Abstract
Viral protein 40 (VP40) of Ebola virus appears equivalent to matrix proteins of other viruses, yet little is known about its role in the viral life cycle. To elucidate the functions of VP40, we investigated its ability to induce the formation of membrane-bound particles when it was expressed apart from other viral proteins. We found that VP40 is indeed able to induce particle formation when it is expressed in mammalian cells, and this process appeared to rely on a conserved N-terminal PPXY motif, as mutation or loss of this motif resulted in markedly reduced particle formation. These findings demonstrate that VP40 alone possesses the information necessary to induce particle formation, and this process most likely requires cellular WW domain-containing proteins that interact with the PPXY motif of VP40. The ability of VP40 to bind cellular membranes was also studied. Flotation gradient analysis indicated that VP40 binds to membranes in a hydrophobic manner, as NaCl at 1 M did not release the protein from the lipid bilayer. Triton X-114 phase-partitioning analysis suggested that VP40 possesses only minor features of an integral membrane protein. We confirmed previous findings that truncation of the 50 C-terminal amino acids of VP40 results in decreased association with cellular membranes and demonstrated that this deletion disrupts hydrophobic interactions of VP40 with the lipid bilayer, as well as abolishing particle formation. Truncation of the 150 C-terminal amino acids or 100 N-terminal amino acids of VP40 enhanced the protein's hydrophobic association with cellular membranes. These data suggest that VP40 binds the lipid bilayer in an efficient yet structurally complex fashion.
Ebola virus is an enveloped, nonsegmented, negative-sense RNA virus of the family Filoviridae in the order Mononegavirales (10). Four subtypes of Ebola virus have been identified to date, namely, Zaire, Sudan, Ivory Coast, and Reston (24). Human infection with subtype Zaire causes a fulminating, febrile, hemorrhagic disease that results in extensive mortality (10). Although recent studies have begun to address the immune response to viral infection (1, 2, 28, 32), as well as the functions of the viral proteins involved in the replicative process (VP30, VP35, NP, L) (2, 20) and the transmembrane glycoprotein (GP) (24, 27, 29, 30, 33–36), little is known about the functions of the viral proteins associated with the membrane, including viral protein 40 (VP40).
The matrix proteins of many nonsegmented, negative-sense RNA viruses play a critical role in viral particle formation and budding (11). Expression of the matrix protein of vesicular stomatitis virus (VSV) in insect and mammalian cells results in evagination of matrix protein-containing vesicles from the plasma membrane surface (16, 19). Matrix proteins interact with membranes in a hydrophobic and/or electrostatic manner, and electron micrographs of nonsegmented, negative-sense RNA viruses have demonstrated that the matrix protein forms a layer associated with the inner leaflet of the lipid bilayer (11). This interaction is thought to be essential for virus assembly and release from infected cells.
VP40, encoded by the third gene in the linear 3′-5′ RNA genome of Ebola virus and 326 amino acids in length, contains a number of hydrophobic regions, represents approximately 38% of the protein in the viral particle, and lines the interior surface of the viral envelope (9, 23). It contains a PPXY motif (X denotes any amino acid) at amino acids 10 to 13 (14) that is also present at amino acids 16 to 19 in Marburg virus, strain Popp (23). This motif has been shown to play an important role in the budding of rabies virus and VSV: when either of the prolines or the tyrosine of this motif is altered in the matrix proteins of these viruses, viral budding is markedly reduced by comparison to findings with wild-type virus (14). Mutation of the PPXY motif in the matrix protein of VSV appears to reduce virus yield by preempting budding of assembled virions at the plasma membrane (15). This motif interacts with the WW domains found in many cellular regulatory and signal transduction proteins (5, 6, 26), and interactions between one or more cellular proteins and the matrix proteins of these viruses are thought to be crucial for efficient virus release from cells (14). Thus, VP40 appears equivalent to the matrix proteins of other nonsegmented, negative-sense RNA viruses.
In the study reported here, we tested the ability of VP40 expressed in mammalian cells to induce the budding of protein-associated vesicles and determined amino acids that make specific and important contributions to this process. The plasma membrane affinity of VP40 expressed in mammalian cells was also studied, with particular attention being paid to the domains involved.
MATERIALS AND METHODS
Cells.
293 and 293T human embryonic kidney cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal calf serum, 2% l-glutamine, and a penicillin-streptomycin solution (Sigma). The cells were grown at 37°C in 5% CO2.
Construction of plasmids.
To generate cDNA constructs encoding the VP40 protein, we used primers binding to the start and stop codons (positions 4479 and 5459 of the positive-sense antigenomic RNA, respectively) to reverse transcribe and PCR amplify purified viral RNA (Titan, reverse transcription-PCR kit; Roche). The PCR product was cloned in the pT7Blue vector (Novagen), resulting in pT7EboZVP40. The cloned Ebola virus VP40 gene was sequenced to ensure that unwanted nucleotide replacements were not present.
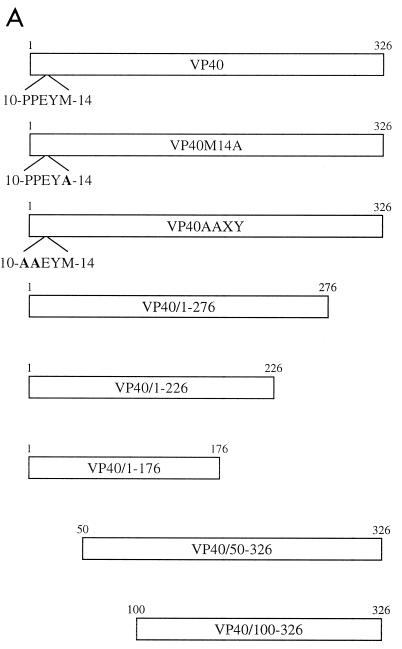
To generate plasmid pETEBoZVP40His for the expression of six-histidine-tagged VP40 in Escherichia coli, we used pT7EboZVP40 as a template for PCR amplification with the appropriate primers. The PCR product was blunt-end ligated into the SmaI-digested site of the vector pM (Clonetech). This construct was digested with NdeI and EcoRI, and the fragment containing VP40 was ligated into the expression vector pET-5a (Promega). To generate plasmids pCEboZVP40, pCEboZVP40AAXY, pCEboZVP40M14A, pCEboZVP40/1–276, pCEboZVP40/1–226, pCEboZVP40/1–176, pCEboZVP40/50–326, and pCEboZVP40/100–326 (proteins expressed from these plasmids are designated VP40, VP40AAXY, etc.) for expression of VP40 and its mutant forms in eukaryotic cells, we amplified the Ebola virus Zaire VP40 gene from pT7EboZVP40 using specific forward primers, each containing an EcoRI site 5′ to the start of the coding region, and specific reverse primers, each containing a BglII site 3′ to the stop codon for each construct, and blunt-end ligated the fragment into the EcoRV-digested site of the vector pT7Blue. Each construct was digested with EcoRI and BglII, and the fragment containing the VP40 gene or modified VP40 gene was cloned into the EcoRI- and BglII-digested eukaryotic expression vector pCAGGS/MCS (expression controlled by the chicken β-actin promoter) (17, 21). Eukaryotic expression constructs employed in this study are schematically presented in Fig. 1A.
FIG. 1.
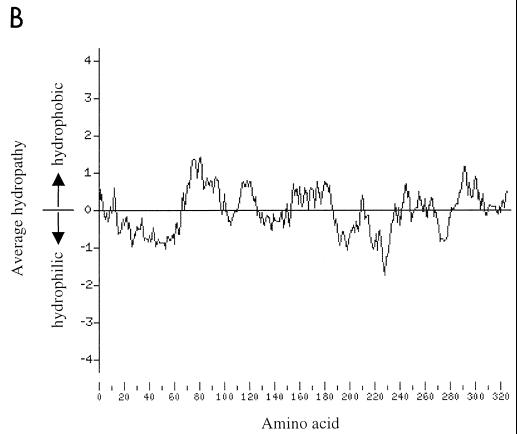
(A) Schematic representation of wild-type VP40 and VP40 mutant proteins reported in this paper. Amino acid residues are represented with the single-letter code. Substituted residues are indicated in boldface type. (B) Kyte-Doolittle hydrophobicity plot of Ebola virus VP40 over a window of 17 amino acids (16).
Antibody.
A polyclonal antibody against Ebola virus Zaire VP40 was produced as follows: BL21 E. coli cells were transformed with plasmid pETEboZVP40His. Expression of the 6-his-tagged VP40 protein was induced with 1 mM IPTG (isopropyl-β-d-thiogalactopyraxoside) for 3 h. The E. coli cells were lysed, and cellular debris was removed by centrifugation. The supernatant was purified over an Ni-nitrilotriacetic acid agarose column (Qiagen). Expression of VP40 was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) followed by Western blotting using a monoclonal antibody against the histidine tag (Kodak). Rabbits were immunized with approximately 0.5 mg of VP40, and antibody against keratin present in the antiserum was removed with a keratin column (13).
Cell transfection for expression of VP40 and its mutants.
293 or 293T cells (60-mm-diameter plates) were transfected with expression vectors with the use of the Trans IT LT-1 liposomal reagent (Panvera) according to the manufacturer's instructions. Briefly, DNA and transfection reagent were mixed (6 μl of Trans IT LT-1 with 3 μg of DNA) in 0.2 ml of OPTI-MEM (Gibco-BRL), incubated for 30 min at room temperature, and added to the cells. Transfected cells were incubated at 37°C until harvest of the supernatant and/or cell monolayer.
Particle formation assay.
Particles were assayed by the method of Li et al. (19) with some modifications. Forty-eight hours after transfection of 293T cells with pCEboZVP40, pCEboZVP40AAXY, pCEboZVP40M14A, or pCEboZVP40/1–276, the culture medium was removed and placed on ice. The cell monolayer was washed with phosphate-buffered saline (PBS), scraped into lysis buffer (0.25 M Tris-HCl [pH 8.0], 0.5% Triton X-100) and kept at 4°C. The culture medium (2 ml) was centrifuged at 2,000 rpm in a microcentrifuge (Sorvall MC 12V; Dupont) for 5 min to remove cellular debris, layered over 20% sucrose in STE buffer (0.01 M Tris-Cl [pH 7.5], 0.01 M NaCl, 0.001 M EDTA [pH 8.0]) (2 ml), and centrifuged at 150,000 × g for 2 h at 4°C. After centrifugation, the supernatant was removed and added to the cell lysate. This mixture was saved for analysis of total protein expression. The pellet was resuspended in 1 ml of STE buffer overnight at 4°C. The resuspended pellet was layered over a 10 to 50% discontinuous sucrose gradient in STE buffer and centrifuged at 150,000 × g for 4 h at 4°C, and fractions (1 ml) were collected from the top of the gradient. Each fraction was mixed with 0.25 ml of 50% trichloroacetic acid (TCA), the fractions were incubated for 30 min on ice, and the precipitated proteins were pelleted by microcentrifugation for 15 min. The pellets were washed once with cold acetone, air dried, and resuspended in 0.05 ml of SDS-PAGE sample buffer. Proteins in the mixture of cell lysate and supernatant from centrifugation through 20% sucrose were precipitated with 10% TCA, washed with acetone, and resuspended in 0.5 ml of SDS-PAGE sample buffer.
Protease protection assay.
293T cells were transfected with pCEboZVP40, and at 48 h posttransfection, the culture medium was removed. The medium was microcentrifuged at 2,000 rpm (Sorvall MC 12V; Dupont) for 5 min to remove cellular debris, layered over a 20% sucrose cushion, and centrifuged at 165,000 × g for 1 h at 4°C. The supernatant was removed, and the pellet was resuspended overnight at 4°C in 0.4 ml of STE buffer. This resuspension was divided into six aliquots and treated according to a protocol previously described (31). Aliquot 1 received no further treatment, aliquot 2 was treated with soybean trypsin inhibitor (Biofluids) to a final concentration of 3 mg/ml, aliquot 3 was treated with Triton X-100 to a final concentration of 1%, aliquot 4 was treated with trypsin (Worthington) to a final concentration of 0.1 mg/ml, aliquot 5 was treated with both Triton X-100 and trypsin, and aliquot 6 was treated with both trypsin inhibitor and trypsin. The samples were incubated at room temperature for 30 min, after which an excess of trypsin inhibitor (5 mg/ml) was added to each aliquot. SDS-PAGE sample buffer (6×) was added to each aliquot.
Membrane association assay.
The method of Bergmann and Fusco (3) was used, with some modifications, to determine the membrane associations of VP40 and its mutant forms. Briefly, 48 h after transfection of 293 cells with pCEboZVP40 or a mutant-VP40 expression plasmid, the culture medium was removed, and the cell monolayer, after a wash with PBS, was scraped into ice-cold sucrose homogenization buffer (10% [wt/wt] sucrose, 10 mM Tris-HCl [pH 7.4], 1 mM EDTA, and 10 mM iodoacetamide). Cells were disrupted with 30 strokes of a Dounce homogenizer on ice and microcentrifuged for 3 min at 2,000 rpm (Sorvall MC 12V; Dupont) to remove nuclei. The resulting supernatant was made to 1 M NaCl or left untreated, incubated at room temperature for 20 min, made to 80% sucrose (wt/vol), placed at the bottom of a Beckman SW41 centrifuge tube, and overlaid with 5 ml of 65% (wt/vol) sucrose and 2.5 ml of 10% sucrose. The gradient was centrifuged to equilibrium at 150,000 × g for 18 h at 4°C. Fractions (1 ml) were collected from the top of the gradient, diluted 1:1 with TBS–Triton X-100 buffer (0.025 M Tris-HCl [pH 7.5], 0.15 M NaCl, 0.5% Triton X-100) or, for experiments involving expression of VP40/100–326, precipitated with TCA (as described for the particle formation assay) owing to the weak signal of this deletion construct in Western analysis, and mixed with SDS-PAGE sample buffer.
Triton X-114 phase-partitioning analysis.
The method used for phase-partitioning analysis was essentially that of Bordier (4). Forty-eight hours posttransfection of 293 cells with pCEboZVP40, pCEboZVP40/1–276, pCEboZVP40/1–226, pCEboZVP40/1–176, pCEboZVP40/50–326, pCEboZVP40/100–326, or, as a control, a vector expressing A/WSN/33 (H1N1) influenza virus hemagglutinin (HA), we scraped cells into cold TN buffer (10 mM Tris-HCl [pH 7.4], 150 mM NaCl), disrupted them with 30 strokes in a Dounce homogenizer, and subjected them to centrifugation at 2,000 rpm for 3 min to remove nuclei. Triton X-114 (Sigma) was added to each supernatant to 1%, and the resulting solution was incubated for 15 min at 4°C with agitation. Unsolubilized material was pelleted by centrifugation in a picofuge for 5 min at 4°C, and the supernatant was heated to 37°C for 5 min. The supernatant was layered onto a 37°C sucrose (6%) cushion in TN buffer containing 0.06% Triton X-114 and centrifuged at 2,000 rpm (Sorvall MC 12V; Dupont) for 3 min at room temperature. The detergent (lower) and aqueous (upper) phases were recovered separately, the aqueous phase was extracted a second time, like phases were pooled, and the detergent phase was diluted in TN buffer. Proteins in each phase were precipitated with 50% acetone and resuspended in SDS-PAGE sample buffer.
Western blotting.
Samples in sample buffer (10 μl) were incubated at 100°C for 5 min and separated on 12% polyacrylamide gels. Resolved proteins were transferred to Western polyvinylidine difluoride membranes (Schleicher & Schuell) and blocked overnight at 4°C with 5% skim milk in PBST (0.05% Tween 20 [Sigma] in PBS). Blots were incubated in primary antibody for 1 h at room temperature, washed three times with PBST, incubated in biotinylated anti-rabbit secondary antibody (Vector Laboratories) for 30 min, washed three times with PBST, incubated in streptavidin-horseradish peroxidase reagent (Vector Laboratories) for 30 min, and washed three times with PBST. Blots were then incubated in Lumi-Light Western blotting substrate (Boehringer Mannheim) for 5 min and exposed to X-ray film (Kodak).
RESULTS
Expression of VP40 in mammalian cells.
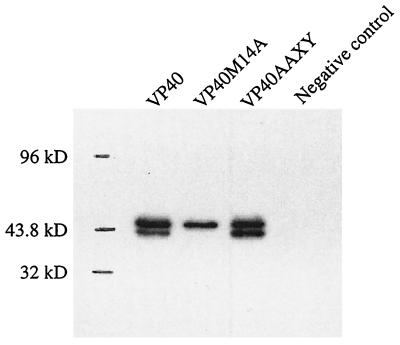
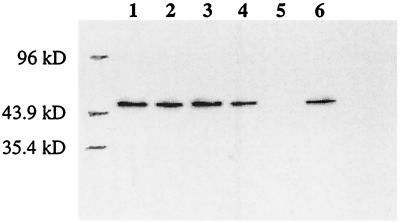
To ensure that VP40 is expressed at efficient levels in human embryonic kidney 293T cells, we analyzed the cell lysate 24 h after transfection with pCEboZVP40 by Western blotting. Two bands reacting with anti-VP40 polyclonal antibody were found, a small distance apart, near 40 kDa (Fig. 2). The lysate from cells transfected with the expression vector alone did not react with the antibody.
FIG. 2.
Expression of VP40, VP40/M14A, and VP40AAXY in 293T cells. Cells were transfected with plasmids expressing VP40 or its mutants. The sample for the negative control was prepared from cells transfected with the empty vector (pCAGGS/MCS). Lysates were harvested 24 h posttransfection, and proteins were separated by SDS-PAGE (12% polyacrylamid gel) and detected by Western blotting.
VP40 contains an internal start codon at nucleotides 40 to 42 (codon 14) that is in frame with the first AUG. To determine whether protein synthesis from this internal start codon was responsible for the faster-migrating band on the gel, we generated a construct, pCEboZVP40M14A, which expresses a mutant VP40 with this second AUG changed to GCG, which encodes alanine, and expressed it as described above. Analysis of the cell lysate revealed a single, larger-sized band (Fig. 2), suggesting that the second AUG is used as a start codon to an appreciable extent in this system.
We then asked whether loss of the PPXY motif at amino acids 10 to 13 of VP40 affects expression of the protein. 293T cells were transfected with pCEboZVP40AAXY, which expresses a mutant VP40 in which the PPEY sequence at amino acids 10 to 13 was changed to AAEY. Two bands corresponding to those seen with the expression of wild-type VP40 were detected (Fig. 2). However, in contrast to the results obtained with wild-type VP40 expression, where the slower-migrating band was the predominate product, pCEboZVP40AAXY expressed the two products at similar levels, indicating that loss of the PPXY motif affects either the translation of VP40 or its stability.
Production of membrane-bound particles.
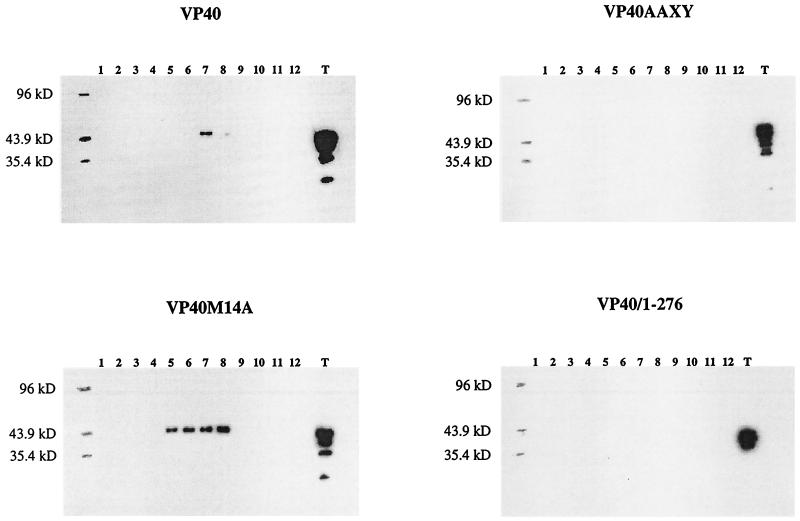
To determine whether VP40-associated vesicles are produced when the protein is expressed in the absence of other viral proteins, we transfected 293T cells with pCEboZVP40 and, after 48 h, collected the supernatant. After removal of cellular debris, the supernatant was subjected to ultracentrifugation over a 20% sucrose cushion. The pellet was resuspended and centrifuged through a 10 to 50% discontinuous sucrose gradient, and fractions were analyzed by Western blotting (Fig. 3). Fractions 6 to 8 contained VP40, with the majority of the protein being found in fraction 7. Only the full-length species of VP40 was detected. The VP40 in fractions 6 to 8 was most likely associated with membrane lipids in a particle-like structure, as the sucrose densities in these fractions ranged from 1.11 to 1.13 g/ml, which corresponds to findings for matrix protein-generated particles of other viruses (12, 25). Bands detected below that of the full-length protein in the total protein fraction were likely degradation products. These data indicate that VP40 expressed in the absence of other viral proteins can produce membrane-bound particles.
FIG. 3.
Particle formation by VP40 and its mutant proteins. The culture medium was harvested 48 h posttransfection with pCEboZVP40, pCEboZVP40AAXY, pCEboZVP40M14A, or pCEboZVP40/1–276, layered over a 20% sucrose cushion, and ultracentrifuged. The pellet resuspended in STE buffer was fractionated through a 10 to 50% discontinuous sucrose gradient. Fractions were collected from the top of the gradient. Fraction proteins were precipitated with 10% TCA and resuspended in SDS-PAGE sample buffer. Total protein from the cell lysate and cleared supernatant (T) was precipitated with 10% TCA and resuspended in SDS-PAGE sample buffer. Proteins were separated by SDS–12% PAGE and detected by Western blotting. Fractions are numbered from the top to the bottom of the gradient.
Protease protection assay.
To confirm the ability of VP40 to produce membrane-bound particles when expressed alone, we employed a trypsin protection assay. Culture supernatant from cells transfected with pCEboZVP40 was centrifuged at 165,000 × g through 20% sucrose, and the pellet was resuspended in STE buffer and divided into six equal aliquots. Aliquots 1 to 3 served as controls (untreated, trypsin inhibitor treated, and Triton X-100 treated), aliquot 4 was treated with trypsin, aliquot 5 was treated with trypsin and Triton X-100, and aliquot 6 was treated with trypsin inhibitor and trypsin. Trypsin degraded VP40 only in the presence of Triton X-100 (Fig. 4), indicating that the viral protein does induce the production of fully membrane-bound particles; that is, trypsin digestion of VP40 required disruption of the lipid bilayer surrounding the protein.
FIG. 4.
Protease protection analysis of VP40-induced particles. 293T cells were transfected with pCEboZVP40, and 48 h posttransfection, the culture medium was collected. VP40-induced particles in the culture supernatant were pelleted through 20% sucrose. The pellet was resuspended in STE buffer and divided into six equal aliquots. Aliquot 1 (lane 1) received no further treatment. Aliquot 2 (lane 2) received soybean trypsin inhibitor to a final concentration of 3 mg/ml. Aliquot 3 (lane 3) received Triton X-100 to a final concentration of 1%. Aliquot 4 (lane 4) received trypsin to a final concentration of 0.1 mg/ml. Aliquot 5 (lane 5) received Triton X-100 to 1% and trypsin to a 0.1-mg/ml final concentration. Aliquot 6 (lane 6) received trypsin inhibitor to 3 mg/ml and trypsin to a 0.1-mg/ml final concentrations. Proteins from each aliquot were separated by SDS–12% PAGE and detected by Western blotting.
VP40 mutants and membrane-bound particle formation.
Does the PPXY motif at amino acids 10 to 13 of VP40 contribute to particle production? To answer this question, we expressed VP40AAXY in 293T cells and assayed for particles as described above for wild-type VP40. VP40AAXY was not detected in fractions corresponding to the sucrose densities to which wild-type VP40 particles migrated (Fig. 3). Since VP40AAXY was synthesized at levels similar to that of wild-type VP40 (Fig. 3), this finding indicates that mutation of the PPXY motif markedly disrupts VP40-generated vesicle formation.
Figure 3 also shows the effect of loss of the second AUG codon on particle formation. A substantial amount of VP40M14A was present in fractions 5 to 8 in the gradient, and the percentage of total VP40M14A expressed in 293T cells that contributed to membrane-bound particle formation was much greater than the percentage of total wild-type VP40 involved in particle formation. This result is consistent with the finding that the PPXY motif present immediately upstream of the second AUG is critical for VP40-associated particle formation (Fig. 3).
To determine whether the C terminus of VP40 is essential for particle formation, we assayed a deletion mutant protein, VP40/1–276, which lacks the final 50 amino acids of VP40, for particle generation. Since this deletion mutant protein was not present at the same sucrose densities that characterized the migration of wild-type VP40, we concluded that the first 276 amino acids of VP40 are not sufficient for particle formation (Fig. 3).
VP40 association with cell membranes and structural requirements for this activity.
Flotation analysis was used to determine if VP40 binds cellular membranes efficiently in mammalian cells. In this method, postnuclear membrane fractions in 80% sucrose are loaded at the bottom of a centrifuge tube and overlaid with 65 and 10% sucrose. During centrifugation, cellular membranes and their associated proteins float to the 10% sucrose-65% sucrose interface while soluble proteins remain in the dense sucrose fractions at the bottom of the tube.
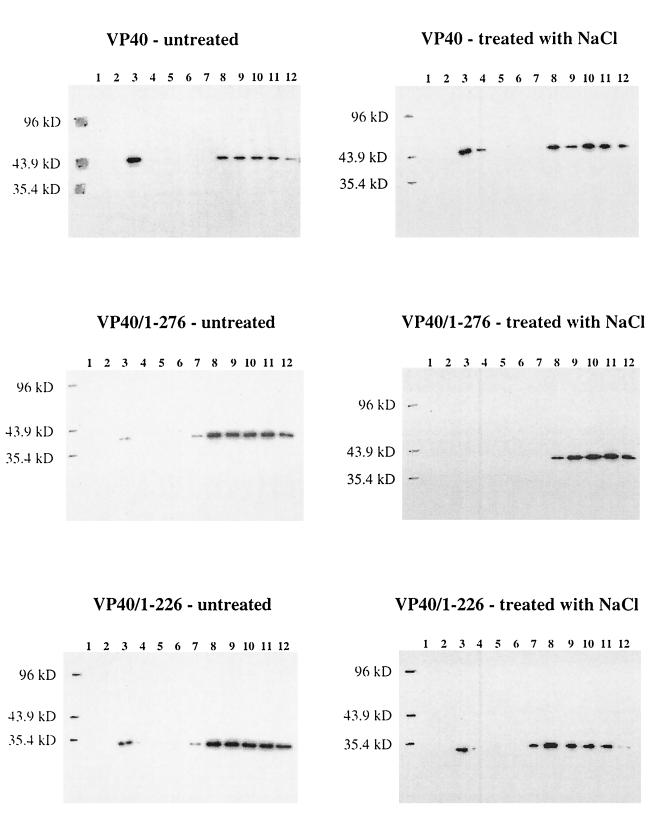
A large percentage of wild-type VP40 was found at the 10% sucrose-65% sucrose interface (fraction 3), while the remaining protein was found in the loading zone (fractions 8 to 12) (Fig. 5), indicating that VP40 does indeed bind cellular membranes. To clarify the interactions involved in this association, we treated VP40-associated membranes with 1 M NaCl to determine whether electrostatic interactions were required for this association and subjected them to flotation analysis. Salt treatment had a negligible affect on the ability of VP40 to associate with membranes (Fig. 5), suggesting that the protein contains at least one hydrophobic domain able to associate with membranes.
FIG. 5.
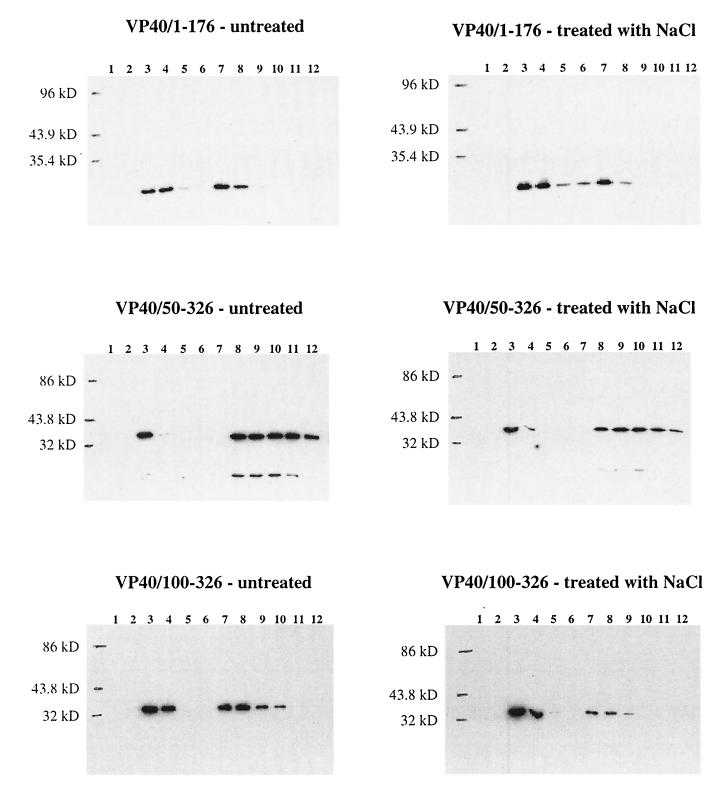
Membrane association analysis of VP40 and its deletion mutant proteins. 293 cells were transfected with the appropriate plasmid DNA, and 48 h posttransfection, the cells were harvested and broken with a Dounce homogenizer in sucrose homogenization buffer. The homogenate was made to 80% sucrose or treated with 1 M NaCl for 20 min and then made to 80% sucrose and overlaid with 65 and 10% sucrose layers. The gradient was centrifuged at 150,000 × g for 18 h, and fractions were collected from the top. Fractions were diluted with an equal volume of TBS–Triton X-100 buffer or, for fractions containing VP40/100–326, precipitated with 10% TCA (see Materials and Methods), and proteins were separated by SDS–12% PAGE and analyzed by Western blotting. Shown are gradients from cells expressing VP40, VP40/1–276, VP40/1–226, VP40/50–176, VP40/50–326, and VP40/100–326. Fractions are numbered from the top to the bottom of the gradient.
To elucidate the domain(s) of VP40 important for membrane association, deletion mutant proteins were generated. Constructs expressing amino acids 50 to 326 (pCEboZVP40/50–326), amino acids 100 to 326 (pCEboZVP40/100–326), amino acids 1 to 176 (pCEboZVP40/1–176), amino acids 1 to 226 (pCEboZVP40/1–226), and amino acids 1 to 276 (pCEboZVP40/1–276) of VP40 were expressed in 293 cells, and their membrane association in the presence or absence of 1 M NaCl was examined (Fig. 5). The mutant proteins with the largest truncations. VP40/1–176 and VP40/100–326, showed the highest level of association with the lipid bilayer. Salt treatment did not affect these interactions. Mutant VP40/1–226 and VP40/50–326 associated with membranes to the extent found with wild-type VP40, and these interactions were also relatively unperturbed by treatment with salt. By contrast, only a small portion of VP40/1–276 associated with the lipid bilayer, and this interaction was eliminated upon treatment with salt. These results indicate that loss of the 50 C-terminal amino acids of VP40 markedly alters the membrane-binding capabilities of VP40, primarily by disrupting hydrophobic interactions. This effect was ameliorated when 50 additional C-terminal amino acids were deleted, and membrane association was promoted when the protein was further truncated to 176 amino acids. Deletion of the 49 N-terminal amino acids of VP40 did not alter the membrane-binding characteristics of the protein, although truncation of 50 additional N-terminal amino acids did enhance protein-membrane association, as seen with VP40/1–176 (Fig. 5).
Since particle formation was markedly reduced with VP40AAXY, we subjected cells expressing this mutant protein to flotation analysis in order to determine whether a decreased ability to bind membranes was involved in this deficiency. As shown in Fig. 6, the loss of the PPXY motif in VP40 did not affect the ability of the protein to bind membranes, indicating that lack of particle production with this mutant protein was not due to the loss of membrane association.
FIG. 6.
Membrane association analysis of the VP40 mutant proteins VP40AAXY and VP40M14A. Forty-eight hours posttransfection of 293 cells with the appropriate plasmid DNA, the cells were harvested and broken with a Dounce homogenizer in sucrose homogenization buffer. The homogenate was made to 80% sucrose and overlaid with 65 and 10% sucrose layers. The gradient was centrifuged at 150,000 × g for 18 h, and fractions were collected from the top. Fractions were diluted with an equal volume of TBS–Triton X-100 buffer, and proteins were separated by SDS–12% PAGE and analyzed by Western blotting. Fractions are numbered from the top to the bottom of the gradient.
Flotation analysis was also used to determine whether the more efficient particle formation induced by VP40M14A, by comparison to that of wild-type VP40, could be attributed, at least in part, to increased membrane binding by this mutant. The percentage of VP40M14A associated with membranes was only slightly greater than that determined for wild-type VP40 (Fig. 6), indicating that this mutant protein relies on another mechanism to increase particle formation.
Triton X-114 phase-partitioning analysis.
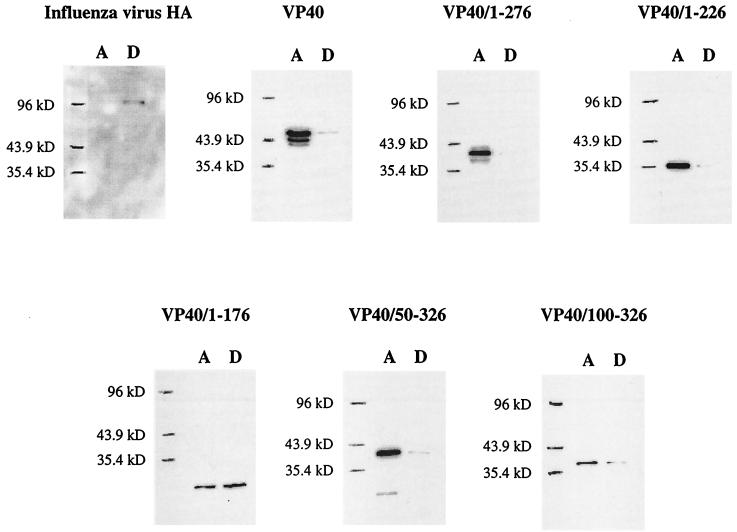
To probe the nature of the VP40-membrane interaction further, we relied on Triton X-114 phase-partitioning analysis, an assay in which integral membrane proteins and lipid-anchored proteins partition in the detergent phase of a protein extraction while peripheral membrane proteins partition in the aqueous phase. Figure 7 shows the results of this analysis for wild-type VP40, the five deletion mutant proteins of VP40, and influenza virus HA. HA, an integral membrane protein, was found entirely in the detergent phase of the extraction, as expected. Only a small portion of total VP40 was found in the detergent phase, while VP40/1–276 was found almost entirely in the aqueous phase. VP40/1–226 and VP40/50–326 partitioned in the detergent phase in proportions similar to that found for wild-type VP40. By contrast, when VP40/1–176 and VP40/100–326 were expressed, large proportions of each partitioned in the detergent phase. These results indicate that wild-type VP40 possesses only minor traits of an integral membrane protein and that deletion of its 50 C-terminal amino acids (VP40/1–276) abrogates these features. Further truncation of the C terminus (VP40/1–226 and VP40/1–176) enhanced the integral membrane character of protein. Deletion of the 49 N-terminal amino acids of VP40 (VP40/50–326) did not alter the general structural features of the protein, while deletion of amino acids 1 to 99 (VP40/100–326) appeared to increase the extent of anchoring to lipids.
FIG. 7.
Triton X-114 phase-partitioning analysis of VP40 and its deletion mutant proteins. 293 cells were transfected with the appropriate plasmid DNA, and at 48 h posttransfection, the cells were collected in TN buffer and disrupted by Dounce homogenization. The homogenate was partitioned into aqueous (A) and detergent (D) phases as described in Materials and Methods. Proteins were separated by SDS–12% PAGE and analyzed by Western blotting.
DISCUSSION
Here we have demonstrated that VP40 of Ebola virus, when it is expressed in the absence of other viral proteins, can induce the formation of membrane-encompassed particles, much in the manner of the matrix proteins of VSV, rabies virus, and simian immunodeficiency virus (12, 14, 15, 16, 19). Cellular proteins containing the WW domain are, in all likelihood, crucial for this process, as VP40 containing an altered version of a PPXY motif at amino acids 10 to 13 induces little or no particle formation. Harty et al. (14) demonstrated that the matrix proteins of VSV and rabies virus, which possess this motif at their N termini, bind the cellular Yes kinase-associated and Nedd4 proteins via a PPXY motif-WW domain interaction and that the loss of this motif results in impaired VSV matrix protein-associated vesicle release from infected cells. Jayakar et al. (15) recently demonstrated that mutation of the PPXY motif in the matrix protein of VSV impedes budding of fully assembled virions at the plasma membrane. Our conclusion is further supported by the facts that only the full-length species of VP40 was detected in the membrane-bound vesicles and that the smaller species of VP40, which is translated from the start codon immediately following the PPXY motif, appeared to be largely excluded (Fig. 3 and 4). This result indicates that VP40 lacking the PPXY motif fails to be efficiently incorporated into membrane-bound particles when it is expressed in the presence of wild-type VP40 as well as when it is expressed in its absence. Because the N terminus of VP40 is not required for efficient membrane association (Fig. 5), this inhibition is most likely the result of disrupted protein-protein interactions.
The efficiency of particle production markedly increased when the second ATG codon of VP40 (codon 14) was changed to GCG (alanine), but the reason for this enhancement remains unclear. Perhaps the faster-migrating version of VP40, which lacks the PPXY motif, interferes with the assembly or budding of full-length VP40 molecules at the cell surface or with the interaction between VP40 and a cellular protein. Whether translation from this second ATG occurs in actual viral infection or is an artifact of the system employed in this study is unknown.
While this paper was being prepared, Ruigrok et al. (22) reported that VP40 expressed in E. coli can bind liposomes in vitro and that this interaction is largely electrostatic. Here we determined the ability of VP40 expressed in mammalian cells to bind the cellular membrane and attempted to elucidate the chemical nature of this interaction and the domains involved. We found that a substantial amount of VP40 bound to the cellular membrane and that this interaction was disrupted negligibly by the presence of 1 M NaCl, indicating that at least one hydrophobic domain is involved in this interaction. A small but appreciable portion of VP40 partitioned with detergent in the manner of an integral membrane or lipid-anchored protein in Triton X-114 phase-partitioning analysis. This result, together with the inability of 1 M NaCl to dissociate VP40 from the lipid bilayer, indicates that the protein has certain properties of an integral membrane protein, as do a number of matrix proteins of negative-stranded RNA viruses (7, 37), even though Ebola virus VP40 does not appear to contain a region of significant length and hydrophobicity to span the cell membrane (Fig. 1B). Short hydrophobic stretches of VP40 may be able to penetrate the lipid bilayer to some extent, lending a modest integral-membrane character to the protein.
Ruigrok et al. (22) also reported that a deletion mutant protein of VP40 containing amino acids 31 to 212 failed to bind liposomes efficiently, indicating that the C terminus of VP40 is absolutely required for membrane binding. We constructed carboxy- and amino-terminal deletion mutant proteins to elucidate the domains involved in the association of VP40 with cellular membranes. VP40 lacking its 50 C-terminal amino acids demonstrated appreciably reduced membrane association. The Kyte-Doolittle hydrophobicity plot (18) of VP40 (Fig. 1B) indicates that amino acids 277 to 326 of the protein are primarily hydrophobic, so that deletion of amino acids 277 to 326 eliminates a substantial hydrophobic region that is likely important for efficient membrane binding by the full-length protein. This hypothesis is supported by the fact that 1 M NaCl completely disrupted this association, suggesting that affinity of this deletion construct with the lipid bilayer depends primarily on electrostatic interactions. When amino acids 227 to 326 of VP40 were deleted, the resulting truncated protein associated with the lipid bilayer as efficiently as wild-type VP40; moreover, C-terminal deletion of amino acids 177 to 326 resulted in a protein with much higher affinity for the lipid bilayer than was found for wild-type VP40. Salt treatment did not perturb membrane association of these truncated versions of VP40, indicating the presence of hydrophobic interactions mediated by the 176 N-terminal amino acids of the protein. The hydrophobicity plot indicates that amino acids 227 to 276, and particularly amino acids 177 to 226, are primarily hydrophilic. Deletion of the hydrophilic residues present in this region of VP40 may allow the truncated protein to fold into a structure capable of strong hydrophobic association with the cell membrane, perhaps by effectively exposing the highly hydrophobic central domain of the protein. These results are consistent with data obtained by Triton X-114 extraction analysis (Fig. 7). Since VP40 lacking its 50 C-terminal amino acids was unable to produce particles (Fig. 3) and these C-terminal residues appear to be required for efficient membrane association of VP40, binding of this highly hydrophobic region to the lipid bilayer may be an essential step in the particle formation process.
The crystal structure of amino acids 31 to 326 of Ebola virus was recently elucidated by Dessen et al. (8). It shows VP40 to be distinct from other viral matrix proteins in that it consists of two similar domains connected by a flexible linker at amino acids 195 to 200. Ruigrok et al. (22) showed that amino acids 31 to 212 of VP40 form hexamers spontaneously in solution. Dessen and associates postulate that, during the life cycle of Ebola virus, VP40 molecules associate with the lipid bilayer through interactions contributed primarily by their C termini. After membrane binding, the molecules undergo a conformational change that frees their N termini for hexamerization. These hexamers then form building blocks for a lattice that underlies the plasma membrane and subsequently may interact with the cytoplasmic tails of viral glycoproteins and/or the ribonucleoprotein complex. This model is based on data demonstrating the hexamerization of VP40 molecules that lack their 30 N-terminal amino acids as well as their 114 C-terminal amino acids. The PPXY motif that appears crucial for membrane-bound particle formation is located at amino acids 10 to 13 of VP40, and this motif most likely interacts with a cellular protein that exhibits a WW domain during virus particle assembly or budding. It has not yet been demonstrated that VP40 with a truncated C terminus can form hexamers when the entire N terminus is present. If hexamerization does occur during virion morphogenesis, the hexamers that form presumably must leave the PPXY motif accessible to cellular proteins that participate in particle formation and/or budding.
ACKNOWLEDGMENTS
We thank Krisna Wells and Martha McGregor for excellent technical assistance and John Gilbert for editing the manuscript.
Support for this work came from National Institute of Allergy and Infectious Diseases and Public Health Service research grants.
REFERENCES
- 1.Baize S, Leroy E M, Georges-Courbot M C, Capron M, Lansoud-Soukate J, Debre P, Fisher-Hoch S P, McCormick J B, Georges A J. Defective humoral responses and extensive intravascular apoptosis are associated with fatal outcome in Ebola virus-infected patients. Nat Med. 1999;5:423–426. doi: 10.1038/7422. [DOI] [PubMed] [Google Scholar]
- 2.Basler C F, Wang X, Muhlberger E, Volchkov V, Paragas J, Klenk H D, Garcia-Sastre A, Palese P. The Ebola virus VP35 protein functions as a type I IFN antagonist. Proc Natl Acad Sci USA. 2000;97:12289–12294. doi: 10.1073/pnas.220398297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergmann J E, Fusco P J. The M protein of vesicular stomatitis virus associates specifically with the basolateral membranes of polarized epithelial cells independently of the G protein. J Cell Biol. 1988;107:1707–1715. doi: 10.1083/jcb.107.5.1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 5.Bork P, Sudol M. The WW domain: a signalling site in dystrophin? Trends Biochem Sci. 1994;19:531–533. doi: 10.1016/0968-0004(94)90053-1. [DOI] [PubMed] [Google Scholar]
- 6.Chen H I, Sudol M. The WW domain of Yes-associated protein binds a proline-rich ligand that differs from the consensus established for Src homology 3-binding modules. Proc Natl Acad Sci USA. 1995;92:7819–7823. doi: 10.1073/pnas.92.17.7819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chong L D, Rose J K. Membrane association of functional vesicular stomatitis virus matrix protein in vivo. J Virol. 1993;67:407–414. doi: 10.1128/jvi.67.1.407-414.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dessen A, Volchkov V, Dolnik O, Klenk H D, Weissenhorn W. Crystal structure of the matrix protein VP40 from Ebola virus. EMBO J. 2000;19:4228–4236. doi: 10.1093/emboj/19.16.4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elliott L H, Kiley M P, McCormick J B. Descriptive analysis of Ebola virus proteins. Virology. 1985;147:169–176. doi: 10.1016/0042-6822(85)90236-3. [DOI] [PubMed] [Google Scholar]
- 10.Feldmann H, Klenk H D, Sanchez A. Molecular biology and evolution of filoviruses. Arch Virol Suppl. 1993;7:81–100. doi: 10.1007/978-3-7091-9300-6_8. [DOI] [PubMed] [Google Scholar]
- 11.Garoff H, Hewson R, Opstelten D E. Virus maturation by budding. Microbiol Mol Biol Rev. 1998;62:1171–1190. doi: 10.1128/mmbr.62.4.1171-1190.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Giddings A M, Ritter G D J, Mulligan M J. The matrix protein of HIV-1 is not sufficient for assembly and release of virus-like particles. Virology. 1998;248:108–116. doi: 10.1006/viro.1998.9284. [DOI] [PubMed] [Google Scholar]
- 13.Girault J A, Gorelick F S, Greengard P. Improving the quality of immunoblots by chromatography of polyclonal antisera on keratin affinity columns. Anal Biochem. 1989;182:193–196. doi: 10.1016/0003-2697(89)90741-0. [DOI] [PubMed] [Google Scholar]
- 14.Harty R N, Paragas J, Sudol M, Palese P. A proline-rich motif within the matrix protein of vesicular stomatitis virus and rabies virus interacts with WW domains of cellular proteins: implications for viral budding. J Virol. 1999;73:2921–2929. doi: 10.1128/jvi.73.4.2921-2929.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jayakar H R, Murti K G, Whitt M A. Mutations in the PPPY motif of vesicular stomatitis virus matrix protein reduce virus budding by inhibiting a late step in virion release. J Virol. 2000;74:9818–9827. doi: 10.1128/jvi.74.21.9818-9827.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Justice P A, Sun W, Li Y, Ye Z, Grigera P R, Wagner R R. Membrane vesiculation function and exocytosis of wild-type and mutant matrix proteins of vesicular stomatitis virus. J Virol. 1995;69:3156–3160. doi: 10.1128/jvi.69.5.3156-3160.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kobasa D, Rodgers M E, Wells K, Kawaoka Y. Neuraminidase hemadsorption activity, conserved in avian influenza A viruses, does not influence viral replication in ducks. J Virol. 1997;71:6706–6713. doi: 10.1128/jvi.71.9.6706-6713.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kyte J, Doolittle R F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Luo L, Schubert M, Wagner R R, Kang C Y. Viral liposomes released from insect cells infected with recombinant baculovirus expressing the matrix protein of vesicular stomatitis virus. J Virol. 1993;67:4415–4420. doi: 10.1128/jvi.67.7.4415-4420.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Muhlberger E, Weik M, Volchkov V E, Klenk H D, Becker S. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J Virol. 1999;73:2333–2342. doi: 10.1128/jvi.73.3.2333-2342.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108:193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- 22.Ruigrok R W, Schoehn G, Dessen A, Forest E, Volchkov V, Dolnik O, Klenk H D, Weissenhorn W. Structural characterization and membrane binding properties of the matrix protein VP40 of Ebola virus. J Mol Biol. 2000;300:103–112. doi: 10.1006/jmbi.2000.3822. [DOI] [PubMed] [Google Scholar]
- 23.Sanchez A, Kiley M P, Holloway B P, Auperin D D. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res. 1993;29:215–240. doi: 10.1016/0168-1702(93)90063-s. [DOI] [PubMed] [Google Scholar]
- 24.Sanchez A, Trappier S G, Mahy B W, Peters C J, Nichol S T. The virion glycoproteins of Ebola viruses are encoded in two reading frames and are expressed through transcriptional editing. Proc Natl Acad Sci USA. 1996;93:3602–3607. doi: 10.1073/pnas.93.8.3602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sandefur S, Varthakavi V, Spearman P. The I domain is required for efficient plasma membrane binding of human immunodeficiency virus type 1 Pr55Gag. J Virol. 1998;72:2723–2732. doi: 10.1128/jvi.72.4.2723-2732.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sudol M, Chen H I, Bougeret C, Einbond A, Bork P. Characterization of a novel protein-binding module—the WW domain. FEBS Lett. 1995;369:67–71. doi: 10.1016/0014-5793(95)00550-s. [DOI] [PubMed] [Google Scholar]
- 27.Takada A, Robison C, Goto H, Sanchez A, Murti K G, Whitt M A, Kawaoka Y. A system for functional analysis of Ebola virus glycoprotein. Proc Natl Acad Sci USA. 1997;94:14764–14769. doi: 10.1073/pnas.94.26.14764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanderzanden L, Bray M, Fuller D, Roberts T, Custer D, Spik K, Jahrling P, Huggins J, Schmaljohn A, Schmaljohn C. DNA vaccines expressing either the GP or NP genes of Ebola virus protect mice from lethal challenge. Virology. 1998;246:134–144. doi: 10.1006/viro.1998.9176. [DOI] [PubMed] [Google Scholar]
- 29.Volchkov V E, Feldmann H, Volchkova V A, Klenk H D. Processing of the Ebola virus glycoprotein by the proprotein convertase furin. Proc Natl Acad Sci USA. 1998;95:5762–5767. doi: 10.1073/pnas.95.10.5762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Volchkov V E, Volchkova V A, Slenczka W, Klenk H D, Feldmann H. Release of viral glycoproteins during Ebola virus infection. Virology. 1998;245:110–119. doi: 10.1006/viro.1998.9143. [DOI] [PubMed] [Google Scholar]
- 31.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wilson J A, Hevey M, Bakken R, Guest S, Bray M, Schmaljohn A L, Hart M K. Epitopes involved in antibody-mediated protection from Ebola virus. Science. 2000;287:1664–1666. doi: 10.1126/science.287.5458.1664. [DOI] [PubMed] [Google Scholar]
- 33.Wool-Lewis R J, Bates P. Characterization of Ebola virus entry by using pseudotyped viruses: identification of receptor-deficient cell lines. J Virol. 1998;72:3155–3160. doi: 10.1128/jvi.72.4.3155-3160.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wool-Lewis R J, Bates P. Endoproteolytic processing of the Ebola virus envelope glycoprotein: cleavage is not required for function. J Virol. 1999;73:1419–1426. doi: 10.1128/jvi.73.2.1419-1426.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Yang Z, Delgado R, Xu L, Todd R F, Nabel E G, Sanchez A, Nabel G J. Distinct cellular interactions of secreted and transmembrane Ebola virus glycoproteins. Science. 1998;279:1034–1037. doi: 10.1126/science.279.5353.1034. [DOI] [PubMed] [Google Scholar]
- 36.Yang Z Y, Duckers H J, Sullivan N J, Sanchez A, Nabel E G, Nabel G J. Identification of the Ebola virus glycoprotein as the main viral determinant of vascular cell cytotoxicity and injury. Nat Med. 2000;6:886–889. doi: 10.1038/78645. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Lamb R A. Characterization of the membrane association of the influenza virus matrix protein in living cells. Virology. 1996;225:255–266. doi: 10.1006/viro.1996.0599. [DOI] [PubMed] [Google Scholar]