Abstract
Phenolic phytometabolites are promising bioactive compounds for management of genomic instability related diseases. Formononetin (FMN) and arbutin (ARB) are found in several plant sources. Our goal was to investigate the safety and efficacy of FMN and ARB using in vitro both standardized and alternative toxicogenetic methods. FMN and ARB were evaluated through the OECD’S guidelines No. 471 (Bacterial Reverse Mutation Test –Salmonella/microsome) and No. 487 (In vitro Mammalian Micronucleus Test – CBMN assay), accordingly to the mentioned recommendations. Also, antimutagenicity of FMN and ARB was assessed in S. Typhimurium strains TA98, TA100 and TA1535, following pre-, co- and post- treatment protocols. Liver human lineages HepG2 and F C3H were assayed for cytotoxicity after exposure to FMN and ARB (24, 48 and 72 h) using in vitro WST-1 test. ARB showed no mutagenicity in the Salmonella/microsome test under both metabolic conditions (in presence or absence of 4 % S9 mix), but FMN was cytotoxic to the TA97 and TA100 strains after metabolic activation. Under this same condition, FMN induced an increase in the mutagenic index of strain TA1535 at two of the highest tested concentrations. Even so, ARB and FMN exhibited protection against the induced alkylation of DNA in multiple action modes. In the antimutagenicity assay, FMN reached the maximum of 80 % of oxidative-provoked mutagenicity reduction in TA98 strain in co-treatment with known mutagen, besides 69 % of reduction in TA100 in the same exposure condition. ARB showed up to reduce induced mutagenicity in strains TA100 and TA1535, reaching percentages from 55 % to 100 % of antimutagenicity in all of the tested exposure models against alkylating agent. In the CBMN assay, no increase in micronuclei formation was observed. The results suggest that FMN and ARB prevent DNA from mutation using multi-targeted antimutagenic roles. Finally, our data suggests that FMN and ARB are not genotoxic and presented encouraging antimutagenicity action in vitro, being promising compounds for use in genomic instability-related diseases therapeutics.
Keywords: Phenolic compounds, Toxicogenetic assessment, Antimutagenicity, Chemoprevention
1. Introduction
Cyrtopodium glutiniferum Raddi is an orchid found in southeastern Brazil, and its bulbs are used in folk medicine to treat skin lesions and infectious abscesses. The Cyrtopodium genus is popularly called “Sumaré”, and the species are frequently confused with each other due to difficulties in visually distinguishing them [1]. The genus has ethnopharmacological relevance reported, and it is used in ointments and juices to treat chest colds, tuberculosis, bacterial infections, and to diminish inflammatory responses, actions mainly related to the presence of self-protection glucomannans and stilbenes, produced as a result of the orchid’s secondary metabolism [2].
A previous investigation of our group [3] presented results on the chemical composition of an aqueous extract of pseudobulbs from C. glutiniferum Raddi. Tóth et al. [4] also highlighted the most abundant molecules from this plant. A genotoxicity assessment and in vitro antiproliferative activity was also reported, suggesting that the plant has promising potential as a phytopharmaceutical [3].
Of the phenols identified by Araujo-Lima et al. [3], phenanthrene was the main class of molecules. However, dihydroformononetin, caffeic acid 4-O-glucoside, and arbutin were detected in more significant quantities, being the most abundantly found secondary metabolites in C. glutiniferum Raddi. Dihydroformonetin is a reduction product of formononetin (FMN), an isoflavone mainly found among polyphenols from the Leguminosae family [5]. Caffeic acid 4-O-glucoside (or caffeic acid) represents the hydro cinnamic class, and its structure comprises an aromatic acid derived from catechol phenols [6]. Arbutin (or β-arbutin, ARB) belongs to the hydroquinone glucoside (β-glucose) group and is widely distributed in the Plantae kingdom [7]. Several remarkable biological effects exerted by these compounds have been described [8], [9], [10], [11], [12], such as anti-inflammatory action and skin protection against UV-induced pigmentation, which is of great importance since they are the main components of the extract from C. glutiniferum Raddi.
Due to the large number of molecules and therapies intended for human use, it is crucial to standardize and set regulations for testing chemicals regarding their toxicological safety and efficacy [13], [14]. Regulatory agencies from countries worldwide make use of recommendations from the Organisation for Economic Co-operation and Development (OECD) to produce experimental protocols on risk assessment, which facilitates commercial transactions between countries with legislation disparities in the field of chemicals in general. These guidelines (e.g., TG471, Bacterial Reverse Mutation Test and TG487, In Vitro Mammalian Cell Micronucleus Test) [15], [16] are employed, for instance, in the pre-clinical stages of pharmaceutical development and provide strong scientific evidence that can be used to determine whether a substance can be made available for consumption or otherwise [17].
International organisations worldwide validate the usage of alternative approaches in genotoxic testing, and have delineated guidelines and parameters for its applications with regulatory character [18]. Such fact could be related to the constantly increasing concern on evaluating the true necessity of applying animal testing in research. This concept was introduced in the 1960’s since the contributions from Russel and Burch, that cemented the Three R’s principles, a very well absorbed notion in genotoxicity assessment [19].
This becomes noticeable in the toxicogenetic field by the use of emerging scientific findings in the field of nanotechnology and computational chemistry to various purposes, with prominent advances in drug discovery, design and delivery research [20], [21].
For the drug discovery area, ligand-based (e.g. molecular docking) and structure-based (e.g. quantitative structure-activity relationship, QSAR) [22] techniques are available for a variety of applications for the goal of describing new leader compounds [23], [24]. The usage of computational predictors for in silico research is becoming a useful tool for screening molecules for genotoxic profile [25]. In silico alternatives, such as QSAR-related prediction for pharmacokinetic properties (absorption, distribution, metabolization, excretion and toxicity, ADMET) of substances also impose great advantage in terms of saving time and financial resources. [26].
Nanotechnology provided new insights into drug delivery systems, in which is very currently seen that natural occurring element (e.g. metals) or substances (e.g. secondary metabolites from plants) are modified through nano systems in order to improve its physicochemical properties [27], [28]. Utilizing nanostructured-based tools (e.g. liposomes), it is possible to increase absorption, solubility and targeted distribution of natural bioactive molecules, and even more, reduce the toxicity of such therapeutic-intended substances to organisms and to the natural environment [29], [30], [31].
Despite the mistaken common belief that medicinal plants or their products are safe and can only pose benefits to health, there is increasing scientific evidence that clarifies this misconception [32]. Thus, the main objective of the present study was to employ both standardized and alternative toxicogenetic methods to assess the in vitro safety and efficacy of FMN and ARB, two of the most present phenolic compounds obtained from C. glutiniferum Raddi, and search for an understanding of how these plant molecules might provide health benefits to humans, especially in the context of diseases related to genomic instability, like cancer.
2. Material and methods
2.1. Chemicals
Formononetin (FMN) (C6H12O4) (CAS #485–72–3) and arbutin (ARB) (C12H16O7) (CAS #497–76–7) were purchased from Sigma Aldrich (St. Louis, MO, USA), as were all the other chemical reagents used in the experiments detailed below.
2.2. Prokaryotic models
2.2.1. Salmonella/microsome assay (Ames Test)
The Salmonella/microsome assay (also referred to as the Ames test) was applied as described by Maron and Ames [33], following recommendation from OECD’s test guideline 471 [15], and with considerations from recently published work from our group [34]. Briefly, a set of five strains of enterobacteria Salmonella enterica serovar Typhimurium (TA97, TA98, TA100, TA102, and TA1535) were submitted to a pre-incubation (20 min at 37º C, under agitation) protocol in the presence and absence of an exogenous metabolic activation system (4 % S9 mix, Moltox Inc., USA). The cells were exposed (20 min at 37º C with agitation) to FMN and ARB (prepared to final concentrations of from 0.001 to 10 µM/plate). Stock solutions of FMN and ARB were prepared based upon maximum solubility in dimethyl sulfoxide (DMSO, CAS #67–68–5), then gradually add sterile water to each stock solution of 50 mM of FMN and 25 mM of arbutin (reaching up 25 % of water in each, kept in freezer). Sterile water (0.0 µM/plate) was the negative control. Positive controls (mutagens) were used as listed: (without metabolic activation, -S9) 4-nitroquinoline 1-oxide (4-NQO, CAS #7608–65–0) for TA97 and TA98 (1.0 μg/plate and 0.5 μg/plate, respectively), sodium azide (NaN3, CAS #26628–22–8) for TA100 and TA1535 (5.0 μg/plate for both strains); and mitomycin C (MMC, CAS #50–07–7) for TA102 (0.5 μg/plate); (with metabolic activation, +S9) 2-aminoanthracene (2-AA, CAS #613–13–8) for TA97 and TA1535 (5.0 μg/plate for both strains), and also for TA98 and TA100 (1.0 μg/plate for both strains); Benzo[a]pyrene (B[a]P, CAS #634–6671–7) (50 μg/plate) for TA102. All the mutagens were purchased from Sigma-Aldrich as mentioned above. After incubation, 2 mL top agar containing traces of essential amino acids (0.05 mol L−1 L-histidine and D-biotin) were added to the reaction mix tubes and then spread on top of minimum glucose agar plates. After 30 minutes, the inverted plates were incubated at 37º C, protected from light, for up to 72 h (2–3 days). Revertant colonies from each strain and treatment group were visually counted (separately). Mutagenicity index (M.I.) was determined as a ratio between the average number of colonies from the sample concentration divided by the average number found in the negative control (spontaneous revertants). The mutagenic criterion was based upon: 1) an increase in M.I. at least twice as high as in the spontaneous revertants (M.I. ≥ 2.0); and 2) statistically significant results. This procedure was repeated at least three independent times (n=3), using triplicates for all concentrations and controls, testing all five strains. The results were reported as the mean number of revertant colonies ± standard deviation (SD), and statistical differences between the groups were determined by a univariate one-way ANOVA (p <0.05) with Tukey’s post hoc pairwise analysis.
2.2.2. Antimutagenicity assay
Antimutagenicity was determined as previously described by our group [35], [36] with a few modifications. The pre-, co-, and post- treatments (without the addition of metabolic activation) were carried out for strains TA98, TA100, and TA1535. A volume of 0.1 mL of bacterial suspension (2 ×108 cells/mL of nutrient broth) was added to reaction tubes containing 0.5 mL of a 0.1 mol/L sodium phosphate buffer (pH 7.4). FMN and ARB were prepared the same way as in the previous assay (final concentrations of from 0.0 to 10 µM/plate). The strains were either submitted to 0.1 mL of methyl methanesulfonate (MMS, CAS #66–27–3, diluted to a final concentration of 250 µg/plate) or 4-nitroquinoline 1-oxide (4-NQO, diluted to a final concentration of 0.5 µg/plate). For co-treatment, FMN or ARB were added concurrently with the chosen mutagen for 1 h (MMS for TA1535 and TA100, 4-NQO for TA98). In the pre-treatment, the strains reacted with FMN or ARB for 30 min before adding the mutagen substance (followed by another 30 min incubation), whereas in the post-treatment, the first incubation time (30 min) was just between the bacterial cells and the mutagen, then followed by 30 min of exposure to FMN or ARB. All treatment formats occurred for a total time of 1 h, and then top agar was added and the mixtures incorporated in plates (as in the Ames test). After 72 h, the revertant colonies were counted in each treatment group. The experiments were carried out at three independent times (n=3), in triplicate for each set of exposed samples. The results were compiled as the mean values ± SD of revertant colonies, and the percent reduction in mutagenicity calculated using a linear regression analysis (MMS or 4-NQO as 0 % of reduction, and negative control – untreated – as 100 %). Statistical differences between groups were determined using one-way ANOVA with Tukey’s post hoc pairwise analysis.
2.3. Eukaryotic models
2.3.1. Cytotoxicity to hepatic cell lineages (WST-1)
The HepG2 (human hepatocellular carcinoma, purchased from the American Type Culture Collection, Manassas, VA, USA, #HB-8065) and F C3H (mouse liver fibroblast, purchased from the Banco de Células do Rio de Janeiro, BCRJ, Brazil, #BCRJ0082) cell lineages were maintained in stock solution (liquid nitrogen, approximately −70ºC), harvested after centrifugation (10000 g for 5 min) and added to fresh culture media (Dulbecco’s Modified Eagle Media, DMEM, Gibco) supplemented with antibiotics (1 % penicillin 100 µg/mL and streptomycin 100 µg/mL) and nutrients from foetal bovine serum (10 % FBS). The cells were allowed to spread and adhere to 75 cm2 flasks (Thermo Fischer, Waltham, MA, USA) in an atmosphere with 95 % humidity and 5 % CO2 at 37ºC for at least 48 h (or until they reached 80–90 % confluency), before being harvested by chemical removal (0.05 % trypsin and 0.02 % EDTA) and prepared for the assay. The live and dead cells were distinguished by the trypan blue exclusion method [37]. Both lineages were assayed using the WST-1 cytotoxicity assay kit (Roche Co., Manheim, Germany) according to the manufacturer’s instructions and the considerations of our group [36]. Shortly after, the cells were seeded in 96-well flat-bottomed plates with 1 ×104 cells/well. The day after seeding, the plates were exposed to FMN and ARB (up to a final concentration of 1000.0 µM on a semi-log scale), to distilled water (negative control), and to the positive control (5 % Triton X-100, CAS #9036–19–5), for every corresponding triplicate for 24 h, 48 h or 72 h. After incubation, the cells were washed with PBS (1X), and replaced with a water-soluble tetrazolium salt (4-[3-(4-iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) solution (90 µl cell media and 10 µl WST-1, per well), and incubated under the same conditions for 3 h. The absorbance signals were detected using a cell plate reader (Polaris, MG, Brazil) at 440 nm. In viable cells, the WST-1 salt undergoes reduction mediated by mitochondrial dehydrogenases, going from clear red to dark yellow when transformed into the formazan salt. The amount of absorbance measured is proportional to the number of metabolically active cells in the culture. The absorbance readings from the negative control were considered as 100 % viable cells, while the positive control was considered as non-viable cells (0 %). All further comparisons were based upon this reference level to determine lethal concentration to 50 % cultured cells (LC50). The nonlinear regression fit of the dose-response curves was calculated using the GraphPad Prism software (version 8.0.2), and the same program was employed to determine the statistical differences between the groups, using one-way ANOVA with Tukey’s post hoc test. The experiments were repeated at least three times (n =3) in triplicate for each exposure setting.
2.3.2. Cytokinesis-block micronucleus assay (CBMN-assay)
For the cytokinesis-block micronucleus assay, the HepG2 cell culture was maintained as previously described and the CBMN-assay carried out including the recommendations of both OCED’s TG487 [16] and Fenech [38], with a few modifications. Briefly, fresh HepG2 cells were harvested from the culture and seeded into 24-well plates (1 mL per well), with a density of 2–3 ×104 cells/mL. A thin glass slide (0.13 mm) treated with HNO3 (0.1 M) was previously added to each well to enhance cell adhesion to the glass surface. After 24 h, of exposure to FMN and ARB, each sample was diluted to 0.0–100 µM/mL (prepared in fresh cell media, negative control), and benzo[a]pyrene (0.1 mg/mL) was used as the positive control. After 2 h of incubation with the samples and controls, the cell media from all wells were replaced with a solution of cytochalasin-B (3 µg/mL) and incubated for another 20–24 h. For all steps, the incubations were carried out under the cell culture conditions mentioned above. The media were then removed, the wells carefully washed with 1X PBS, 1 mL of freshly prepared fixative solution (methanol-glacial and acetic acid, 3:1) added, and incubated for a further 15 min. The fixed cells were placed on coverslips and stained with 0.6 % (w/w) Giemsa dye (Merck, Darmstad, Germany) (1:5) for 1 h. Finally, the coverslips were collected, excess dye removed with distilled water and allowed to dry for a few minutes before preparing the slides with Entellan-New (Merck). The slides were observed under a light microscope (Olympus CX31) with X40 magnification. Together with binucleated cells, other classical CBMN presentations (e.g., nuclear buds, nucleoplasmatic bridges, multinucleated cells) and death events (apoptosis and necrosis) were scored. The nuclear division indexes (NDI) and frequency of micronuclei in once-divided cells (MNi) were calculated by counting at least 2000 binucleated cells per slide for each exposure set (approximately 666 binucleated cells per replicate). Mathematical models for the acquisition of the cytokinesis block proliferation index were calculated as suggested by Fenech [39] and statistical differences between the groups were determined by a univariate one-way ANOVA (p<0.05) with Tukey’s post hoc pairwise analysis.
3. Results
3.1. Mutagenicity
Without metabolic activation, neither FMN nor ARB induced mutagenicity in S. Typhimurium strains. However, to the contrary, after activation with the S9 mix, FMN increased the mutagenicity index (M.I. = 2.3) for TA1535 at 10 µM/plate, although no significant differences in comparison to the negative control were detected in the statistical analysis. FMN also showed cytotoxicity (cell viability ≤70 % as related to the negative control revertants, data not shown) for TA97 and TA100 (up to 0.1 µM/plate and 1.0 µM/plate, respectively) (Table 1). ARB did not induce cytotoxicity in any of the strains tested, neither in the presence nor absence of metabolic activation.
Table 1.
Mean values ± SD resulting from His+ revertants of S. Typhimurium in the Salmonella/microsome assay facing phenolic compounds from C. glutiniferum Raddi.
| FMN | ARB | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| -S9a | +S9b | -S9 | +S9 | |||||||||
| Strain | µM/plate | M.I.c | His+± SDd | M.I. | His+± SD | M.I. | His+± SD | M.I | His+± SD | |||
| TA97 | 0 | 1.0 | 54.0 ± 9.5 | 1.0 | 148.7 ± 17.0 | 1.0 | 54.0 ± 9.5 | 1.0 | 148.7 ± 17.0 | |||
| 0.001 | 1.0 | 51.3 ± 1.2 | 0.9 | 129.0 ± 21.1 | 0.8 | 41.0 ± 12.5 | 1.0 | 156.0 ± 4.2 | ||||
| 0.01 | 1.1 | 59.0 ± 3.6 | 0.8 | 121.3 ± 16.6 | 0.8 | 41.0 ± 8.5 | 0.9 | 140.3 ± 16.7 | ||||
| 0.1 | 1.1 | 58.7 ± 2.1 | — | cytotoxic | 1.1 | 61.7 ± 10.2 | 1.0 | 143.7 ± 23.7 | ||||
| 1 | 1.1 | 57.3 ± 10.5 | — | cytotoxic | 1.0 | 55.7 ± 10.0 | 0.9 | 136.3 ± 11.0 | ||||
| 10 | 0.9 | 50.0 ± 3.0 | — | cytotoxic | 1.0 | 56,0 ± 4.0 | 1.4 | 201.0 ± 9.5 | ||||
| TA98 | 0 | 1.0 | 25.3 ± 2.3 | 1.0 | 37.3 ± 21.9 | 1.0 | 25.3 ± 2.3 | 1.0 | 37.3 ± 21.9 | |||
| 0.001 | 1.1 | 28.0 ± 5.3 | 1.1 | 40.0 ± 17.0 | 1.1 | 28.7 ± 7.6 | 0.7 | 26.0 ± 6.0 | ||||
| 0.01 | 0.9 | 24.0 ± 2;8 | 0.9 | 34.0 ± 8.5 | 0.9 | 22.3 ± 4.7 | 0.9 | 34.0 ± 14.1 | ||||
| 0.1 | 1.0 | 25.7 ± 3.5 | 1.0 | 37.3 ± 8.3 | 0.7 | 18.5 ± 7.8 | 1.3 | 46.7 ± 5.8 | ||||
| 1 | 1.2 | 31.3 ± 3.1 | 1.1 | 41.0 ± 9.9 | 0.8 | 21.3 ± 2.1 | 1.1 | 42.0 ± 5.3 | ||||
| 10 | 1.1 | 28.0 ± 3.0 | 0.9 | 32.0 ± 5.7 | 1.0 | 24.7 ± 1.5 | 1.3 | 49.3 ± 8.1 | ||||
| TA100 | 0 | 1.0 | 124.3 ± 4.5 | 1.0 | 127.0 ± 12.7 | 1.0 | 124.3 ± 4.5 | 1.0 | 127.0 ± 12.7 | |||
| 0.001 | 1.0 | 143.0 ± 4.2 | 0.4 | 50.0 ± 8.5 | 1.1 | 134.3 ± 20.8 | 0.6 | 75.3 ± 14.2 | ||||
| 0.01 | 1.0 | 122.0 ± 15.6 | 0.7 | 83.3 ± 21.9 | 1.2 | 154.3 ± 20.3 | 1.1 | 144.7 ± 17.9 | ||||
| 0.1 | 1.0 | 128.3 ± 8.1 | 0.5 | 65.0 ± 15.6 | 1.1 | 141.7 ± 13.6 | 0.6 | 78.0 ± 36.8 | ||||
| 1 | 1.0 | 119.0 ± 3.6 | — | cytotoxic | 1.4 | 169.3 ± 19.1 | 1.2 | 149.0 ± 1.2 | ||||
| 10 | 1.0 | 119.5 ± 6.4 | — | cytotoxic | 0.9 | 108.0 ± 7.1 | 0.7 | 90.0 ± 23.1 | ||||
| TA102 | 0 | 1.0 | 209.0 ± 14.1 | 1.0 | 235.0 ± 1.7 | 1.0 | 209.0 ± 14.1 | 1.0 | 235.0 ± 1.7 | |||
| 0.001 | 1.0 | 235.0 ± 15.1 | 1.0 | 224.0 ± 9.5 | 1.0 | 211.3 ± 17.0 | 0.9 | 221.7 ± 6.7 | ||||
| 0.01 | 1.0 | 201.0 ± 1.4 | 0.9 | 209.5 ± 2.1 | 1.0 | 200.5 ± 4.9 | 1.2 | 277.5 ± 40.3 | ||||
| 0.1 | 1.0 | 210.7 ± 15.3 | 0.9 | 221.0 ± 21.7 | 1.1 | 232.0 ± 14.1 | 1.2 | 272.0 ± 19.8 | ||||
| 1 | 1.1 | 230.7 ± 8.1 | 1.0 | 227.7 ± 17.0 | 1.1 | 222.0 ± 26.5 | 1.0 | 225.5 ± 6.4 | ||||
| 10 | 1.0 | 214.5 ± 9.2 | 1.0 | 231.7 ± 19.6 | 1.0 | 215.5 ± 2.1 | 1.1 | 253.3 ± 35.7 | ||||
| TA1535 | 0 | 1.0 | 10.0 ± 2.8 | 1.0 | 20.0 ± 5.7 | 1.0 | 10.0 ± 2.8 | 1.0 | 20.0 ± 5.7 | |||
| 0.001 | 0.6 | 6.3 ± 2.3 | 1.3 | 26.0 ± 3.5 | 0.8 | 8.3 ± 1.5 | 1.5 | 30.0 ± 2.8 | ||||
| 0.01 | 0.9 | 9.3 ± 2.5 | 1.4 | 28.0 ± 8.7 | 0.6 | 6.0 ± 1.0 | 0.7 | 14.0 ± 2.0 | ||||
| 0.1 | 0.7 | 7.3 ± 1.2 | 1.4 | 27.0 ± 4.2 | 0.5 | 4.7 ± 0.6 | 0.7 | 14.0 ± 0.0 | ||||
| 1 | 1.1 | 11.0 ± 4.2 | 1.7 | 34.7 ± 11.7 | 0.6 | 5.5 ± 0.7 | 1.5 | 30.0 ± 2.8 | ||||
| 10 | 1.2 | 10.3 ± 1.5 | 2.3 | 46.7 ± 4.2 | 0.8 | 7.3 ± 4.0 | 1.5 | 30.7 ± 2.3 | ||||
Different of negative control (one-way ANOVA followed by Tukey’s post hoc test, *p <0.05).
Experiment done in triplicate plates, repeated at least three times (n=3).
Positive controls without S9 (-S9): 4-NQO (1.0 μg/plate) for TA97, 1333.0 ± 309.7 revertants; 4NQO (0.5 μg/plate) for TA98, 133.7 ± 16.3 revertants; AS (5.0 μg/plate) for TA100, 1290.7 ± 30.0 revertants; AS (5.0 μg/plate) for TA1535, 1109.3 ± 28.6 revertants; MMC (0.5 μg/plate) for TA102, 1157.3 ± 305.1 revertants.
Positive controls with S9 (+S9): 2-AA (5.0 μg/plate) for TA97 and TA1535, 205.5 ± 18.0 and 38.0 ± 6.6 revertants, respectively; 2-AA (1 μg/plate) for TA98 and TA100, 245.3 ± 61.2 and 332.0 ± 84.9 revertants, respectively; B[a]P (50 μg/plate) for TA102, 311.0 ± 53.1 revertants.
Experiment done in absence of metabolic activation (-S9).
Experiment done in the presence of metabolic activation (+S9).
Mutagenicity index: No. of His+ induced revertants in the sample/number of spontaneous His+ spontaneous revertants in the negative control (sterile water).
SD: standard deviation.
3.2. Antimutagenicity
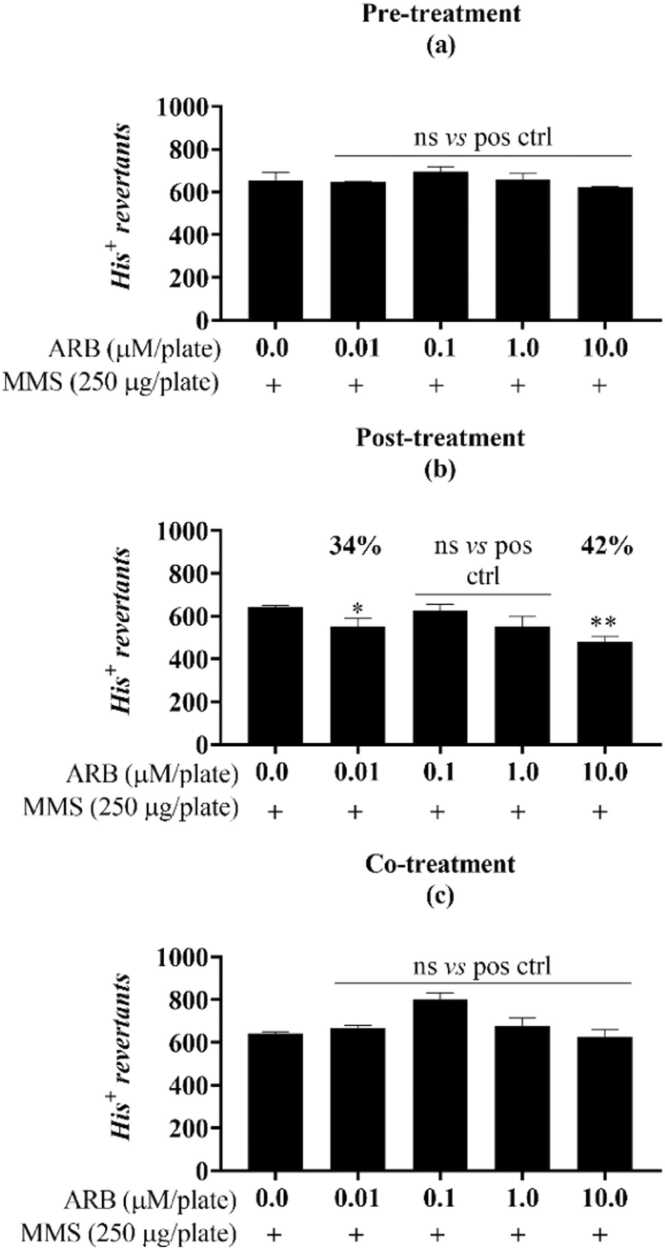
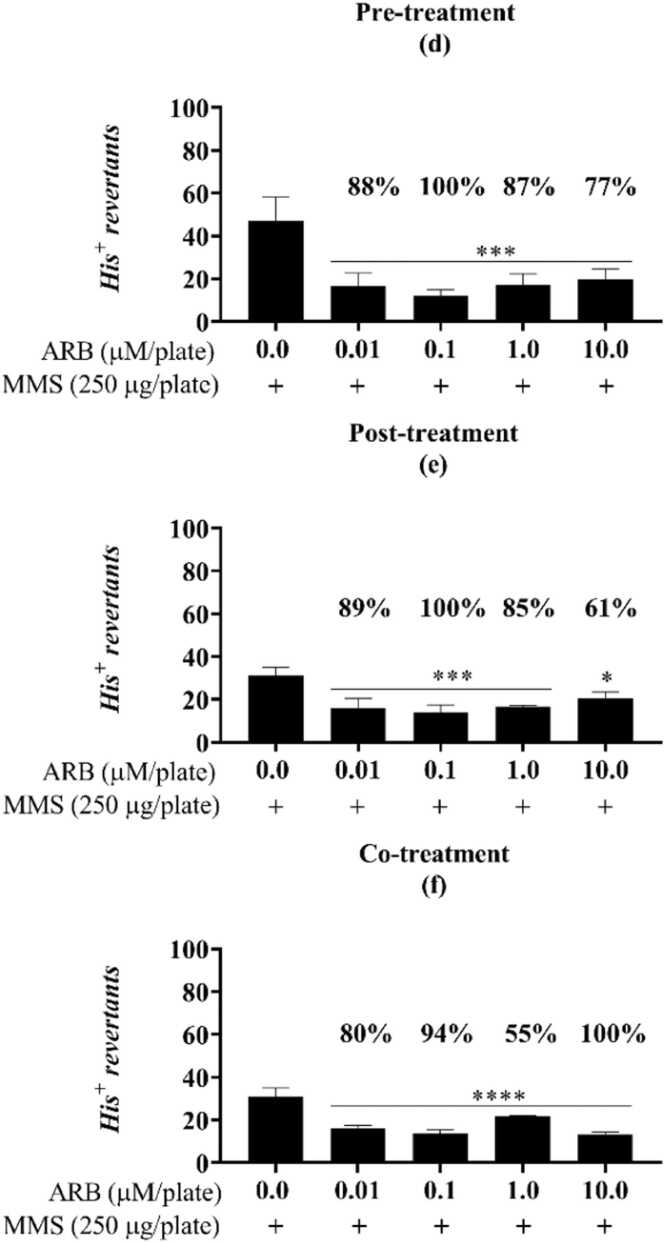
Both FMN and ARB were shown to protect DNA from chemical attack, significantly reducing the number of chemically-induced point mutations (Fig. 1, Fig. 2, Fig. 3, Fig. 4, Fig. 5). For FMN, this outcome was present for the TA98 strain in the co-treatment with mutagen, yielding an 80 % reduction in the mutagenicity caused by 4-nitroquinoline 1-oxide (4-NQO) at 1 µM/plate (Fig. 1c). For the TA100 strain, the protective response of FMN was observed in the post and co-treatments, reducing 54 % and 69 % of the methyl methane sulfonate (MMS) induced damage, respectively (Fig. 2e, f). For TA1535, the percent reduction caused by FMN reached 98 % (0.01 µM/plate) when in co-incubation with MMS and 100 % in the pre-treatment condition (0.1 µM/plate) (Fig. 3g, i). Similarly, ARB induced a reduction in MMS-mediated mutagenicity for both the TA100 and TA1535 strains. For TA100, a reduction of 42 % was observed only in the post-treatment and was marked at the higher concentration tested (10.0 µM/plate) (Fig. 4b). Finally, the results caused by ARB for TA1535 stood out when compared to the effects of FMN for the same strain, obtaining antimutagenicity percentages above 55 % for all exposure models and concentrations assessed (Fig. 5d, e, f).
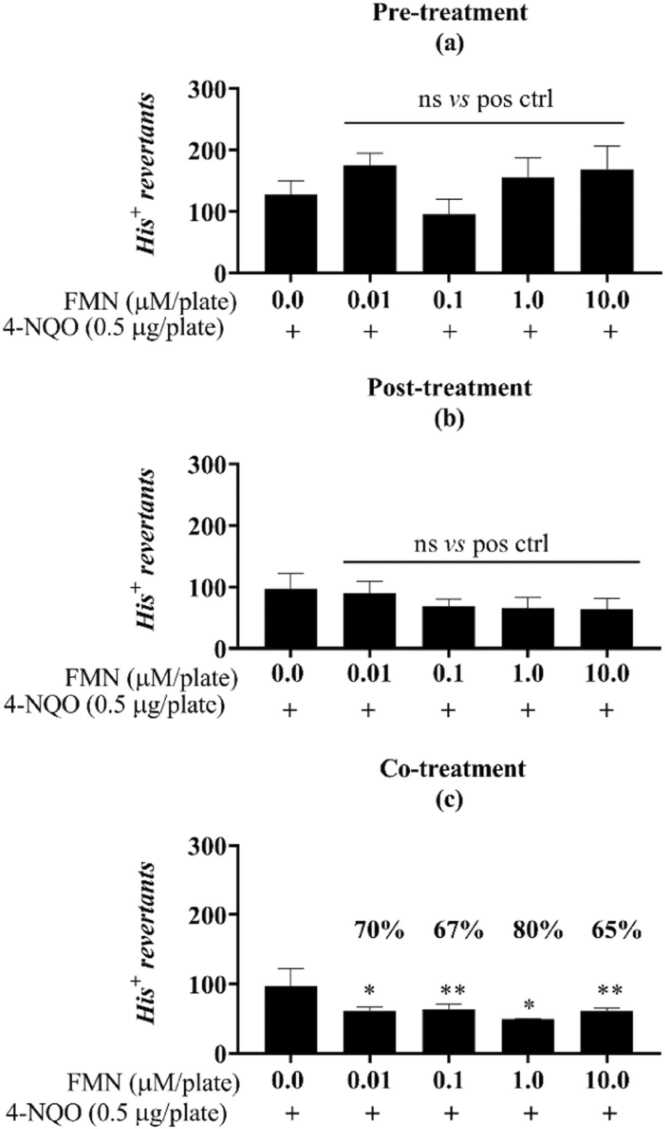
Fig. 1.
FMN protective response against the oxidative damage of 4-NQO in the antimutagenicity assay for the S.Typhimurium strain TA98. Different from the positive control (one-way ANOVA followed by Tukey’s post hoc test; *p < 0.01, **p < 0.001, ***p =0.0001, ****p <0.0001). Pre- (a), post- (b) and co- (c) incubation of FMN with the mutagen 4-nitroquinoline 1-oxide (4-NQO, “pos ctrl”, 0.5 µg/plate). His+ revertant colonies. ns: not statistically different from the untreated or positive controls. Reduction percentages are indicated along with the corresponding concentration.
Fig. 2.
FMN protective response against the alkylation damage of MMS in the antimutagenicity assay for the S.Typhimurium strain TA100. Different from the positive control (one-way ANOVA followed by Tukey’s post hoc test; *p <0.01, **p <0.001, ***p =0.001, ****p <0.0001; n=3). Pre- (d), post- (e) and co- (f) incubation of FMN with the mutagen MMS (methyl methanesulfonate, “pos ctrl”, 250 µg/plate). His+ revertant colonies. ns: not statistically different from the untreated or positive controls. Reduction percentages are indicated along with the corresponding concentration.
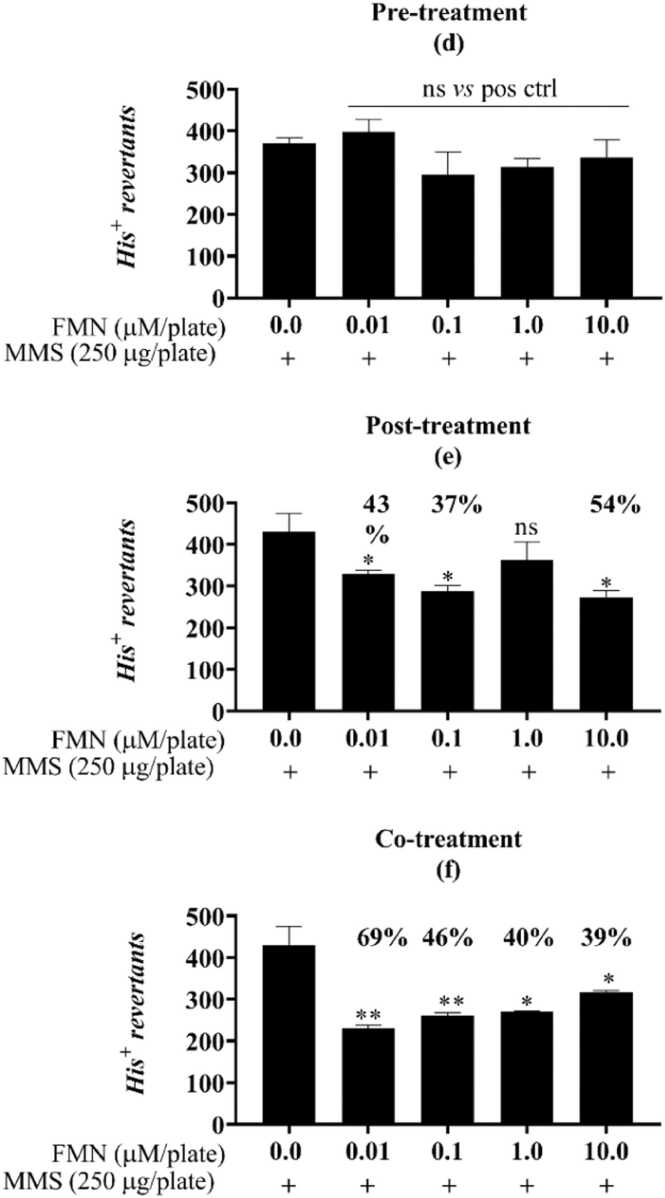
Fig. 3.
FMN protective response against the alkylation damage of MMS in the antimutagenicity assay for the S.Typhimurium strain TA1535. Different from the positive control (one-way ANOVA followed by Tukey’s post hoc test; *p < 0.01; n=3). Pre- (g), post- (h) and co- (i) incubation of FMN with the mutagen MMS (methyl methanesulfonate, “pos ctrl”, 250 µg/plate). His+ revertant colonies. Ns: not statistically different from the untreated or positive controls. Reduction percentages are indicated along with the corresponding concentration.
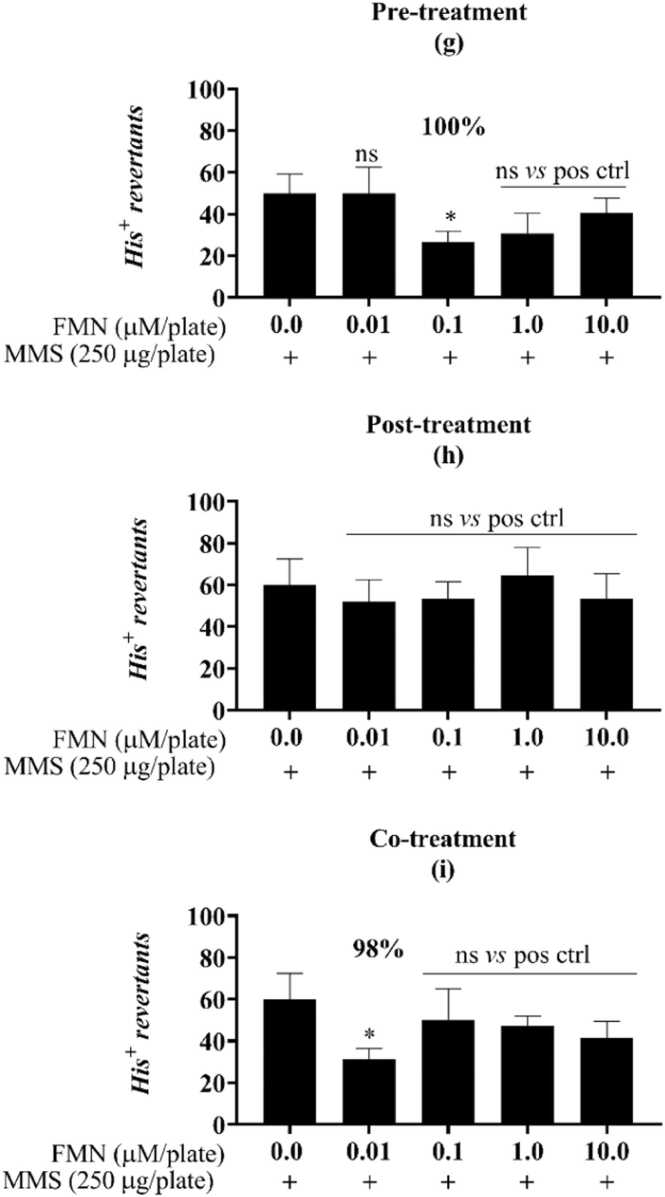
Fig. 4.
ARB protective response against the alkylation damage of MMS in the antimutagenicity assay for the S.Typhimurium strain TA100. Different from the positive control (one-way ANOVA followed by Tukey’s post hoc test; *p <0.01, **p <0.001, ***p = 0.001, ****p <0.0001; n=3). Pre- (a), post- (b) and co- (c) incubation of ARB with the mutagen MMS (methyl methanesulfonate, “pos ctrl”, 250 µg/plate). His+: revertant colonies. ns: not statistically different from the untreated or positive controls. Reduction percentages are indicated along with the corresponding concentration.
Fig. 5.
ARB protective response against the alkylation damage of MMS in the antimutagenicity assay for the S.Typhimurium strain TA1535. Different from the positive control (one-way ANOVA followed by Tukey’s post hoc test; *p< 0.01, **p< 0.001, ***p= 0.001, ****p< 0.0001; n=3). Pre- (d), post- (e), and co- (f) incubation of ARB with the mutagen MMS (methyl methanesulfonate, “pos ctrl”, 250 µg/plate). His+: revertant colonies. ns: not statistically different from the untreated or positive controls. Reduction percentages are indicated along with the corresponding concentration.
3.3. Micronuclei formation and genotoxicity in eukaryotic cells
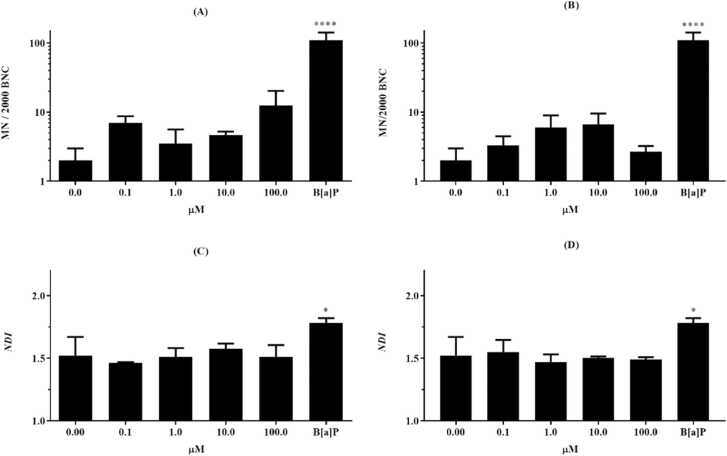
Considering that the results of the WST-1 assay for FMN and ARB suggested that none of the compounds resulted in cytotoxicity for HepG2 or F C3H at any of the concentrations tested during 24 h, 48 h or 72 h (Table 2), safe concentrations for viability were established to carry out the in vitro genotoxicity testing. In the CBMN assay, no FMN or ARB concentration tested induced micronuclei (MNi) formation in HepG2 cells (Fig. 6, a, b), and no cytostatic or cytotoxic markers were observed. The nuclear division indexes (NDI) for FMN and ARB showed no statistical significance when compared to the NDI of the negative control (untreated binucleated cells) (Fig. 6, c, d).
Table 2.
Lack of cytotoxicity of FMN and ARB after 24, 48 and 72 h of exposure to HepG2 and F C3H cells.
|
LC50(µM) |
|||||||
|---|---|---|---|---|---|---|---|
| Compound | HepG2 | F C3H | |||||
| 24 h | 48 h | 72 h | 24 h | 48 h | 72 h | ||
| FMN | >1000.0 | >1000.0 | >1000.0 | >1000.0 | >1000.0 | >1000.0 | |
| ARB | >1000.0 | >1000.0 | >1000.0 | >1000.0 | >1000.0 | >1000.0 |
LC50: lethal concentration of 50 % of exposed HepG2 cells. FMN: formononetin. ARB: arbutin. T-X100: Triton X-100 (positive control).
Fig. 6.
Genotoxicity in vitro panel of FMN and ARB in the HepG2 human lineage. FMN and ARB induction of micronuclei in HepG2 cells. To determine the number of micronuclei (MN/2000 BNC) a sum of 2000 binucleated cells per slide was counted. The NDI was obtained from mono-, bi-, tri- and polynucleated cells, scored from the same parameter. FMN is presented on the left (a and c), ARB on the right (b and d). B[a]P: benzo[a]pyrene (0.1 mg/mL), positive control. Sterile water (0): negative control. Different from the negative control (one-way ANOVA, with Tukey’s post hoc analysis. *p <0.01, **p <0.001, ***p =0.0001, ****p <0.0001, n=1).
4. Discussion
Species from all over the Plantae kingdom produce substances as a result of enduring abiotic and biotic threats imposed by natural conditions (e.g., herbivores, bacterial or fungal attacks, UV radiation from sunlight, or a scarcity of nutrients and water), besides unnatural impositions (e.g., climate changes imposed by human industrial activities) [40]. These compounds are referred to as secondary metabolites, and phenolic compounds are representatives of these molecules and of great interest to the field of phytochemistry [41], [42], [43].
The contribution given by orally transmitted information on the use of plants to oppose various diseases must be addressed. However, the natural path of perpetuating this traditional knowledge presumes it is undoubtedly liable to modifications with time [44]. Nevertheless, it is imperative to challenge these misconceptions or popular beliefs that a product extracted from plants cannot cause adverse effects on health [32], [45].
As part of the genotoxic approach, the Ames test is recommended for assessing the ability of a substance to provoke a point mutation in the DNA of Salmonella Typhimurium strains, therefore enabling the resulting events to be counted visually [15]. This methodology is useful to assess the mutagenicity potential of a wide variety of chemicals, since genetically bioengineered strains are sensitive to very specific base pair or frameshift mutations [33], [46]. However, some substances, referred to as pro-mutagens, do not pose a direct mutagenic risk to organisms, and depend on biotransformation (such as metabolic enzymes from the P450 cytochrome family) to attack the DNA structure, as well as other macromolecules (e.g., lipids) [47]. To overcome this obstacle, an exogenous metabolic system was proposed for use in the assay, since bacteria do not possess this endogenous machinery [48]. In this work, we applied the Salmonella/microsome assay in the absence and presence of metabolic activation using the S9 mix. Only FMN induced an increase in mutagenicity index for TA1535 (10 µM/plate) after metabolic activation, even though no statistical relevance was found for this event (p > 0.05). However, we observed a cytotoxic effect on the strains TA97 and TA100 promoted by FMN, starting at 0.1 µM/plate for TA97 and 1.0 µM/plate for TA100. This outcome may have been a response of cellular units facing accumulative mutagenic events leading to cell death. TA100 is specifically susceptible to base pair substitution inducers, whereas TA97 detects mutagens that can alter the reading frame of DNA, and both strains were designed to be responsive to various known mutagens [33]. In this specific case, it is possible that FMN was chemically transformed into a more reactive intermediate by the CYP enzymes (mainly CYP 1 A) present in the S9 mix, therefore becoming mutagenic and hence cytotoxic for these strains. As previously reported by Bartholomew and Ryan [49], eight other isoflavones, including FMN, were found to be not mutagenic to strains TA1538, TA98 and TA100 in the Salmonella/microsome evaluation. Even more, phytoestrogen isoflavones tested in the presence and in absence of S9 mix were not able to induce cytotoxicity to the bacterial cells, in comparison to 2-anthramine (in the presence of exogenous metabolization) and quercetin (either presence or absence of exogenous metabolization) which are also naturally occurring estrogen-like substances [49]. The discussed mutagenic mechanism proposed for isoflavones, such as quercetin, rely on the chemical structure of the substance where a free hydroxyl group is available in C-3 position, therefore capable of inducing nucleophilic attack to the DNA structure [50]. This behaviour could be related to the increase in revertant colonies (M.I. = 2.3) in TA1535 that we observed for FMN, after metabolic activation.
As recommended by OECD, the assessment of genotoxicity must comprise a combination of alternative bacterial and mammalian in vitro approaches to be considered consistent [16]. Thus, we applied the CBMN-test in HepG2 cells to investigate the occurrence of morphological events in the nuclei related to clastogenic or aneugenic features, when facing exposure to FMN or ARB [38]. Using the CBMN methodology, mammalian cells can be examined regarding alterations in their DNA (via whole chromosome loss or smaller breakages) that correspond to dividing cells when in the presence of genotoxins or under a scarcity of nutrients (e.g., folic acid) [51]. Besides MNi, the CBMN also provides results from particular cellular events during cell division: the extrusion of excessive amplificated DNA through small nuclear buds (NBUDs) [52]; visualization of nucleoplasmic bridges (NPBs), formed when centromeres from dicentric chromosomes are pulled to opposite sides during the anaphase, which accumulate since cytokinesis is inhibited by cytochalasin B [39]. Finally, this method allows for the observation of cell death (apoptosis or necrosis), and the calculation of the nuclear division index (NDI), which is related to cell proliferation, and it suggests whether, for instance, a chemical substance is preventing cells from entering the cell cycle or being an enhancer of this event [53].
Our findings using CBMN methodology showed that FMN and ARB do not induce micronuclei formation in binucleated cells after 24 h of exposure at any of the concentrations tested for either of the compounds (Fig. 6, a, b). Not only were no NBUDs or NPBs formed (Fig. 6, c, d), but dead cells were not detected, significantly different from the untreated cells (data not shown). Altogether, the results of the CBMN-assay with HepG2 cells suggest that both compounds are safe in a genotoxic context with metabolically competent eukaryotic cells, since no losses or chromosome breaks from dividing cells were perceived. Hazman and colleagues [54], reported previously that β-arbutin (in comparison to α-arbutin) is capable of inducing DNA breaks and increase micronuclei formation in treatment-resistant breast cancer cell lineage in vitro (MCF-7). HepG2, a transformed liver cancer lineage, is capable of endogenously expressing phase I and II metabolic enzymes, and this characteristic is applicable to various assays aiming to unveil indirect and direct mutagenic substances [55], [56]. Benzo[a]pyrene, the mutagen chosen for this test, is a well-known polycyclic aromatic hydrocarbon that is considered a promutagen [57]. Thus, the mutagenic potential of this substance was perceived by the metabolically competent cell model used.
Contrary to our results, other reports indicate that FMN is involved in molecular effects related to the cell cycle, causing overall disturbance and death in cancer cells. Previous studies have reported that FMN promoted cell apoptosis in human colon carcinoma cells (HCT) after 48 h of exposure with an EC50 of 50 µM [58]. Cell cycle arrest was observed at G0/G1 in MCF-7 breast cancer cells when treated with FMN, besides inhibition of cell proliferation and the growth of tumours in nude mouse xenografts [8]. The findings of Gyémant et al. [59] revealed that FMN could act synergistically with epirubicin and enhance an antiproliferative effect on multidrug-resistant cell models, such as MDA-MB-231 human breast cancer cells. FMN has gained a significant relevance in the field of cancer therapy research and disease prevention based on chemical entities found in Nature [60]. Pre-clinical studies reported in vitro cytotoxicity evaluated in many cell lineages, including breast cancer (MCF-7 and MDA-MB-231), ovarian cancer (OV90, ES2), prostate cells (PC3, DU145), colon cancer (HCT116) [60]. The anti-tumorigenic potential of FMN is attributed mainly to its estrogenic activity, which allows the interaction with nuclear mediators (such as pro- and anti-apoptotic proteins) to regulate endogenous signalling pathways (e.g. p38, MAPK, PI3K/Akt, STAT3, EFGR) involved in important biological processes in cancer, like cell death and division [61]. Antitumor in vitro activity of FMN has been described in a range from 10 to 300 µM in inhibition for 50 % (IC50) of tested cancer cell lineages [60]. FMN shows prominent results on antitumor therapy strategies [62]. However, as many other benzene-derived phenolic compounds from plants, it has limited solubility in water, which results in poor rates of absorption, low bioavailability, being rapidly cleared from the organism [61]. To overcome this fact and ensure pharmacological activity, researches worldwide have been dedicated to produce nanoparticle-based (NPs) systems with FMN [62]. Drug delivery systems loaded with FMN are being designed based upon a plethora of carriers, including polymeric forms combined with dextrose (e.g. poly [lactic-co-glycolic acid] PLGA-cyclodextrin complex); single-walled carbon nanotubes conjugated with dextrose; D-α-tocopherol polyethylene glycol 1000 succinate (TPGS) micelles of FMN [62]. Pre-clinical studies in animals of a water-soluble derivatives of FMN (e.g. sodium-formononetin-3’-sulfonate, Sul-F, and 7-O-phosphate formononetin) evaluated acute toxicity in rats and dog models. Results indicate that these compounds should be further studied owing their toxicity, but provided safe concentrations to be applied for this purpose [60], [61]. In comparison to other isoflavones, FMN is known to possess very concise biologic effect in terms of chemoprevention and exhibits less toxicity. Not only, FMN is also an anti-inflammatory and microbial agent, among other soybean-derived compounds [63].
Formononetin belongs to the isoflavonoid group, and this class is composed mainly of secondary metabolites that act through estrogenic-like responses [5]. For this reason, isoflavonoids are referred to as phytoestrogens and are known in the fields of nutrition, cosmetology, and medicine, which recommend the ingestion of flavonoid-rich food for fighting metabolic syndrome symptoms such as those of the menopause in women [64]. It is found in many species of the Fabaceae family, such as red propolis (Dalbergia ecastophyllum), red clover (Trifolium pratense L.) [58], and Astragalus membranaceus (Fisch) [65]. FMN is not present directly in the human diet, but it is known that the isoflavones from soybeans are metabolized by the gut microflora and CYP enzymes into estrogenic intermediates such as genistein [5]. Genistein is one of the principal metabolites from soybeans, along with daidzein, and was reported to induce apoptosis in HepG2 and Hep3B hepatic tumour cells via endoplasmic reticulum stress and mitochondrial injury [66], [67].
Aligned to anticancer therapies using phenolic compounds, the antimutagenicity assay is a reliable means for assessing substances capable of reverting or blocking DNA damage caused by mutagens [68], [69]. The test is employed as a risk-benefit assessment of substances that could be considered to integrate chemo preventive therapies. Diseases related to mutation propagation (e.g., chronic degenerative heart diseases) can be prevented by either diminishing exposure to the carcinogenic agents or by exploiting the self-defence mechanisms enhanced by substances that improve these tools [70], [71]. Mutagenic agents possess different modes of action to pose injuries to DNA, and are classified according to the assigned specific mechanism [68]. Alkylating agents, such as methyl methane sulfonate (MMS), act by transferring an alkyl group to purines and pyrimidines forming adducts, which, in turn, produce double-strand breaks (DSB) and consequently mispairing of sequences [72]. Guanine is a frequent location for the formation of adducts, and MMS damage produces O6-methylguanine, involved in the mispairing of purines [35]. The compound 4-nitroquinoline 1-oxide (4-NQO) is known to cause both frameshift and base pair mutations in a direct manner, and is classified as an oxidative mutagen using nucleophilic reactions [72]. In the present study, we assayed the TA98, TA100 and TA1535 strains of the Salmonella enterica serovar Typhimurium, using pre-, post- and co- incubation models without metabolic activation. The strain TA98 detects frameshift alterations in GC hotspots of the hisD3052 region, whilst the strains TA1535 and its correspondent TA100 are sensitive to base pair substitutions (transition from G:C to A:T) in the hisG46 operon location. The TA98 and TA100 strains have the addition of a pKM101 plasmid which enhances induced and spontaneous mutations through an error-prone DNA repair system. This plasmid is absent in TA1535, making the derivative strain TA100 more sensitive to mutagens detected by TA1535 [33]. Thus, the use of this set of strains to assess FMN (TA98, TA100 and TA1535) and ARB (TA100 and TA1535) is feasible, to observe a variety of specific mutations caused by different modes of action. The results indicated antimutagenic effects in all three exposure models proposed in this work. For TA98, FMN showed a 60 % reduction in induced mutations (Fig. 1). Such a response may be classified as a blocking activity of FMN against 4-NQO, preventing the cells from oxidative damage by inactivating this direct electrophilic mutagen outside the cellular space [72]. Antimutagens that pose this effect are named desmutagens, since they protect cells from mutational damage before it occurs, as it was represented in the co-incubation exposure [69]. When facing MMS exposure, FMN also featured better reduction results with co-incubation, both for TA100 and TA1535, reaching an almost complete reduction (98 %) with a concentration of 0.1 µM in TA1535 (Fig. 2, Fig. 3). Similar to the previous strain, the effect exerted by FMN in the co-incubation model suggests blocking of the action of mutagens extracellularly via chemical interactions between the sample and the mutagen. As in the case of the results with ARB, a promising antimutagenic profile was presented. This compound markedly reduced over 40 % of the alkyl transfer mediated by MMS in TA100, and even higher values appeared for TA1535 (Fig. 4, Fig. 5). With this bacterial strain, ARB was antimutagenic in all three sets of exposure testing and reached a complete reduction of mutagenicity in cells already attacked by MMS. This response was attributed to substances named bioantimutagens, which interfere in the fixation of a mutation already implied to the cell (intracellular), altering the steps in the DNA replication and repair system triggered to stop the mutation from being established [72], [73]. Bioantimutagenic agents are distinguished according to whether they are found in pre-treatment exposure (when the test sample already penetrated bacterial cells) or post-incubation exposure (where the antimutagen removes damage about to be fixed [74]. Therefore, applying different modes of exposure in the antimutagenicity evaluation is a feasible tool to clarify the mechanistic involved in the protection responses of DNA facing mutagens [75], [76].
Antioxidant substances from natural compounds have been reported as potent antimutagens and are found in many vegetable species [77]. Several antioxidant mechanisms can be exerted by these molecules, resulting in interactions with various free radical species and preventing injuries to macromolecules from happening. Eliminating these reactive intermediates (such as peroxyl radicals and O2— ions) is extensively applied as adjuvant therapy for various disease outcomes [71], [77]. Besides the inactivation of free radicals, either through hydrogen atom transfer or by scavenging an electron from the oxidant substance, antioxidants also play an indirect action when related to the redox balance mediated by endogenous enzymes [75], [77].
Arbutin exhibits well-known antioxidant properties assigned to its hydroquinone skeleton. It has been proposed that this substance acts as a direct scavenger of hydroxyl radicals and other free forms of reactive species, thus neutralizing their oxidative effect. Furthermore, ARB has been described as interacting with mediators from self-antioxidant defence mechanisms to promote an indirect antioxidant effect, a role it plays in melanin-related diseases. The synthesis of melanin mediated by tyrosine forms reactive oxygen species (ROS), and the accumulation of these intermediates is related to the death of melanocytes and to hyper or hypopigmentation pathogenesis [78]. Nrf2 (nuclear factor erythroid 2-related factor 2) illustrates this since it binds to antioxidant-responsive elements (ARE, such as ARB) on promoter regions, leading to the expression of self-antioxidant molecules, such as glutathione. When stimulated by polyphenols from the diet, the Nrf2-ARE pathway also suppresses melanogenesis induced by UVR light [11]. ARB is highly soluble in water and is commonly conjugated to glycosides to enhance its resistance to oxidation and light degradation [12]. These glycosylated forms of ARB – be they the alpha or beta isoforms - are often found in nature, such as in the Rosaceae family (Pyrus pyrifolia, an Asian pear), or blueberries (Vaccinium sp.) amongst many other foods [7]. In summary, our observations of the antimutagenic properties of ARB as shown in the antimutagenicity assays, reveal that this widely produced natural compound is likely to present a multi-target antimutagen effect, indicating that further investigations of this molecule are of interest, above all with respect to the pathogenesis of mutation-related diseases.
5. Conclusions
The present study assessed the toxicogenetic aspects of formononetin (FMN) and arbutin (ARB), the metabolites found in C. glutiniferum Raddi, a traditional orchid used in Brazilian folk medicine. Mutagenicity assays suggest that both FMN and ARB are considered safe from the risk of inducing point mutations. Also, FMN and ARB did not provoke structural damage to chromosomes in the in vitro CBMN test. Finally, the data from antimutagenicity assays presented remarkable endpoints with respect to protecting DNA from chemically induced alkylation. To our knowledge, this the first report discussing pre-clinical aspects of FMN and ARB through the landscape of a standardized toxicogenetic assessment, encompassing a DNA protection efficacy. Seen from this point of view, here we reported evidence-based prospects of possible applications of FMN and ARB in a variety of therapy strategies to confront diseases related to the occurrence and perpetuation of mutations. The results suggest that FMN and ARB play multi-targeted antimutagenic roles in protecting DNA from mutagens, blocking the alkylation action of MMS through chemical interactions before the DNA is injured, and reverting any mutation about to occur. Therefore, our future perspectives are that further investigations should mind on novel strategies that can challenge such substances in different contexts of diseases, since our screening describes efficacy of FMN and ARB against very common DNA damage agents (e.g. reactive oxygen species inducers). It is imperative to consider and make use of innovative alternatives to delineate more green-conscious tools for the discovery of drugs. In conclusion, our findings show that FMN and ARB should be considered for subsequent investigations aimed at chemo preventive approaches and therapy strategies for diseases related to genomic instability and mutation, like cancer.
Funding
This work was supported by the Coordination for Superior Level Staff Improvement (CAPES) [Code 0001]; by the Carlos Chagas Filho Foundation for Research Support of the State of Rio de Janeiro (FAPERJ) [E-26/202.759/2017, E-26/200839/2021, E-26/210.096/2023, E-26/010.002119/2019, E-26/010.100956/2018, E26/202.256/2018]; by the National Council for Scientific and Technological Development (CNPq) [302345/2017–5, 305600/2021–4].
CRediT authorship contribution statement
Israel Felzenszwalb: Writing – review & editing, Visualization, Validation, Supervision, Project administration, Funding acquisition. Carlos Araujo Lima: Writing – review & editing, Validation, Supervision, Project administration, Funding acquisition, Conceptualization. Barbara Verena Dias Galvão: Methodology, Investigation, Formal analysis. Lizandra Vitória de Souza Santos: Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Conceptualization. Andreia da Silva Fernandes: Writing – review & editing, Validation, Supervision, Resources, Funding acquisition, Conceptualization. Lays Souza: Methodology, Investigation, Data curation.
Declaration of Competing Interest
The authors report no competing interests.
Contributor Information
Lizandra Vitoria de Souza Santos, Email: lizabiomed@gmail.com.
Barbara Verena Dias Galvão, Email: barbaradiasbiomed@gmail.com.
Lays Souza, Email: souza.lays@hotmail.com.
Andreia da Silva Fernandes, Email: andreiadasilvaf@gmail.com.
Carlos Fernando Araujo-Lima, Email: biomed.carlos@gmail.com.
Israel Felzenszwalb, Email: uerj.felzen@gmail.com.
Data availability
Data will be made available on request.
References
- 1.Romero-González G.A., Batista J.A.N., de Bem Bianchetti L. A synopsis of the genus Cyrtopodium (Catasetinae: Orchidaceae) Harv. Pap. Bot. 2008;13(1):189–206. doi: 10.3100/1043-4534(2008)13[189:ASOTGC]2.0.CO;2. [DOI] [Google Scholar]
- 2.Araujo-Lima C.F., Felzenszwalb I., Macedo A.F. In: Orchids Phytochemistry, Biology and Horticulture. Reference Series in Phytochemistry. Merillon J.M., Kodja H., editors. Springer; Cham: 2021. Cyrtopodium glutiniferum, an example of orchid used in folk medicine: phytochemical and biological aspects; pp. 1–16. (place unknown) (place unknown) [DOI] [Google Scholar]
- 3.Araujo-Lima C.F., Oliveira J.P.S., Coscarella I.L., Aiub C.A.F., Felzenszwalb I., Evaristo G.P.C., Macedo A.F. Metabolomic analysis of Cyrtopodium glutiniferum extract by UHPLC-MS/MS and in vitro antiproliferative and genotoxicity assessment. J. Ethnophamacol. 2020;253(10):10 p. doi: 10.1016/j.jep.2020.112607. [DOI] [PubMed] [Google Scholar]
- 4.Tóth B., Hohmann J., Vasas A. Phenanthrenes: a promising group of plant secondary metabolites. J. Nat. Prod. 2016;81:661–678. doi: 10.1021/acs.jnatprod.7b00619. [DOI] [PubMed] [Google Scholar]
- 5.Heinonen S.M., Wähälä K., Adlercreutz H. Identification of urinary metabolites of the red clover isoflavones formononetin and biochanin A in human subjects. J. Agric. Food Chem. 2004;52:6802–6809. doi: 10.1021/jf0492767. [DOI] [PubMed] [Google Scholar]
- 6.Khan F.A., Maalik A., Murtaza G. Inhibitory mechanism against oxidative stress of caffeic acid. J. Food Drug. Anal. 2016;24(4):695–702. doi: 10.1016/j.jfda.2016.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nahar L., Al-Groshi A., Kumar A., Sarker S.D. Arbutin: Occurrence in Plants, and Its Potential as an Anticancer Agent. Molecules. 2022;27(24):22 p. doi: 10.3390/molecules27248786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen J., Zeng J., Xin M., Huang W., Chen X. Formononetin induces cell cycle arrest of human breast cancer cells via IGF1/PI3K/Akt pathways in vitro and in vivo. Horm. Metab. Res. 2011;43:681–686. doi: 10.1055/s-0031-1286306. [DOI] [PubMed] [Google Scholar]
- 9.Brautigan D.L., Gielata M., Heo J., Kubicka E., Wilkins L.R. Selective toxicity of caffeic acid in hepatocellular carcinoma cells. Biochem. Biophis. Res. Commun. 2018;505(2):612–617. doi: 10.1016/j.bbrc.2018.09.155. [DOI] [PubMed] [Google Scholar]
- 10.Choi H.G., Tran P.T., Lee J.H., Min B.S., Kim J.A. Anti-inflammatory activity of caffeic acid derivatives isolated from the roots of Salvia miltiorrhiza Bunge. Arch. Pharm. Res. 2018;41:64–70. doi: 10.1007/s12272-017-0983-1. [DOI] [PubMed] [Google Scholar]
- 11.Boo Y.C. Arbutin as a skin depigmenting agent with antimelanogenic and antioxidant properties. Antioxidants. 2021;10(7):22. doi: 10.3390/antiox10071129. https://doi.org/0.3390/antiox10071129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saeedi M., Khezri K., Zakaryaei A.S., Mohammadamini H. A comprehensive review of the therapeutic potential of α-arbutin. Phytother. Res. 2021:1–19. doi: 10.1002/ptr.7076. [DOI] [PubMed] [Google Scholar]
- 13.Vanhaecke T., Snykers S., Rogiers V., Garthoff B., Castell J.V., Hengstler J.G. EU research activities in alternative testing strategies: current status and future perspectives. Arch. Toxicol. 2009;83:1037–1042. doi: 10.1007/s00204-009-0484-1. [DOI] [PubMed] [Google Scholar]
- 14.Carrão-Dantas E.K., Araujo-Lima C.F., Ferreira C.L.S., Goldstein A.C., Aiub C.A.F., Felzenszwalb I. Toxicogenetic assessment of a pre-workout supplement: In vitro mutagenicity, cytotoxicity, genotoxicity and glutathione determination in liver cell lines and in silico ADMET approaches. Mut. Res. Gen. Toxicol. Environ. Mutagen. 2022:11 p. doi: 10.1016/j.mrgentox.2022.503517. [DOI] [PubMed] [Google Scholar]
- 15.OECD] Organization for Economic Co-operation and Development. 2020. Test no. 471 Bacterial reverse mutation test. OECD guidelines for testing of chemicals, Section 4. Paris: OECD Publishing. 10.1787/9789264071247-en. [DOI]
- 16.OECD] Organization for Economic Co-operation and Development. 2023. Test no. 487 In Vitro Mammalian Cell Micronucleus Test. OECD guidelines for testing of chemicals, Section 4. Paris: OECD Publishing. https://doi.org/10.1787/9789264264861-en.
- 17.Kirkland D.J., Henderson L., Marizn D., Muller L., Parry J.M., Speit G., Tweats D.J., Williams G.M. Testing strategies in mutagenicity and genetic toxicology: An appraisal of the guidelines of the European Scientific Committee for Cosmetics and Non-Food Products for the evaluation of hair dyes. Mut. Res. 2005;588:88–105. doi: 10.1016/j.mrgentox.2005.09.006. [DOI] [PubMed] [Google Scholar]
- 18.Groff K., Evans S.J., Doak S.H., Pfuhler S., Corvi R., Saunders S., Stoddart G. In vitro and integrated in vivo strategies to reduce animal use in genotoxicity testing. Mutagenesis. 2021;36:389–400. doi: 10.1093/mutage/geab035. [DOI] [PubMed] [Google Scholar]
- 19.Hubrecht R.C., Carter E. The 3Rs and human experimental technique: implementing change. Animals. 2023;9:754. doi: 10.3390/ani9100754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hsieh C., Giannakoulias S., Petersson E.J., Mach R.H. Computational chemistry for the identification of lead compounds for radiotracer development. Pharmaceuticals. 2023;16:317. doi: 10.3390/ph16020317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Araujo N.G.R., Araujo-Lima C.F., Oliveira R.T., Macedo A.F., Felzenszwalb I. In vitro cytotoxicity and genotoxicity assessment of methanolic extracts of vanillas from Brazilian biodiversity with commercial potential. Tox. Rep. 2024;13 doi: 10.1016/j.toxrep.2024.101693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Atanasov, A., Zotchev, S.B., Dirsch, V.M., the International Natural Product Sciences Taskforce and Supuran, C. T. 2021. Natural products in drug discovery: advances and opportunities. Nat. Rev.20: 200-216. 10.1038/s41573-020-0014-2. [DOI] [PMC free article] [PubMed]
- 23.Muratov E.N., Bajorath J., Sherida R.P., Tetko I.V., Filimonov D., Poroikov V., Oprea T.I., Baskin I.I., Varnek A., Roitberg A., Isayev O., Curtalolo S., Fourches D., Cohen Y., Aspuru-Guzik A., Winkler D.A., Agrafiotis D., Cherkasov Y., Topsha A. QSAR without borders. Chem. Soc. Rev. 2020;49:3525. doi: 10.1039/d0cs00098a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim S., Lim S.W., Choi J. Drug discovery inspired by bioactive small molecules from nature. Anim. Cell. Sys. 2022;(6):254–265. doi: 10.1080/19768354.2022.2157480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oliveira R.T., Oliveira J.P.S., Silva A.L.M., Dantas E.K.C., Koblitz M.G.B., Bello M.L., Felzenszwalb I., Araújo-Lima C.F., Macedo A.F. Vanilla from Atlantic Forest: In vitro and in silico toxicity assessment and high-resolution metabolomic analysis of Vanilla spp. Ethanolic extracts. Food Chem. 2024 doi: 10.1016/j.foodchem.2024.139948. [DOI] [PubMed] [Google Scholar]
- 26.Dantas E.K.C., Ferreira C.L.S., Goldstein A.C., Fernandes A.S., Ferraz E.R.A., Felzenszwalb I., Araújo-Lima C.F. Marketable 1,3-dimethylamylamine and caffeine-based thermogenic supplements: Regulatory genotoxicity assessment through in vitro and in silico approaches. J. Tox. Environ. Health Part A. 2024;87(6):245–265. doi: 10.1080/15287394.2023.2294925. [DOI] [PubMed] [Google Scholar]
- 27.Ajabshir Z.Z., Ajabshir S.Z. Preparation and characterization of curcumin niosomal nanoparticles via a simple and eco-friendly route. J. Nanostruct. 2019;9(4):784–790. doi: 10.22052/JNS.2019.04.020. [DOI] [Google Scholar]
- 28.Yadav S.J., Thakor S., Raval A.V., Sharma P., Shah D., Rana V.A. Synthesis, characterization, and unveiling dielectric spectroscopy insights of Fe3Se4 and Fe7Se8 nanoparticles. J. Mater. Sci. Mater. Electron. 2024;35:1323. doi: 10.1007/s10854-024-13035-z. [DOI] [Google Scholar]
- 29.Qazvini N.T., Zinatloo S. Synthesis and characterization of gelatin nanoparticles using CDI/NHS as a non-toxic cross-linking system. J. Mater. Sci. Mater. Med. 2011;22:63–69. doi: 10.1007/s10856-010-4178-2. https://doi.or/10.1007/s10856-010-4178-2. [DOI] [PubMed] [Google Scholar]
- 30.Ajabshir-Zinatloo S., Mahmoudi-Moghaddam H., Amiri M., Javar H.A. A green Route for the sysnthesis of sponge-like Pr6O11 nanoparticles and their application for the development of chlorambucil sensor. Measurement. 2024;235 doi: 10.1016/j.measurement.2024.114924. [DOI] [Google Scholar]
- 31.Cardoso R.V., Pereira P.R., Freitas C.S., Paschoalin V.M.F. Trends in drug delivery systems for natural bioactive molecules to treat health disorders: the importance of nano-liposomes. Pharmaceutics. 2024;14:2808. doi: 10.3390/pharmaceutics14122808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sponchiado G., Adam M.L., Silva C.D., Soley B.S., Mello-Sampaio C., Cabrini D.A., Correr C.J., Otuki M.F. Quantitative genotoxicity assays for analysis of medicinal plants: A systematic review. J. Ethnopharmacol. 2016;178:289–296. doi: 10.1016/j.jep.2015.10.026. [DOI] [PubMed] [Google Scholar]
- 33.Maron D.M., Ames B.N. Revised methods for the Salmonella mutagenicity test. Mut. Res. Gen. Toxicol. Environ. Mutagen. 1983;113(3-4):173–215. doi: 10.1016/0165-1161(83)90010-9. https://doi.org/0.1016/0165-1161(83)90010-9. [DOI] [PubMed] [Google Scholar]
- 34.Goldstein A.C., Araujo-Lima C.F., Fernandes A.S., Santos-Oliveira R., Felzenszwalb I. In vitro genotoxicity assessment of graphene quantum dots nanoparticles: A metabolism-dependent response. Mut. Res. Gen. Toxicol. Environ. Mutagen. 2023;885:8. doi: 10.1016/j.mrgentox.2022.503563. https://doi.org/0.1016/j.mrgentox.2022.503563. [DOI] [PubMed] [Google Scholar]
- 35.Araujo-Lima C.F., Christoni L.S.A., Justo G., Soeiro M.N.C., Aiub C.A.F., Felzenszwalb I. Atorvastatin downregulates in vitro methyl methanesulfonate and cyclophosphamide alkylation-mediated cellular and DNA injuries. Oxid. Med. Cell. 2018;2018:11. doi: 10.1155/2018/7820890. https://doi.org/0.1155/2018/7820890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Scharf R.M., Gonçalves C.O., Fernandes A.S., Mazzei J.L., Ferraz E.R.A., Lima C.F.A., Felzenszwalb I. Antimutagenic and antitumor activities of a water soluble fraction of soursop (syn Graviola, Annona muricata L.) fruit pulp. J. Tox. Environ. Health Part A. 2024 doi: 10.1080/15287394.2024.2309335. [DOI] [PubMed] [Google Scholar]
- 37.Chan L.L.Y., Rice W.L., Qiu J. Observation and quantification of the morphological effect of trypan blue rupturing dead or dying cells. PLoS ONE. 2020;15(1):17. doi: 10.1371/journal.pone.0227950. p. https://do.org/10.1371/journal.pone.0227950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fenech M. Cytokinesis-block micronucleus assay evolves into a "cytome" assay of chromosomal instability, mitotic dysfunction and cell death. Mut. Res. 2006;600:58–66. doi: 10.1016/j.mrfmmm.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 39.Fenech M. Cytokinesis-block micronucleus cytome assay. Nat. Protoc. 2007;2(5):1084–1104. doi: 10.1038/nprot.2007.77. [DOI] [PubMed] [Google Scholar]
- 40.Stiller A., Garrison K., Gurdyumov K., Kenner J., Yasmin F., Yates P., Song B.H. From fighting critters to saving lives: Polyphenols in plant defense and human health. Int. J. Mol. Sci. 2021;22(16):21 p. doi: 10.3390/ijms22168995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cragg G.M., Newman D.J. Natural products: A continuing source of novel drug leads. Bihophis. Bhiochem. Act. 2013;1830(6):3670–3695. doi: 10.1016/j.bbagen.2013.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Baltz R.H. Natural product drug discovery in the genomic era: realities, conjectures, misconceptions, and opportunities. J. Ind. Microbiol. Biotech. 2019;46:281–299. doi: 10.1007/s10295-018-2115-4. [DOI] [PubMed] [Google Scholar]
- 43.Alara O.R., Abdurahman N.H., Ukaegbu C.I. Extraction of phenolic compounds: A review. Curr. Res. Food Sci. 2021;4:200–214. doi: 10.1016/J.CRFS.2021.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pirintsos S., Panagiotopoulos A., Bariotakis M., Daskalakis V., Lionis C., Sourvinos G., Karakasiliotis I., Kampa M., Castanas E. From Traditional Ethnopharmacology to Modern Natural Drug Discovery: A Methodology Discussion and Specific Examples. Molecules. 2022;27(13):18 p. doi: 10.3390/molecules27134060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Petroski W., Minich D.M. Is there such a thing as “anti-nutrients”? A narrative review of perceived problematic plant compounds. Nutrients. 2020;12(10):32 p. doi: 10.3390/nu12102929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sugiyama K., Yamada M., Awogi T., Hakura A. The strains recommended for use in the bacterial reverse mutation test (OECD guideline 471) can be certified as non-genetically modified organisms. Gen. Environ. 2016;38(2):3 p. doi: 10.1186/s41021-016-0030-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mccann J., Spingarn N.E., Kobori J., Ames B.N. Detection of Carcinogens as Mutagens: Bacterial Tester Strains with R Factor Plasmids (error-prone recombinational repair/7,12-dimethylbenzanthrecene/aflatoxin/nitrofurans) Proc. Nat. Acd. Sci. 1975;72(3):979–983. doi: 10.1073/pnas.72.3.979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mortelmans K., Zeiger E. The Ames Salmonella/microsome mutagenicity assay. Mut. Res. 2000;455:29–60. doi: 10.1016/s0027-5107(00)00064-6. (https:/) [DOI] [PubMed] [Google Scholar]
- 49.Bartholomew R.M., Ryan D.S. Lack of mutagenicity of some phytoestrogens in the Salmonella/mammalian microsome assay. Mut. Res. 1980;78:317–321. doi: 10.1016/0165-1218(80)90036-1. [DOI] [PubMed] [Google Scholar]
- 50.MacGregor J.T., Jurd L. Mutagenicity of plant flavonoids: structural requirements for mutagenic activity in Salmonella typhimurium. Mut. Res. 1978;54:297–309. doi: 10.1016/0165-1161(78)90020-1. [DOI] [PubMed] [Google Scholar]
- 51.Fenech M., Ferguson L.R. Vitamins/minerals and genomic instability in humans. Mut. Res. 2001;475:1–6. doi: 10.1016/S0027-5107(01)00069-0. [DOI] [PubMed] [Google Scholar]
- 52.Shimizu N., Itoh N., Utiyama H., Wahl G.M. Selective Entrapment of Extrachromosomally Amplified DNA by Nuclear Budding and Micronucleation during S Phase. J. Cell Biol. 1998;140(6):1307–1320. doi: 10.1083/jcb.140.6.1307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Fenech M. The advantages and disadvantages of the cytokinesis-block micronucleus method. Mut. Res. 1997;392:11–18. doi: 10.1016/s0165-1218(97)00041-4. [DOI] [PubMed] [Google Scholar]
- 54.Hazman O., Sariova A., Bozkurt M.F., Cigerci I.H. The anticarcinogen activity of [b]-arbutin on MCF-7 cells: Stimulation of apoptosis though estrogen receptor-[a] signal pathway, inflammation and genotoxicity. Mol. Cell. Biochem. 2021;476:349–360. doi: 10.1007/s11010-020-03911-7. [DOI] [PubMed] [Google Scholar]
- 55.Gerets H.H.J., Tilmant K., Gerin B., Chanteux H., Depelchin B.O., Dhalluin S., Atienzar F.A. Characterization of primary human hepatocytes, HepG2 cells, and HepaRG cells at the mRNA level and CYP activity in response to inducers and their predictivity for the detection of human hepatotoxins. Cell. Biol. Toxicol. 2012;28:69–87. doi: 10.1007/s10565-011-9208-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Choi J.M., Oh S.J., Lee S.Y., Im J.H., Oh J.M., Ryu C.S., Kwak H.C., Lee J.Y., Kang K.W., Kim S.K. HepG2 cells as an in vitro model for evaluation of cytochrome P450 induction by xenobiotics. Arc. Pharm. Res. 2015;38:691–704. doi: 10.1007/s12272-014-0502-6. [DOI] [PubMed] [Google Scholar]
- 57.Valentin-Severin I., Hegarat L.L., Lhuguenot J.C., le Bon A.M., Chagnon M.C. Use of HepG2 cell line for direct or indirect mutagens screening: comparative investigation between comet and micronucleus assays. Mut. Res. 2003;536:79–90. doi: 10.1016/S1383-5718(03)00031-7. [DOI] [PubMed] [Google Scholar]
- 58.Auyeung K.K.W., Ko J.K.S. Novel herbal flavonoids promote apoptosis but differentially induce cell cycle arrest in human colon cancer cell. Invest. New. Drugs. 2010;28:1–13. doi: 10.1007/s10637-008-9207-3. [DOI] [PubMed] [Google Scholar]
- 59.Gyemánt N., Tanaka M., Antus S., Hohmann J., Csuka O., Mandoky L., Molnar J. In Vitro Search for Synergy Between Flavonoids and Epirubicin on Multidrug-resistant Cancer. Cells. 2005;19:367–374. [PubMed] [Google Scholar]
- 60.Aliya S., Alhammadi M., Park U., Tiwari J.N., Lee J., Han Y., Huh Y.S. The potential role of formononetin in cancer treatment: An updated review. Biomed. Pharmacother. 2024;168 doi: 10.1016/j.biopha.2023.115811. [DOI] [PubMed] [Google Scholar]
- 61.Almatroodi S.A., Almatroodi A., Khan A.A., Rahmani A.H. 2024. Potential therapeutic targets of formononetin, a type of methoxylated isoflavone, and its role in cancer therapy through the modulation of signal transduction pathways. Int. J. Mol. Sci. 2023;24(9719)) doi: 10.3390/ijmas24119719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tat K., Tan L.T., Chan C.K., Hong S.L., Chan K., Yap W.S., Pusparajah P., Lee L., Goh B. Formononetin: a review of its anticancer potentials and mechanisms. Front. Pharmacol. 2019;10:820. doi: 10.3389/fphar.2019.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jiang D., Rasul A., Batool R., Sarfraz I., Hussain G., Tahir M.M., Qin T., Selamoglu Z., Ali M., Li J., Li X. Potential anticancer properties and mechanisms of action of formononetin. BioMed. Res Int. 2019;2019(5854315) doi: 10.1155/2019/5854315. 11 p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Machado-Dutra J., Espitia P.J.P., Batista R.A. Formononetin: Biological effects and uses – A review. Food Chem. 2021;359:10 p. doi: 10.1016/j.foodchem.2021.129975. [DOI] [PubMed] [Google Scholar]
- 65.Tay K.C., Tan L.T.H., Chan C.K., Hong S.L., Chan K.G., Yap W.H., Pusparajah P., Lee L.H., Goh B.H. Formononetin: A Review of Its Anticancer Potentials and Mechanisms. Front. Pharmacol. 2019;10:19 p. doi: 10.3389/fphar.2019.00820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yeh T.C., Chiang P.C., Li T.K., Hsu J.L., Lin C.J., Wang S.W., Peng C.Y., Gu J.H. Genistein induces apoptosis in human hepatocellular carcinomas via interaction of endoplasmic reticulum stress and mitochondrial insult. Biochem. Pharmacol. 2007;73(6):782–792. doi: 10.1016/j.bcp.2006.11.027. [DOI] [PubMed] [Google Scholar]
- 67.Fang S.C., Hsu C.L., Lin H.T., Yen C.G. Anticancer effects of flavonoid derivatives isolated from Millettia reticulata benth in sk-hep-1 human hepatocellular carcinoma cells. J. Agric. Food Chem. 2010;58:817–820. doi: 10.1021/jf903216r. [DOI] [PubMed] [Google Scholar]
- 68.De Flora S., Camoirano A., D’Agostini F., Balansky R. Modulation of the mutagenic response in prokaryotes. Mut. Res. 1992;267:183–192. doi: 10.1016/0027-5107(92)90062-. [DOI] [PubMed] [Google Scholar]
- 69.De Flora S. Mechanisms of inhibitors of mutagenesis and carcinogenesis. Mut. Res. 1998;402:151–158. doi: 10.1016/S0027-5107(97)00292-3. [DOI] [PubMed] [Google Scholar]
- 70.De Flora S., Izzoti A., D’Agostini F., Balansky R.M., Noonan D., Albini A. Multiple points of intervention in the prevention of cancer and other mutation-related diseases. Mut. Res. 2001;9(22):480–481. doi: 10.1016/S0027-5107(01)00165-8. [DOI] [PubMed] [Google Scholar]
- 71.Patra S., Pradhan B., Nayak R., Behera C., Das S., Patra S.K., Efferth T., Jena M., Bhutia S.K. Dietary polyphenols in chemoprevention and synergistic effect in cancer: Clinical evidences and molecular mechanisms of action. Phytochemicals. 2021;90:13 p. doi: 10.1016/j.phymed.2021.153554. [DOI] [PubMed] [Google Scholar]
- 72.Słoczyńska K., Powroźnik B., Pekala E., Waszkielewicz A.M. Antimutagenic compounds and their possible mechanisms of action. J. Appl. Genet. 2014;55:273–285. doi: 10.1007/s13353-014-0198-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kada T., Shimoi T. Desmutagens and bio-antimutagens-their modes of action. Bioessays. 1987;7(3):113–115. doi: 10.1002/bies.950070305. [DOI] [PubMed] [Google Scholar]
- 74.Kaezer A.R., Aiub C.A.F., Mazzei J.L., Ribeiro-Pinto L.F., Felzenszwalb I. Antimutagenic effect and phenolic content of green and roasted yerba mate beverages in different packages available in the Brazilian market. CyTA– J. Food. 2012:144–151. doi: 10.1080/19476337.2011.601429. [DOI] [Google Scholar]
- 75.Helvacioǧlu S., Charehsaz M., Guzelmeiç E., Oçjun M.A., Ayran I., Kirmizibekmez H., Kan Y., Aydin A., Yeșilada E. Protective effect of Nigella damascene fized oils against aflatoxin induced mutagenicity in the classical and modified Ames test. Chem. Biodivers. 2021;18(10) doi: 10.1002/cbdv.202000936. [DOI] [PubMed] [Google Scholar]
- 76.Makhafola T.J., Elgoraschi E.E., McGaw L.J., Verschaeve L., Eloff J.N. The correlation between atimutagenic activity and total phenolic content of extracts of 31 plant species with high antioxidant activity. BMC Complement. Altern. Med. 2016;16(490) doi: 10.1186/s1290-16-1437-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Oroian M., Escriche I. Antioxidants: Characterization, natural sources, extraction and analysis. Food Res. Int. 2015;74:10–36. doi: 10.1016/j.foodres.2015.04.018. [DOI] [PubMed] [Google Scholar]
- 78.Feng D., Fang Z., Zhang P. The melanin inhibitory effect of plants and phytochemicals: A systematic review. Phytomedicine. 2022;107:19 p. doi: 10.1016/j.phymed.2022.154449. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data will be made available on request.