metabolites
Fecal samples from 10-week-old Apcmin/+ and Apcmin/+Gsdmd−/− mice were prepared for Liquid chromatography-mass spectrometry (LC-MS) at BioNovoGene (Suzhou, China). In brief, samples were vortexed for 30s in a 2 ml centrifuge tube containing 600 µl MeOH. Steel balls were added to the tubes and placed in a tissue grinder for 120s at 50 Hz. The samples were treated with ultrasound at room temperature for 10 min, and then centrifuged at 12,000×g for 10 min at 4 ℃. The supernatants were filtered with 0.22-µm membrane and transferred into the detection bottle for LC-MS detection. Samples were randomized, data acquisitions were done in one batch to eliminate system errors, and the metabolites were identified based on their molecular weight, mass spectra, and retention time. The raw data were converted to mzXML format by MSConvert in ProteoWizard software package (v3.0.8789) and processed with R XCMS (v3.12.0) for feature detection, retention time correction and alignment. Annotation of metabolite using LC-MS data was done with the Compound Discover 3.3 (Thermo Scientific, Waltham, MA, USA) and referenced to the mzCloud and HMDB database. Differently expressed metabolites between two classes of samples were conducted using a statistically significant threshold of Variable Importance in Projection (VIP) value (VIP > 1) and p ≤ 0.05 using Student’s t-test analysis. The partial least squares discriminant analysis (PLS-DA) was performed with R package ropls (v1.22.0). Heatmaps were constructed using the “pheatmap” package in R language. Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis and network diagram were performed with MetaboAnalyst (www.metaboanalyst.ca.). Correlated heatmaps were carried out with R package cor (v4.0.3). P value < 0.05 was set as the cut-off and activation z-scores were calculated.
Statistical analysis
GraphPad Prism Software was used to perform statistical analysis and graph development. Differences between groups were evaluated using a two-tailed Student’s t-test. Correlations between GSDMD expression and clinicopathological characteristics were assessed using Fisher’s exact test. Statistical significance was set at p < 0.05. Pearson correlation analysis was used to evaluate relationships between microbiota and metabolite data.
Results
GSDMD is upregulated in intestinal cancer
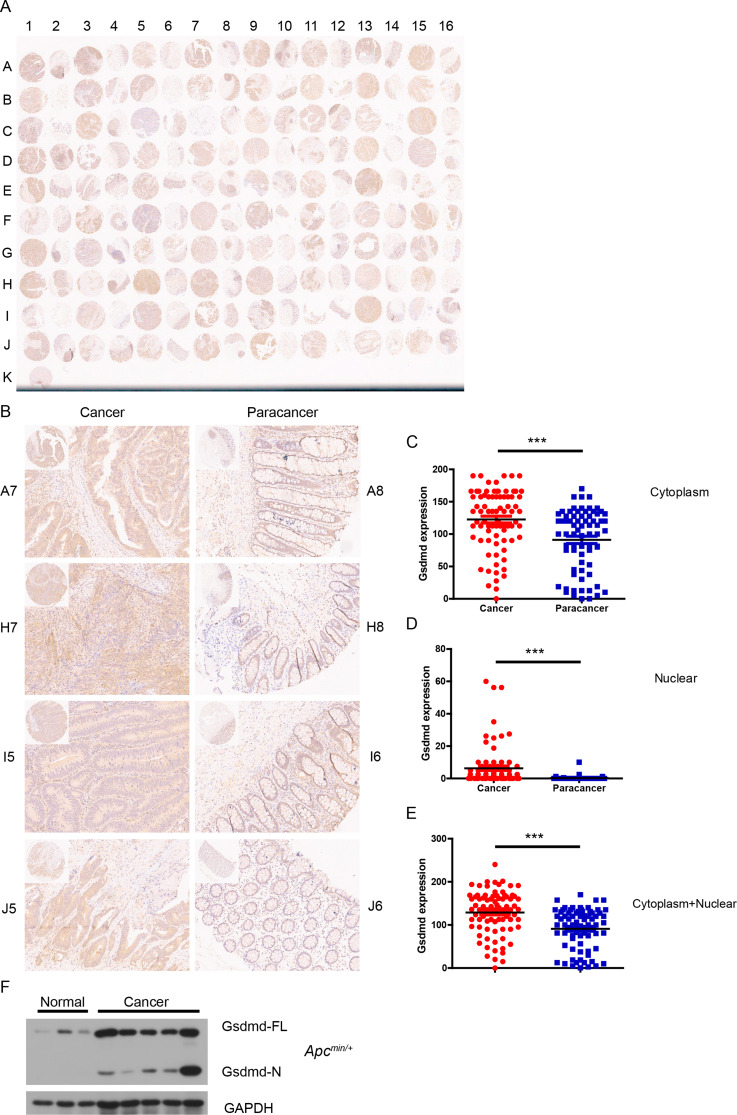
To investigate GSDMD expression in intestinal cancer, a human tissue microarray consisting of 80 colorectal cancer samples and their corresponding adjacent non-tumor specimens was analyzed for GSDMD protein levels using immunohistochemistry (IHC). Compared to adjacent non-tumor tissues, GSDMD expression was significantly elevated in intestinal cancer (Fig. 1A-E). Although GSDMD was predominantly localized in the cytoplasm (Fig. 1C), nuclear expression was detected in 57.5% (46/80) of tumor samples, whereas only 6.25% (5/80) of adjacent tissues exhibited GSDMD expression in the nucleus (Fig. 1D). Additionally, GSDMD expression was higher in the cytoplasm of tumor cells than in the nucleus (Fig. 1C and D). These findings suggest that GSDMD may translocate from the cytoplasm to the nucleus during the development of intestinal cancer. Fisher’s exact test revealed a significant positive correlation between GSDMD expression and both tumor grade and tumor size, while no significant associations were found with patient sex, age, or tumor stage (Table 1).
Fig. 1.
GSDMD is upregulated in intestinal cancer. (A) IHC analysis of GSDMD protein level in a human tissue microarray, including 80 colorectal cancer and matched adjacent tumor specimens. (B) IHC analysis of GSDMD protein level in four colorectal cancer tissue and matched adjacent cancer tissue from (A) (200× magnification). (C-E) GSDMD expression score in cytoplasm (C), nuclear (D), and cytoplasm + nuclear (E) as calculated by multiplying the intensity and positive percentage scores according to (A). (F) Western blot analysis of GSDMD in in small intestine tumor tissue from Apcmin/+ mice (n = 5) and normal tissue from WT mice (n = 3). Data are representative of at least three independent experiments (mean ± SEM). ***p < 0.001 by Student’s t test. FL means full length
Table 1.
Correlation between GSDMD expression and clinicopathological characteristics
| variables | GSDMD expression | total | p value | r value | ||
|---|---|---|---|---|---|---|
| low | high | |||||
| Sex | 1 | 0.024 | ||||
| Male | 30 | 13 | 43 | |||
| Female | 25 | 12 | 37 | |||
| Age (year) | 0.453 | -0.089 | ||||
| ≤ 60 | 17 | 10 | 27 | |||
| > 60 | 38 | 15 | 53 | |||
| Grade | 0.032 | 0.27 | ||||
| I/II | 48 | 16 | 64 | |||
| III | 7 | 9 | 16 | |||
| Tumor size | 0.026 | 0.259 | ||||
| ≤ 4.7 cm | 26 | 5 | 31 | |||
| > 4.7 cm | 29 | 20 | 49 | |||
| T stage | 0.635 | -0.061 | ||||
| T1-T3 | 25 | 13 | 38 | |||
| T4 | 30 | 12 | 42 | |||
| N stage | 0.453 | 0.089 | ||||
| N0 | 38 | 15 | 53 | |||
| N1/N2 | 17 | 10 | 27 | |||
| TNM stage | 0.314 | 0.127 | ||||
| I/II | 38 | 14 | 52 | |||
| III/IV | 17 | 11 | 28 | |||
In addition to human intestinal cancer, GSDMD expression was also increased in small intestinal tumor of Apcmin/+ mice, a model of spontaneous intestinal carcinogenesis (Fig. 1F). To determine whether GSDMD is activated in intestinal cancer, normal small intestine tissue and intestinal tumor tissue from Apcmin/+ mice were analyzed by immunoblot analysis, and we found that GSDMD was significantly activated in tumor tissue but not in normal tissue (Fig. 1F), suggesting that GSDMD is activated in intestinal cancer. Thus, GSDMD is upregulated in both human and mouse intestinal cancer tissues.
GSDMD promotes spontaneous intestinal cancer progression
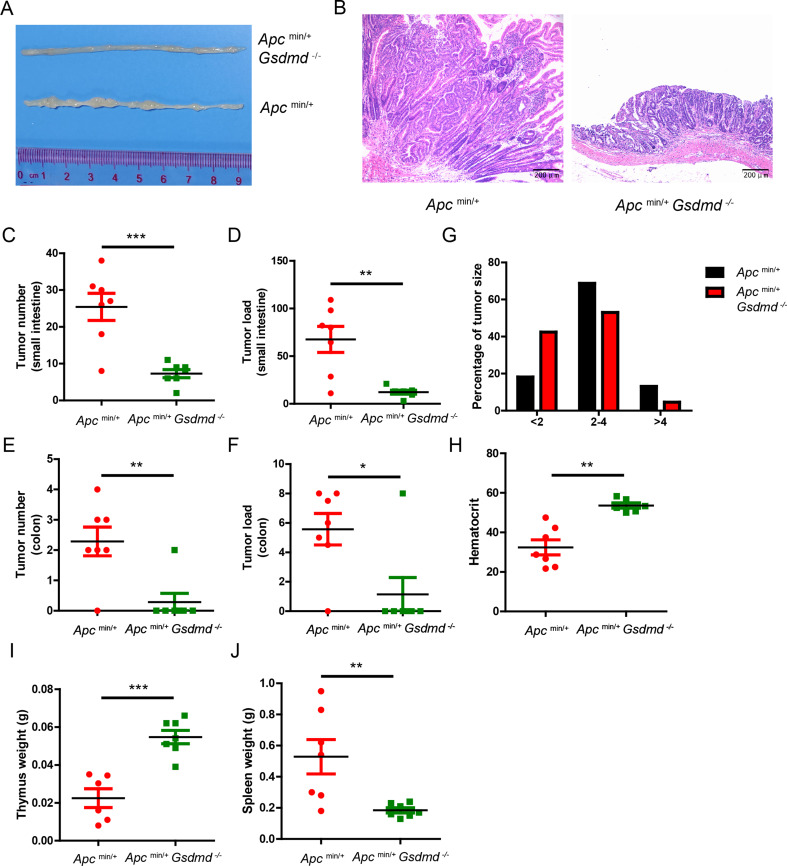
To explore the functional role of GSDMD in spontaneous intestinal cancer, Gsdmd-deficient mice were crossed with Apcmin/+ mice to generate Apcmin/+Gsdmd−/− mice. Deficiency of Gsdmd dramatically inhibited the formation of small intestinal tumors (Fig. 2A). Histological analysis revealed a marked reduction in tumor size in Apcmin/+Gsdmd−/− mice (Fig. 2B). Both tumor number (Fig. 2C and E) and tumor load (Fig. 2D and F) were substantially reduced in the small intestines and colons of Apcmin/+Gsdmd−/− mice. Tumor size distribution analysis further demonstrated that Apcmin/+Gsdmd−/− mice had a higher proportion of smaller tumors (Fig. 2G). Consistent with severely decreased intestinal tumor development, compared to Apcmin/+ mice, the anemia and thymus atrophy were notably improved in Apcmin/+Gsdmd−/− mice (Fig. 2H and I). Additionally, Apcmin/+ mice had much larger spleen than Apcmin/+Gsdmd−/− mice (Fig. 2J). These data suggest that GSDMD is critical for the development of spontaneous intestinal cancer.
Fig. 2.
Deficiency of GSDMD suppresses spontaneous intestinal cancer development. (A) Macroscopic view of representative small intestine from 20-week old Apcmin/+ and Apcmin/+Gsdmd−/− mice. (B) Hematoxylin and eosin (H&E) staining of the representative intestinal tumor from the Apcmin/+ and Apcmin/+Gsdmd−/− mice as in (A) (100× magnification). (C-F) Tumor number (C and E) and tumor load (D and F) from the small intestines or colons of 20-week-old Apcmin/+ (n = 7) and Apcmin/+Gsdmd−/− (n = 7) mice. (G) Histogram showing the size distribution of tumors from the small intestines of 20-week-old Apcmin/+ (n = 7) and Apcmin/+Gsdmd−/− (n = 7) mice. (H-J) Hematocrit (H), thymus weight (I) and spleen weight (J) of 20-week-old Apcmin/+ (n = 6–7) and Apcmin/+Gsdmd−/− (n = 6–7) mice. Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test
GSDMD promotes intestinal tumorigenesis via IL-1β release
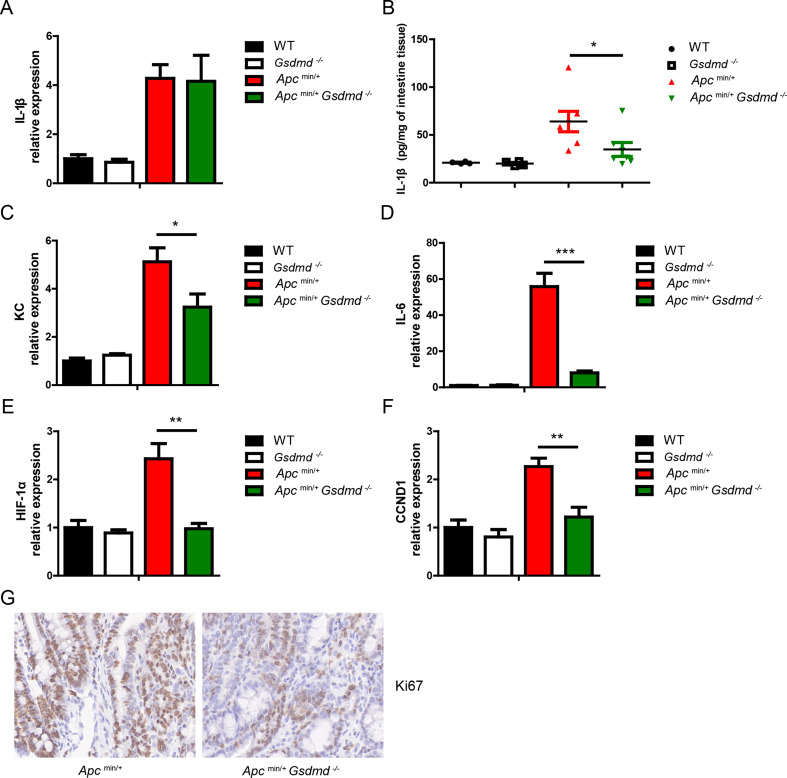
IL-1β, a cytokine known to promote intestinal cancer, is released upon inflammasome activation, a process regulated by GSDMD [16]. To determine whether GSDMD influences IL-1β release during spontaneous intestinal cancer development, we compared Apcmin/+ and Apcmin/+Gsdmd−/− mice. While the mRNA levels of IL-1β were similar in tumors from both genotypes (Fig. 3A), the protein levels of IL-1β were significantly higher in tumors from Apcmin/+ mice (Fig. 3B). These findings suggest that GSDMD facilitates IL-1β release from small intestinal tumors. Some studies reported that IL-1β could promote intestinal cancer progression by promoting colon cancer growth and regulating tumor microenvironment [9–12]. Further analysis revealed that chemokine KC, HIF-1α, and pro-proliferative factors (IL-6 and CCND1) were markedly increased in Apcmin/+ tumors compared to Apcmin/+Gsdmd−/− tumors (Fig. 3C-F). The number of Ki67-positive cells, indicative of proliferating cells, was also significantly higher in Apcmin/+ tumors (Fig. 3G), suggesting an increased proliferation in the small intestinal tumor from Apcmin/+ mice. These data demonstrate that GSDMD promotes IL-1β release from intestinal tumor, and suggest that IL-1β may promote tumor progression by increasing KC, HIF-1α expression and epithelial cells proliferation.
Fig. 3.
Deficiency of GSDMD suppresses IL-1β release in Apcmin/+ mice. (A) Quantitative mRNA expression of GSDMD from small intestines of WT (n = 4), Gsdmd−/− (n = 4), Apcmin/+ (n = 7) and Apcmin/+Gsdmd−/− (n = 6) mice. (B) ELISA analysis of GSDMD protein level from small intestines of WT (n = 4), Gsdmd−/− (n = 4), Apcmin/+ (n = 7) and Apcmin/+Gsdmd−/− (n = 7) mice. (C-F) Quantitative mRNA expression of KC (C), IL-6 (D), HIF-1α (E), and CCND1 (F) from small intestines of WT (n = 4), Gsdmd−/− (n = 4), Apcmin/+ (n = 7) and Apcmin/+Gsdmd−/− (n = 6) mice. (G) Ki67 staining of small intestine from above mice as in (A) (400× magnification). Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test
Exogenous IL-1β partially rescues intestinal tumor formation in Apcmin/+Gsdmd−/− mice
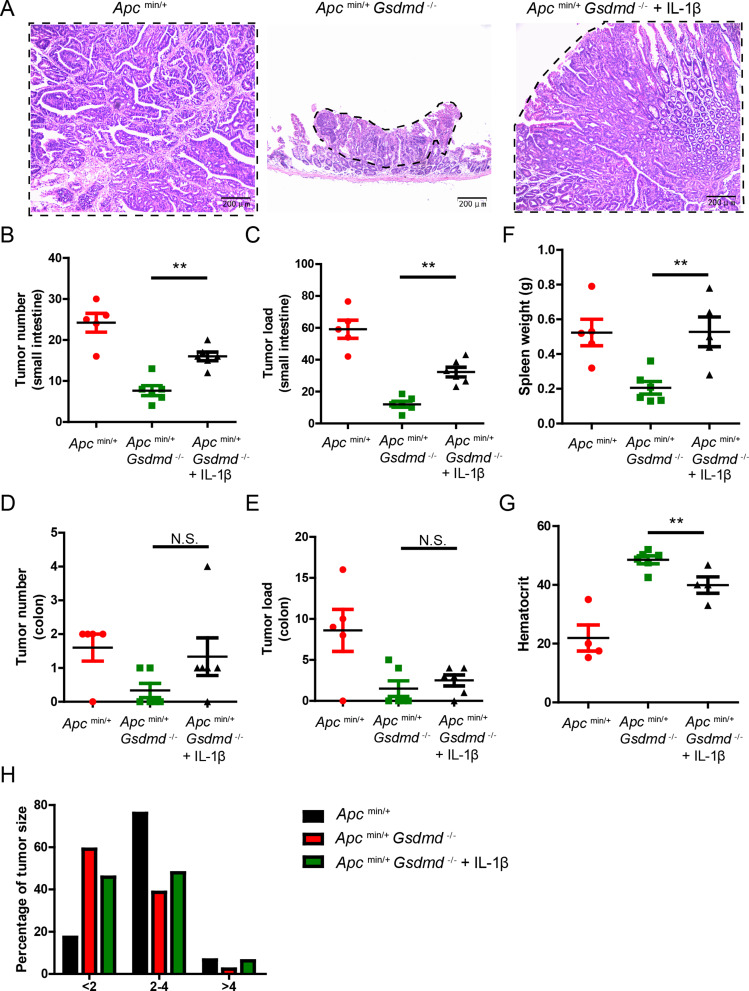
To determine whether GSDMD promotes intestinal tumorigenesis through IL-1β, we administrated recombinant mouse IL-1β (0.5 µg) intraperitoneally twice weekly to 8-week old Apcmin/+Gsdmd−/− mice. We found that exogenous IL-1β significantly increased small intestinal tumor size in Apcmin/+Gsdmd−/− mice (Fig. 4A). The IL-1β-treated Apcmin/+Gsdmd−/− mice exhibited increased tumor number and tumor load of small intestine (Fig. 4B and C). However, both the tumor number and tumor load were lower than those in Apcmin/+ mice. Notably, exogenous IL-1β did not increase the number or load of colon tumors (Fig. 4D and E), but it did increase spleen weight (Fig. 4F) and exacerbate anemia (Fig. 4G). Tumor size distribution analysis showed that exogenous IL-1β reduced the number of small tumors in Apcmin/+Gsdmd−/− mice (Fig. 4H). The data suggest that exogenous IL-1β partially restores intestinal tumor formation in Apcmin/+Gsdmd−/− mice. Furthermore, exogenous IL-1β reinstated KC, HIF-1α, IL-6, and CCND1 expression in these mice (Figure S1A-S1D), and the number of Ki67-positive cells was also increased (Figure S1E). Together, these findings suggest that GSDMD promotes intestinal cancer progression in part by inducing IL-1β release.
Fig. 4.
Exogenous IL-1β promotes intestinal tumor development in Apcmin/+Gsdmd−/− mice. (A) 8-week-old Apcmin/+Gsdmd−/− mice simultaneously received injection of IL-1β or PBS twice a week, while age and sex-matched Apcmin/+ mice were injected with PBS (n = 5–6/group). H&E staining of the representative intestinal tumor from the 20-week old above mice (100× magnification). Tumors are circled with dashed lines. (B-E) Tumor number (B and D) and tumor load (C and E) from the small intestines or colons of 20-week-old above mice as in (A). (F-G) Spleen weight (F), and hematocrit (G) of 20-week-old above mice as in (A). (H) Histogram showing the size distribution of tumors from the small intestines of 20-week-old above mice as in (A). Data are representative of at least three independent experiments (mean ± SEM). **p < 0.01 by Student’s t test. N.S. means no significance
Apcmin/+Gsdmd−/− mice exhibit increased Lactobacillus abundance
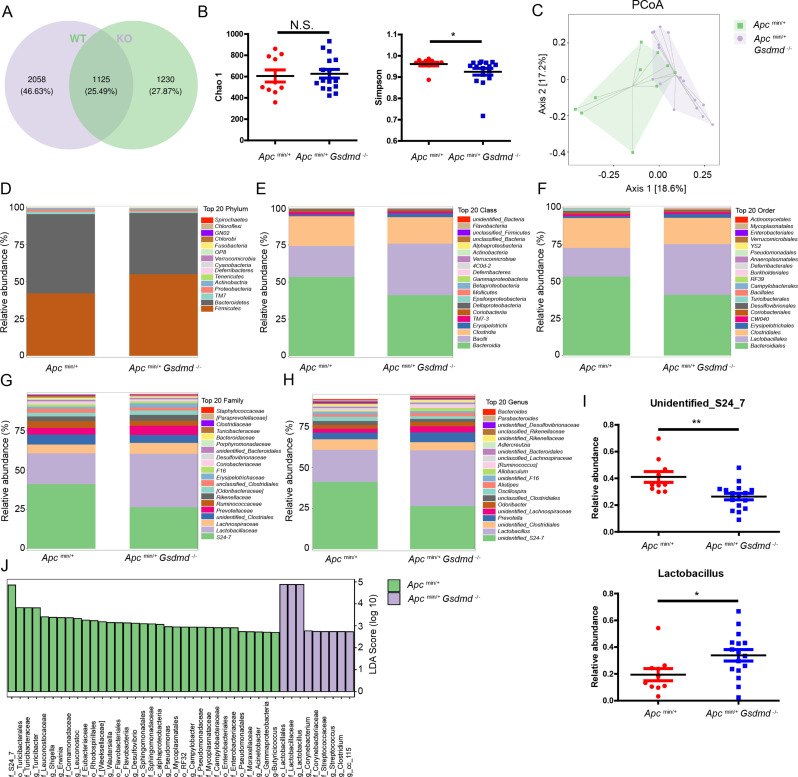
Previous studies have shown that gene modifications can alter the composition of the gut microbiota in mice, and some of these alterations are associated with intestinal cancer development [5]. To investigate the differences in the gut microbiota between Apcmin/+ mice and Apcmin/+Gsdmd−/− mice, we performed 16S ribosomal RNA gene sequencing on fecal samples. A Venn diagram indicated that Apcmin/+ mice had 2,058 specific operational taxonomic units (OTUs), while Apcmin/+Gsdmd−/− mice had 1,230 specific OTUs, with 1,125 OTUs shared between both groups (Fig. 5A). The Simpson index revealed altered alpha diversity in Apcmin/+Gsdmd−/− mice compared to Apcmin/+ mice, although the Chao 1 index was similar between the two groups (Fig. 5B). The community structure of the fecal microbiota also differed markedly between Apcmin/+Gsdmd−/− mice and Apcmin/+ mice (Fig. 5C). A heatmap illustrated differences in the gut microbiota composition between the two groups (Figure S2), which were further analyzed at the phylum, class, order, family, and genus levels (Fig. 5D-I). At the phylum level, Firmicutes were significantly increased, while Bacteroidetes were decreased in Apcmin/+Gsdmd−/− mice compared to Apcmin/+ mice (Fig. 5D). Consistent with these changes, Bacilli (class level), Lactobacillales (order level), Lactobacillaceae (family level), and Lactobacillus (genus level) were significantly enriched in Apcmin/+Gsdmd−/− mice, while Bacteroidia (class level), Bacteroidiales (order level), and Lachnospiraceae (family level) were reduced (Fig. 5E- G). At the genus level, Apcmin/+Gsdmd−/− mice exhibited significantly higher levels of Lactobacillus compared to Apcmin/+ mice (Fig. 5H and I). Linear discriminant analysis effect size (LEfSe) further confirmed the enrichment of Lactobacillales, Lactobacillaceae, and Lactobacillus in Apcmin/+Gsdmd−/− mice (Fig. 5J). These findings suggest that GSDMD deficiency alters the gut microbiota composition, as Apcmin/+Gsdmd−/− mice display an increased abundance of Lactobacillus.
Fig. 5.
Deficiency of GSDMD alters gut microbiota composition of Apcmin/+ mice. (A) Gut microbiota composition of fecal samples from 10-week-old Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice were assessed with 16S rRNA sequencing. Venn diagram of 16S rRNA sequencing data. WT means Apcmin/+ mice; KO means Apcmin/+Gsdmd−/− mice. (B) Chao 1 and Simpson index of 16S rRNA sequencing data as in (A). (C) PCoA analysis of 16S rRNA sequencing data as in (A). (D-H) Relative abundance of top 20 phylum (D), class (E), order (F), family (G), and genus (H) from the gut microbiota of Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. (I) Relative abundance of unidentified_S24_7 and Lactobacillus from the gut microbiota of Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. (J) LEfSe analysis of the relative abundance from the gut microbiota of Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01 by Student’s t test
Apcmin/+Gsdmd−/− mice exhibit reduced kynurenine levels
To determine whether changes in the microbiota composition contribute to intestinal tumor development of Apcmin/+Gsdmd−/− mice through microbiota-associated metabolites, we further investigated the metabolites in the feces of Apcmin/+Gsdmd−/− mice and Apcmin/+ mice using mass spectrometry. Partial least squares discriminant analysis (PLS-DA) revealed distinct metabolomic profiles between Apcmin/+Gsdmd−/− mice and Apcmin/+ mice (Fig. 6A). A total of 781 metabolites were identified in the feces, with 25 downregulated metabolites and 61 upregulated metabolites in Apcmin/+ mice compared to Apcmin/+Gsdmd−/− mice (Fig. 6B). The upregulated metabolites (Fig. 6C) and downregulated metaboiltes (Figure S3) were showed using heatmaps. The correlation of different metaboiltes was showed with correlated heatmap (Figure S4). Z-score analysis highlighted significant differences in metabolite abundance, with L-Kynurenine (Kyn) being particularly elevated (Fig. 6D). Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis identified tryptophan (Trp) metabolism as the pathway most affected by GSDMD deficiency (Fig. 6E). A network diagram revealed that Kyn, indolepyruvate, quinolinic acid, and 3-methoxyanthranilate were elevated, while 3-hydroxyanthranilate and 2-aminobenzoic acid were reduced in Apcmin/+ mice compared to Apcmin/+Gsdmd−/− mice (Fig. 6F), which was confirmed by intensity analysis (Fig. 6G). Kyn has been implicated in intestinal cancer development [21, 22], and its reduction in Apcmin/+Gsdmd−/− mice may contribute to the reduced tumor formation observed in these animals.
Fig. 6.
Deficiency of GSDMD alters gut metabolites of Apcmin/+ mice. (A) PLS-DA scores for fecal metabolites of Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. (B) Statistics of differentially expressed fecal metabolites in Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. (C) Heatmap analysis of upregulated metabolites in Apcmin/+ (n = 10) mice compared to Apcmin/+Gsdmd−/− (n = 16) mice. WT means Apcmin/+ mice; KO means Apcmin/+Gsdmd−/− mice. (D) Top 15 metabolites of Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice based on Z-score. (E) Top 20 enriched KEGG pathways of differentially expressed fecal metabolites in Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. (F) Network of KEGG enriched pathway. Blue dot: pathway, red dot: upregulated metabolite, purple dot: downregulated metabolite. Metabolites from Apcmin/+ mice vs. metabolites from Apcmin/+Gsdmd−/− mice. (G) Log2(intensity) of Trp pathway metabolites in Apcmin/+ (n = 10) and Apcmin/+Gsdmd−/− (n = 16) mice. Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01 by Student’s t test. N.S. means no significance
Kynurenine levels negatively correlate with Lactobacillus abundance
To determine the correlation of microbiota and metabolites, correlated heatmap was carried out with Pearson correlation analysis. We found that Kyn was positively correlated with Bacteroidia (class level) and negatively correlated with Bacilli (class level) (Figure S5A). Consistent with the class level, Kyn was positively correlated with Bacteroidiales (order level), Lachnospiraceae (family level), and unidentified_S24-7, but negatively correlated with Lactobacillales (order level), Lactobacillaceae (family level), and Lactobacillus (genus level) (Figure S5B-5D). Correlation scatter plots showed that fecal microbiota from Apcmin/+Gsdmd−/− mice had more Bacilli (class level) and fewer Bacteroidia (class level), which were negatively and positively correlated with Kyn respectively (Figure S5E). These trends were consistent across the order, family, and genus levels (Figure S5F-5H). The data suggest that increased Lactobacillus may promote intestinal tumor formation through suppressing Kyn production in Apcmin/+Gsdmd−/− mice.
Exogenous kynurenine promotes colon cancer development in Apcmin/+Gsdmd−/− mice
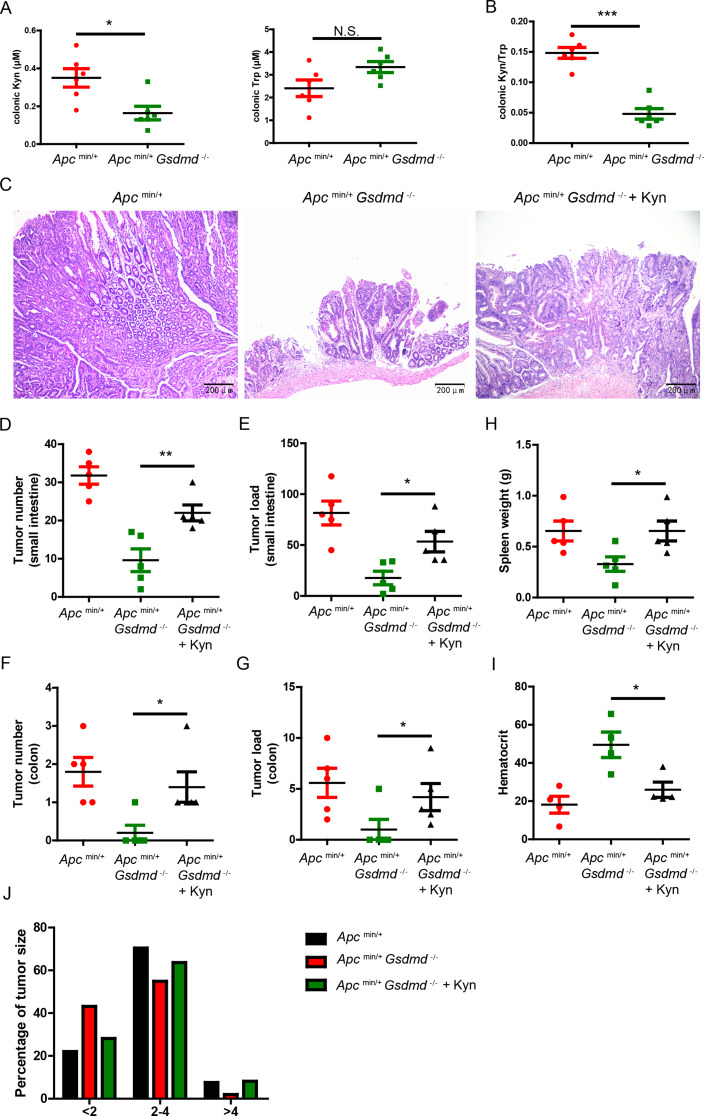
We observed reduced Kyn in the feces of Apcmin/+Gsdmd−/− mice. As is known, Kyn is a metabolite derived from Trp metabolism, which includes endogenous (host) Trp metabolism and bacterial Trp metabolism. Kyn is produced by endogenous Trp metabolism, but not by bacterial Trp metabolism [28]. To confirm this in our setting, we measured Kyn and Trp levels in the colon tissues of mice. Consistent with a previous report [22], we found that Apcmin/+Gsdmd−/− mice exhibited lower Kyn but not Trp levels in colon tissue compared to Apcmin/+ mice (Fig. 7A). The ratio of Kyn/Trp was significantly reduced in the colon of Apcmin/+ mice (Fig. 7B). To determine whether reduced tumor formation in Apcmin/+Gsdmd−/− mice was due to lower Kyn levels, we intraperitoneally injected Kyn into 8-week-old Apcmin/+Gsdmd−/− mice and analyzed tumor development after 12 weeks. We found that the administration of exogenous Kyn increased Kyn levels in the serum and feces of Apcmin/+Gsdmd−/− mice (Figure S6). As expected, exogenous Kyn significantly increased tumor size in the small intestines of Apcmin/+Gsdmd−/− mice (Fig. 7C). Additionally, Kyn-treated Apcmin/+Gsdmd−/− mice exhibited an increased tumor number (Fig. 7D and F) and tumor load (Fig. 7E and G) in both the small intestine and colon. Meanwhile, Kyn increased spleen weight (Fig. 7H) and exacerbated anemia (Fig. 7I) in Apcmin/+Gsdmd−/− mice. Tumor size distribution analysis further indicated that exogenous Kyn increased tumor size in Apcmin/+Gsdmd−/− mice (Fig. 7J). These results suggest that exogenous Kyn partially restores intestinal tumor formation in Apcmin/+Gsdmd−/− mice.
Fig. 7.
Exogenous Kyn promotes intestinal tumor development in Apcmin/+Gsdmd−/− mice. (A) ELISA analysis of Kyn and Trp from colon of Apcmin/+ (n = 6) and Apcmin/+Gsdmd−/− (n = 6) mice. (B) Kyn/Trp was determined as in (A). (C) 8-week-old Apcmin/+Gsdmd−/− mice simultaneously received injection of Kyn or PBS once a week, while age and sex-matched Apcmin/+ mice were injected with PBS (n = 5/group). H&E staining of the representative intestinal tumor from the 20-week old above mice (100× magnification). (D-G) Tumor number (D and F) and tumor load (E and G) from the small intestines or colons of 20-week-old above mice as in (C). (H-I) Spleen weight (H), and hematocrit (I) of 20-week-old above mice as in (C). (J) Histogram showing the size distribution of tumors from the small intestines of 20-week-old above mice as in (C). Data are representative of at least three independent experiments (mean ± SEM). *p < 0.05, **p < 0.01, ***p < 0.001 by Student’s t test. N.S. means no significance
Kyn is known to promote colon cancer by reducing CD8+ T cell numbers and increasing regulatory T (Treg) cell numbers [22, 24]. In Apcmin/+Gsdmd−/− mice, CD8+ T cell numbers were higher in intestinal tumors, and exogenous Kyn reduced these numbers (Fig. 8A and B). Treg cells, which were reduced in Apcmin/+Gsdmd−/− mice, were increased by Kyn treatment (Fig. 8C and D). These data suggest that Kyn promotes tumor development in Apcmin/+Gsdmd−/− mice, likely by enhancing immunosuppression.
Fig. 8.
Exogenous Kyn increases Treg cell number in Apcmin/+Gsdmd−/− mice. (A) 8-week-old Apcmin/+Gsdmd−/− mice simultaneously received injection of Kyn or PBS once a week, while age and sex-matched Apcmin/+ mice were injected with PBS (n = 4/group). Flow cytometry analysis of CD4+ and CD8+ T cell number from intestinal tumor of the 20-week old above mice. (B) Total leukocytes were determined by CD45+ cell. CD4+ T cells % of total leukocyte and CD8+ T cells % of total leukocyte were determined as in (A). (C) Flow cytometry analysis of Treg (CD4+ Foxp3+) cell number from intestinal tumor of the 20-week old mice as in (A). (D) Treg % of CD4+ cells was determined as in (C). Data are representative of at least three independent experiments (mean ± SEM). **p < 0.01, ***p < 0.001 by Student’s t test. N.S. means no significance
Discussion
The interaction between gut microbiota and the host plays a pivotal role in maintaining gut homeostasis. Gut microbiota is critically involved in the development of colitis and colorectal cancer (CRC). However, the molecular mechanisms by which gut microbiota mediates CRC progression remain largely unexplored. In this study, we demonstrate that GSDMD promotes intestinal tumor development by increasing IL-1β release and Kyn production, and GSDMD increases Kyn production might through suppressing Lactobacillus abundance in the gut.
GSDMD is a key executor of the inflammasome, responsible for the release of mature IL-1β and IL-18, as well as mediating pyroptotic cell death [6]. It has been reported that GSDMD sensitized colorectal cancer to chemotherapy in pyroptosis dependent and independent manner [18, 29]. A recent paper reported that GSDMD suppressed inflammation-induced colon cancer by promoting STAT1-dependent apoptosis in azomethanze plus dextran sulfate sodium-induced CRC mice model [19]. In contrast, another paper found that a point mutation in leucine-rich repeat kinase 2 (LRRK2 G2019S) promoted colon cancer progression via LRRK2-GSDMD mediated gut inflammation in the same model [20]. These findings suggest that the role of GSDMD in inflammation-associated colon cancer is complex and context-dependent. In our study, we investigated the role of GSDMD in spontaneous colon cancer development using Apcmin/+ mice, a widely used model for CRC. We found that GSDMD deficiency reduced spontaneous intestinal tumor development, likely through modulation of IL-1β release and Kyn production. While previous studies reported decreased GSDMD expression in human colon cancer tissues compared to peritumoral tissues [19, 29], we observed that GSDMD expression was significantly higher in colon cancer tissues than in adjacent non-tumor tissues. Additionally, GSDMD expression positively correlated with tumor grade and tumor size. Differences in patient specimens, antibodies, and experimental conditions may account for these seemingly contradictory results. Larger studies involving more patient samples are required to confirm GSDMD protein levels in CRC.
It is well-established that IL-1β is an effector cytokine produced by activated inflammasomes and promotes colon cancer development through various mechanisms, including enhancing tumor cell proliferation and modulating the tumor microenvironment [9–12]. Consistent with a previous report [30], we observed that IL-1β mRNA levels were comparable between Apcmin/+ mice and Apcmin/+Gsdmd−/− mice. However, the protein level of IL-1β was significantly elevated in Apcmin/+ mice and reduced in Apcmin/+Gsdmd−/− mice. These results suggest that GSDMD promotes IL-1β release in Apcmin/+ mice. Furthermore, we found exogenous recombinant IL-1β partially rescued intestinal tumor development in Apcmin/+Gsdmd−/− mice. However, IL-1β produced by which type of cell in Apcmin/+ mice still remains unclear, and we speculated that IL-1β may be produced by myeloid cells. To answer the question, future studies should utilize tissue- or cell-specific Gsdmd knockout mice, generated through the Cre/LoxP recombination system, to elucidate the cellular sources of IL-1β [31].
Previous work from our group showed that Gsdmd−/− mice exhibited reduced levels of intestinal Firmicutes and no change in Lactobacillus levels compared to controls [26]. However, in the present study, we observed an increase in Firmicutes and a decrease in Bacteroidetes in Apcmin/+Gsdmd−/− mice compared to Apcmin/+ mice. Consistent with the changes in phylum level, Bacilli (class level), Lactobacillales (order level), Lactobacillaceae (family level), and Lactobacillus (genus level) significantly increased in the gut of Apcmin/+Gsdmd−/− mice. The data suggest that GSDMD may increase Firmicutes levels under normal condition, while it may decrease Firmicutes levels during spontaneous intestinal cancer development. This indicates that GSDMD has varying effects on gut microbiota composition under different physiological and pathological conditions.
Trp metabolism consists of endogenous (host) and bacterial pathways [28]. Kyn is a key metabolite derived from endogenous Trp metabolism but not bacterial Trp metabolism. Indoleamine 2,3-dioxygenase (IDO) is a critical enzyme for digesting Trp to Kyn [25]. Kyn is considered to be an oncometabolite in CRC [21]. In our study, we observed a marked reduction in Kyn levels in the feces of Apcmin/+Gsdmd−/− mice. Moreover, exogenous Kyn promoted intestinal tumor development in these mice, demonstrating that Kyn is crucial for spontaneous intestinal tumor progression. Interestingly, we found a strong inverse correlation between Kyn levels and Lactobacillus abundance. A previous report found that colonization with Lactobacillus johnsonii in rats decreased serum Kyn levels [32]. L.johnsonii reduced IDO1 activity in HT-29 cells, a human colon cancer cell line [33]. Based on these observations, we propose that Lactobacillus may suppress endogenous Trp metabolism in Apcmin/+Gsdmd−/− mice. To test this hypothesis, future studies need to be investigated whether Lactobacillus transplantation into SPF wild-type C57/BL6 or germ-free mice can suppress endogenous Trp metabolism. Additionally, it would be of interest to explore whether Kyn itself influences gut microbiota composition.
In conclusion, our study identifies GSDMD as a key promoter of colon cancer development, likely through enhancing IL-1β release and Kyn production. We propose that GSDMD serves as a critical link between the host and gut microbiota, and GSDMD-mediated suppression of Lactobacillus abundance may enhance Kyn production from host cells. Therefore, targeting GSDMD represents a promising therapeutic strategy for CRC.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Author contributions
Hanchao Gao, Mengtao Cao, and Pengfei Chen designed the experiments. Hanchao Gao and Pengfei Chen wrote the manuscript. Hanchao Gao, Weilong Li, and Shi Xu conducted the experiments and analyzed the data. Zigan Xu, Wenjun Hu, Litao Pan, Ting Xie, Yeye Yu, Huimin Sun, Liwen Huang, Peishan Chen, Jinmei Wu, Dexing Yang and Lian Li helped with experiments. Litao Pan, and Kewang Luo provided reagents and technical support. Hanchao Gao and Shaodong Luan supervised the study. All authors reviewed and approved the submitted version.
Funding
This work was supported by grants from Guangdong Basic and Applied Basic Research Foundation (2024A1515030207), Open Project of Guangdong Provincial Key Laboratory of Tropical Disease Research (KLTDR202002), Shenzhen Foundation of Science and Technology (JCYJ20220531092614032, JCYJ20220531092617039), Shenzhen Longhua District Science and Technology Innovation Special Fund Project (11501A20220923BF59236, 11501A20220923BE5B6B3, 11501A20220923BD5F291).
Data availability
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information repository (no. PRJNA1109541; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1109541).
Declarations
Ethics approval and consent to participate
The animal study was reviewed and approved by Institutional Biomedical Research Ethics Committee of the Guangdong Medical University.
Consent for publication
All the authors give their consent for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Hanchao Gao, Weilong Li and Shi Xu contributed equally to this work.
Contributor Information
Hanchao Gao, Email: hcgao@foxmail.com.
Shaodong Luan, Email: szkidney3@163.com.
Mengtao Cao, Email: cmtbnu@163.com.
Pengfei Chen, Email: pfchen@sibs.ac.cn.
References
- 1.Schmitt M, Greten FR. The inflammatory pathogenesis of colorectal cancer. Nat Rev Immunol. 2021;21:653–67. [DOI] [PubMed] [Google Scholar]
- 2.Okayasu I, Ohkusa T, Kajiura K, Kanno J, Sakamoto S. Promotion of colorectal neoplasia in experimental murine ulcerative colitis. Gut. 1996;39:87–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rakoff-Nahoum S, Medzhitov R. Regulation of spontaneous intestinal tumorigenesis through the adaptor protein MyD88. Science. 2007;317:124–7. [DOI] [PubMed] [Google Scholar]
- 4.Penny HL, Prestwood TR, Bhattacharya N, Sun F, Kenkel JA, Davidson MG, Shen L, Zuniga LA, Seeley ES, Pai R, Choi O, Tolentino L, Wang J, Napoli JL. Engleman, restoring retinoic acid attenuates intestinal inflammation and Tumorigenesis in APCMin/+ mice. Cancer Immunol Res. 2016;4:917–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wong CC, Yu J. Gut microbiota in colorectal cancer development and therapy. Nat Rev Clin Oncol. 2023;20:429–52. [DOI] [PubMed] [Google Scholar]
- 6.Elias EE, Lyons B, Muruve DA. Gasdermins and pyroptosis in the kidney. Nat Rev Nephrol. 2023;19:337–50. [DOI] [PubMed] [Google Scholar]
- 7.Wei X, Xie F, Zhou X, Wu Y, Yan H, Liu T, Huang J, Wang F, Zhou F, Zhang L. Role of pyroptosis in inflammation and cancer. Cell Mol Immunol. 2022;19:971–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharma BR, Kanneganti TD. Inflammasome signaling in colorectal cancer. Transl Res. 2023;252:45–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Y, Wang L, Pappan L, Galliher-Beckley A, Shi J. IL-1beta promotes stemness and invasiveness of colon cancer cells through Zeb1 activation. Mol Cancer. 2012;11:87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rebe C, Ghiringhelli F. Interleukin-1beta and Cancer. Cancers (Basel). 2020;12(7). [DOI] [PMC free article] [PubMed]
- 11.Zhu Y, Zhu M, Lance P. IL1beta-mediated stromal COX-2 signaling mediates proliferation and invasiveness of colonic epithelial cancer cells. Exp Cell Res. 2012;318:2520–30. [DOI] [PubMed] [Google Scholar]
- 12.Voronov E, Apte RN. IL-1 in Colon inflammation, Colon carcinogenesis and invasiveness of Colon cancer. Cancer Microenviron. 2015;8:187–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shi J, Zhao Y, Wang K, Shi X, Wang Y, Huang H, Zhuang Y, Cai T, Wang F, Shao F. Cleavage of GSDMD by inflammatory caspases determines pyroptotic cell death. Nature. 2015;526:660–5. [DOI] [PubMed] [Google Scholar]
- 14.Liu X, Zhang Z, Ruan J, Pan Y, Magupalli VG, Wu H, Lieberman J. Inflammasome-activated gasdermin D causes pyroptosis by forming membrane pores. Nature. 2016;535:153–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kuang S, Zheng J, Yang H, Li S, Duan S, Shen Y, Ji C, Gan J, Xu XW, Li J. Structure insight of GSDMD reveals the basis of GSDMD autoinhibition in cell pyroptosis. Proc Natl Acad Sci U S A. 2017;114:10642–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vasudevan SO, Behl B, Rathinam VA. Pyroptosis-induced inflammation and tissue damage. Semin Immunol. 2023;69:101781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang S, Chang CW, Huang J, Zeng S, Zhang X, Hung MC, Hou J. Gasdermin C sensitizes tumor cells to PARP inhibitor therapy in cancer models. J Clin Invest. 2024;134(1). [DOI] [PMC free article] [PubMed]
- 18.Wu LS, Liu Y, Wang XW, Xu B, Lin YL, Song Y, Dong Y, Liu JL, Wang XJ, Liu S, Kong P, Han M, Li BH. LPS enhances the chemosensitivity of oxaliplatin in HT29 cells via GSDMD-Mediated pyroptosis. Cancer Manag Res. 2020;12:10397–409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tanaka S, Orita H, Kataoka T, Miyazaki M, Saeki H, Wada R, Brock MV, Fukunaga T, Amano T, Shiroishi T. Gasdermin D represses inflammation-induced colon cancer development by regulating apoptosis. Carcinogenesis. 2023;44:341–9. [DOI] [PubMed] [Google Scholar]
- 20.Wang Y, Gao JZ, Sakaguchi T, Maretzky T, Gurung P, Narayanan NS, Short S, Xiong Y, Kang Z. LRRK2 G2019S Promotes Colon Cancer Potentially via LRRK2-GSDMD Axis-Mediated Gut Inflammation. Cells. 2024;13(7). [DOI] [PMC free article] [PubMed]
- 21.Venkateswaran N, Conacci-Sorrell M. Kynurenine: an oncometabolite in colon cancer. Cell Stress. 2020;4:24–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang X, Liu X, Zhou W, Du Q, Yang M, Ding Y, Hu R. Blockade of IDO-Kynurenine-AhR Axis ameliorated Colitis-Associated Colon Cancer via Inhibiting Immune Tolerance. Cell Mol Gastroenterol Hepatol. 2021;12:1179–99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venkateswaran N, Lafita-Navarro MC, Hao YH, Kilgore JA, Perez-Castro L, Braverman J, Borenstein-Auerbach N, Kim M, Lesner NP, Mishra P, Brabletz T, Shay JW, DeBerardinis RJ, Williams NS, Yilmaz OH, Conacci-Sorrell M. MYC promotes tryptophan uptake and metabolism by the kynurenine pathway in colon cancer. Genes Dev. 2019;33:1236–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wu D, Zhu Y. Role of kynurenine in promoting the generation of exhausted CD8(+) T cells in colorectal cancer. Am J Transl Res. 2021;13:1535–47. [PMC free article] [PubMed] [Google Scholar]
- 25.Bishnupuri KS, Alvarado DM, Khouri AN, Shabsovich M, Chen B, Dieckgraefe BK, Ciorba MA. IDO1 and kynurenine pathway metabolites activate PI3K-Akt signaling in the neoplastic Colon epithelium to Promote Cancer Cell Proliferation and inhibit apoptosis. Cancer Res. 2019;79:1138–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gao H, Cao M, Yao Y, Hu W, Sun H, Zhang Y, Zeng C, Tang J, Luan S, Chen P. Dysregulated microbiota-driven gasdermin D activation promotes Colitis Development by mediating IL-18 release. Front Immunol. 2021;12:750841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Song X, Gao H, Lin Y, Yao Y, Zhu S, Wang J, Liu Y, Yao X, Meng G, Shen N, Shi Y, Iwakura Y, Qian Y. Alterations in the microbiota drive interleukin-17 C production from intestinal epithelial cells to promote tumorigenesis. Immunity. 2014;40:140–52. [DOI] [PubMed] [Google Scholar]
- 28.Gao J, Xu K, Liu H, Liu G, Bai M, Peng C, Li T, Yin Y. Impact of the gut microbiota on intestinal immunity mediated by Tryptophan Metabolism. Front Cell Infect Microbiol. 2018;8:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng X, Na R, Zhou W, Meng X, Yang Y, Amini S, Song L. Nuclear translocation of gasdermin D sensitizes colorectal cancer to chemotherapy in a pyroptosis-independent manner. Oncogene. 2022;41:5092–106. [DOI] [PubMed] [Google Scholar]
- 30.Dmitrieva-Posocco O, Dzutsev A, Posocco DF, Hou V, Yuan W, Thovarai V, Mufazalov IA, Gunzer M, Shilovskiy IP, Khaitov MR, Trinchieri G, Waisman A, Grivennikov SI. Cell-type-specific responses to Interleukin-1 Control Microbial Invasion and Tumor-elicited inflammation in Colorectal Cancer. Immunity. 2019;50:166–80. e167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lu ZY, Chen PB, Xu QY, Li B, Jiang SD, Jiang LS, Zheng XF. Constitutive and conditional gene knockout mice for the study of intervertebral disc degeneration: current status, decision considerations, and future possibilities. JOR Spine. 2023;6:e1242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Valladares R, Bojilova L, Potts AH, Cameron E, Gardner C, Lorca G, Gonzalez CF. Lactobacillus johnsonii inhibits indoleamine 2,3-dioxygenase and alters tryptophan metabolite levels in BioBreeding rats. FASEB J. 2013;27:1711–20. [DOI] [PubMed] [Google Scholar]
- 33.Freewan M, Rees MD, Plaza TS, Glaros E, Lim YJ, Wang XS, Yeung AW, Witting PK, Terentis AC, Thomas SR. Human indoleamine 2,3-dioxygenase is a catalyst of physiological heme peroxidase reactions: implications for the inhibition of dioxygenase activity by hydrogen peroxide. J Biol Chem. 2013;288:1548–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets generated and/or analyzed during the current study are available in the National Center for Biotechnology Information repository (no. PRJNA1109541; https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1109541).