Abstract
Klebsiella pneumoniae is a common pathogen capable of causing a wide range of infections. Antibiotic resistance complicates treatment of these infections significantly. We are comparing resistance levels and genotypes among two collections of K. pneumoniae clinical isolates from Alexandria Main University Hospital (AMUH). We used disc diffusion and Minimum Inhibitory Concentration (MIC) by microbroth dilution to assess resistance levels and performed whole genome sequencing (WGS) to describe multilocus sequence types (MLST) and resistance gene presence. Among a collection of 56 K. pneumoniae clinical isolates (19 from 2019 to 37 from 2021), multidrug resistance (MDR) was 33% and 10%, extended drug resistance (XDR) was 24% and 46% and pan-drug resistance (PDR) was 43% and 43%, respectively. We identified 15 MLST STs including two novel types (ST-6118 and ST-6119 ). ST-101 and ST-383 were common between the two collections; ST-101 was the most common genotype in 2019 (28.6%) and ST-147 was most common in 2021 (25%). Ampicillin/sulbactam, amikacin, cefepime, ceftriaxone and ertapenem MICs were significantly higher in 2021. Prevalence of aph(3’) – Ia, aph(3’)-VI, mphA was significantly higher in 2021. The increasing resistance levels and the persistence of some MDR/XDR genotypes is concerning. Understanding mechanisms of resistance will inform infection control and antimicrobial stewardship plans to prevent evolution and spread of XDR and PDR strains.
Supplementary Information
The online version contains supplementary material available at 10.1186/s12864-024-10661-z.
Keywords: Multidrug resistance (MDR), ESBLs, Carbapenem resistance, Whole-genome sequencing (WGS), Multi-locus sequence typing (MLST)
Introduction
Antimicrobial resistance (AMR) is a complex multidimensional problem. AMR can be intrinsic or acquired. Intrinsic resistance is the inherent ability of some microorganisms to tolerate the action of certain antimicrobials [1]. On the other hand, microbes can acquire the ability to grow in the presence of therapeutic levels of the antimicrobials that kill their sensitive counterparts [2]. The unwise usage of antibiotics is directly connected to the increasing frequency of acquired AMR [3]. The increasing rates of resistance development have resulted in the emergence of multidrug-resistant (MDR), extensively drug-resistant (XDR), and even pan-drug resistant (PDR) strains [4].
Klebsiella pneumoniae is a Gram-negative bacterium that is considered one of the most common Enterobacteriaceae associated with hospital-acquired infections including septicemia, pneumonia, urinary tract infections (UTIs) and soft tissue infection [5].The emergence of antibiotic-resistant strains of K. pneumoniae worldwide, including extended-spectrum β-lactamase (ESBL) and carbapenemase-producing strains has become a cause for concern [6]. ESBL-producing strains are resistant to all penicillins, cephalosporins and aztreonam, resulting in a decline in available choices for proper therapy and increasing patient mortality.
A significant rise in the rates of resistance of clinical K. pneumoniae isolates to different drugs has been noticed lately [7]. The main reason is the ability of the bacterium to acquire resistance genes vertically or horizontally. Various carbapenemases, such as Klebsiella pneumoniae carbapenemase (KPC), New Delhi metallo β-lactamase (NDM), Verona Integron encoded metallo β-lactamase (VIM), Imipenemase (IMP), and OXA-48, have been identified in K. pneumoniae [8, 9], some of which appear to be clone-specific and such clones form a reservoir for infection [10].
Globally, the estimated prevalence of AMR in 2015 was 66.9% for third-generation cephalosporin-resistant Klebsiella pneumoniae, and 23.4% for carbapenem-resistant (CR) K. pneumoniae, with projected AMR prevalence in 2030 at 58.2% and 52.8% respectively [11]. This model suggests that third-generation cephalosporins and carbapenems may be ineffective against the majority of K. pneumoniae infections in most parts of the world by 2030, emphasizing the importance of improving stewardship efforts as well as prioritizing research and development of new antibiotics for resistant Enterobacteriaceae [11].
In Egypt, K. pneumoniae is increasingly recognized as an emerging pathogen, showing high levels of antibiotic resistance [12]. A few reports are available on the antibiotic resistance profiles of CR K. pneumoniae and the molecular basis of antibiotic resistance or the clonal diversity of the drug-resistant strains in Egypt. [13, 14] However, there is a paucity of data on the clonality of CR K. pneumoniae [15]. Phenotypic surveillance of antibiotic resistance can benefit from the addition of molecular surveillance by whole-genome sequencing (WGS). WGS gives insights into the genetic basis of resistance mechanisms, pathogen evolution and population dynamics at various geographical and temporal dimensions [16]. Understanding molecular epidemiology would improve the management of Klebsiella infections, guide infection control interventions and support effective stewardship programs. In this study, we are comparing resistance levels, clonal diversity and genetic profiles of a population of K. pneumoniae isolates from patients admitted to Alexandria Main University Hospital (AMUH), Egypt in 2019 and 2021 by using a combination of phenotypic and genomic tools.
Materials and methods
Bacterial isolates
A total of 56 consecutive K. pneumoniae isolates were included in this study. They were obtained from different clinical specimens from inpatients presenting at AMUH in 2019 (September to December) and 2021 (January to April). AMUH is a tertiary hospital and is considered as the largest teaching hospital and a main referral center in northern Egypt with four satellite hospitals and a maximum capacity of 3500 beds. These isolates represented all K. pneumoniae isolates collected at AMUH during the study period. The identity of the isolates was confirmed using conventional methods, including microscopical examination and biochemical tests, and the Micro VITEK® 2 microbial identification instrument (bioMérieux).For long term preservation, the isolates were maintained in 15%glycerol broth at -80 °C [15, 17].
Antibiotic susceptibility screening of the clinical isolates
The susceptibility of the tested isolates, as well as that of appropriate standard microorganisms, to the studied antibiotics was determined by the standard disc diffusion technique according to the Clinical and Laboratory Standards Institute (CLSI) guidelines [18]. All antibiotic discs were the product of Oxoid Ltd, England. The tested antibiotics were Amikacin (AK 30 µg ); Gentamicin (GMN 10 µg); Meropenem (MEM 10 µg); Ertapenem(ETP10µg); Imipenem (IMP10µg); Ceftazidime/avibactam (CZA 30/20µg); Ceftazidime (CAZ 30 µg); Ceftriaxone (CRO 30 µg); Ciprofloxacin (CIP 5 µg); Doxycycline (DOX 30 µg); Trimethoprim/Sulfamethoxazole(SXT1.25/23.75 µg); Ampicillin/Sulbactam (SAM 10/10µg); Cefoperazone/Sulbactam(SCF 75/10µg); Cefotaxime (CTX 30 µg); Cefepime (FEP 30 µg); Piperacillin/Tazobactam(TZP100/10µg); Nitrofurantoin(NTF 300 µg); Azithromycin (AZM 15 µg); Fosfomycin (FOS 200 µg); Ampicillin (AMP10µg); Amoxicillin/Clavulanate (AMC 20/10µg); Norfloxacin (NOR10µg). The average diameter was recorded and interpreted as susceptible (S), intermediate (I) or resistant (R) according to the susceptibility tables of CLSI 2021 [18]. Results were confirmed using a Micro VITEK® 2 microbial identification instrument (bioMérieux France).
Multidrug -resistance (MDR) was defined as nonsusceptibility to ≥ 1 agent in ≥ 3 antimicrobial categories; extensively drug-resistant (XDR) as susceptibility limited to ≤ 2 categories; pan drug resistance (PDR), as nonsusceptibility to all agents in all antimicrobial categories [4]. The resistance score (R score), corresponding to the number of antibiotics the isolate was resistant to, was determined. Each resistant call was given a score of 1, intermediate resistance calls were given a score of 0.5 [19].
The susceptibility results were confirmed by determining the antibiotics’ MIC using the broth microdilution method according to CLSI guidelines [18].The tested antibiotics were amikacin (concentration range : 64 –2 µg/ml), gentamicin (concentration range : 16 –1 µg/ml), meropenem (concentration range : 16–0.25 µg/ml), ertapenem (concentration range : 8–0.5 µg/ml), imipenem (concentration range : 16–0.25 µg/ml), ceftazidime (concentration range : 64 –2 µg/ml), ceftriaxone (concentration range : 32,768 –512 µg/ml), ciprofloxacin (concentration range : 4–0.25 µg/ml), doxycycline (concentration range : 512–1 µg/ml), trimethoprim/sulfamethoxazole (concentration range : 320 –20 µg/ml), ampicillin/Sulbactam (concentration range : 8192/4096–4/2 µg/ml), cefoperazone/sulbactam (concentration range : 16,384 –64 µg/ml), cefotaxime (concentration range : 64 –1 µg/ml), cefepime (concentration range : 64 –1 µg/ml), nitrofurantoin (concentration range : 512 –16 µg/ml), azithromycin (concentration range : 4096–16 µg/ml), fosfomycin (concentration range : 256 –16 µg/ml), ampicillin (concentration range : 32–0.25 µg/ml), amoxicillin/clavulanate (concentration range : 32/16–2/1 µg/ml), norfloxacin (concentration range : 16–0.5 µg/ml). The tested antibiotics were of pharmaceutical grade, and they were purchased on the Egyptian market. The antibiotic solutions were two-fold serially diluted several times. One hundred µl of the prepared antibiotic concentrations were added to the wells of 96-well polypropylene microtiter plates (Ranon, China) serially from left to right in decreasing concentrations. Each well then received 100 µl of double strength Mueller-Hinton broth (CAMHB) (Oxoid, UK) containing 105 CFU/ml of the selected isolate such that each row represents one selected isolate experiment. The first well in each row was a positive control containing 100 µl of sterile CAMHB instead of antibiotic to control the adequacy of the culture medium to support the growth of the organism. The last well was a negative control to check the sterility of the procedure. The microtiter plate was incubated at 37ºC overnight, and then the plate was visually examined for microbial growth to determine the MIC values. MIC was defined as the least concentration of the antibiotic which inhibited visible growth of the organism. K. pneumoniae ATCC® 700,603 was included as a quality control strain as recommended by CLSI 2021 [18]. Results were confirmed using a Micro VITEK® 2 microbial identification instrument (bioMérieux, France). Ceftazidime/avibactam (concentration range: 0.016–256 µg/mL) MIC was determined using E-test (bioMérieux, France) according to the manufacturer’s instructions.
DNA extraction
The isolates were grown on tryptone soy agar (TSA) plates, the colonies were harvested then washed in 1 mL phosphate-buffered saline (PBS) and resuspended in 0.5 mL SET (75 mM NaCl, 25 mM EDTA, 20 mM Tris [pH 7.5]), to which 50 µL of fresh 20 mg/mL lysozyme in PBS and 30 µL Mutanolysin were added; the mixture was then incubated for 60 min at 37 °C. The cells were then treated with 60 µL 10% sodium dodecyl sulfate and 20 µL proteinase K and incubated at 55 °C for 2 h with gentle inversion. The suspension was gently mixed with 210 µL of 6 M NaCl, and 700 µL phenol: chloroform was added, then incubated at room temperature for 30 to 60 min, using a rotating wheel for gentle mixing. This was followed by centrifugation at maximum speed for 10 min, the aqueous phase was separated in a new microcentrifuge tube and gently mixed with an equal amount of isopropanol. DNA was pelleted by centrifugation and the pellet was washed with 70% ethanol, which was left to evaporate overnight. The pellets were resuspended in 50 µL double-distilled water (ddH2O) and stored at − 20 °C for further processing.
Whole genome sequencing of the isolates
For whole-genome library preparation, the Illumina Nextera kit was used. The Illumina MiSeq System was used to sequence each isolate, producing paired-end 2 × 250 bp reads. The onboard MiSeq Control software and MiSeq Reporter v3.1 were used for quality control and demultiplexing of sequence data. Trimmomatic v0.39 was used to trim the raw reads using the following parameters (LEADING:30, TRAILING: 30, SLIDINGWINDOW: 4:15 and MINLEN:36). SPAdes v3.13.0 was then used to assemble the trimmed reads, using the “only-assembler” option for k values of 55, 77, 99, and 127 [20]. BBMap v38.47 (https://sourceforge.net/projects/bbmap/) was used to calculate genome coverage. Bioawk (https://github.com/lh3/bioawk) was used to remove contigs that were less than 500 bp long from the assemblies. PATRIC v3.3.18 was used to annotate genome assemblies, confirm species, and assess assembly quality [21]. PATRIC performs checkM [22] as part of the annotation process. Only genomes with a completeness of ≥95 and contamination ≤5 were included in further analysis. PATRIC results were used to confirm species designation, genome completeness and contamination. The presence of OXA genes was confirmed by blasting them against the beta-lactamase database (Sequence Server: Custom BLAST Server (bldb.eu)) [23]. ResFinder online tool was available from the Center for Genomic Epidemiology [24, 25]. Genome assemblies and raw sequence data from SRA were deposited in NCBI’s Assembly database under BioProject accession number PRJNA1071125.The NCBI Prokaryotic Genome Annotation Pipeline (PGAP) v5.0 was used to annotate the genomes that were deposited [26]. CARD database was used for AMR gene prediction. Unless previously noted, each software tool’s default parameters were used.
Multilocus sequence typing, allelic diversity and population structure analysis
ARIBA v2.14.6 [27]was used to predict the Multilocus sequence typing (MLST) type of the isolates based on their genomic data; novel sequence types (STs) were assigned through the InstitutPasteur(Paris, France)database(http://bigsdb.pasteur.fr/klebsiella/klebsiella.html).DIVEIN was used to study the diversity of individual locus sequences as well as the concatenated sequences of all seven MLST loci per isolate [28]. Clonal grouping/clustering was determined using eBURST [29]. eBURST was also used to generate a population snapshot to assess the clonal relationship between these STs and those in the Institut Pasteur database. A Call SNP and Infer (CSI) Phylogeny was built using the Center for Genomic Epidemiology online tool [30]. Default parameters were used, namely 10x as min. depth at SNP positions, 10% as min. relative depth at SNP positions, 10 bp as minimum distance between SNPs, 30 as min. SNP quality, 25 as min. read mapping quality and 1.96 as min. Z-score. The MDR strain genome of Klebsiella pneumoniae subsp. pneumoniae HS11286 was used as a reference but was not included in the final phylogeny. Interactive Tree of Life (iTol) v.6.9.1 was used as the tree viewer.
Statistical analysis
Data were analysed using a statistical package for the Social Sciences (SPSS) software package version 20.0 (Armonk, NY: IMP Corp). Quantitative data were presented as mean and standard deviation (SD) for normally distributed data, median for skewed data, or as number and percentage for categorical data. Paired t test for normally distributed quantitative variables, to compare between two dependents variables [31]. Results were statistically significant at P-value ≤ 0.05. The level of agreement between two or more isolates within the same ST was expressed a percentage agreement and was calculated by dividing the number of isolates displaying the same phenotypic resistance picture or same content of resistance genes over the total number of isolates within that ST.
Ethics approval
All methods were carried out in compliance with the corresponding regulations and guidelines. The study was approved by the ethics committee at the Faculty of Medicine, Alexandria University (IRB No.: 00012098).The informed consent was waived by the ethics committee at the Faculty of Medicine, Alexandria University (IRB No.: 00012098), since all isolates were obtained as part of the routine care of the patients at the discretion of the treating physician. The isolates were included in the study anonymously and no patient intervention was decided based on the results of the study.
Results
A total of 56 K. pneumoniae isolates were included in the study. Of these 19 were collected consecutively in 2019 and the remaining 37 were obtained consecutively in 2021 (Supplementary File 1).Since these isolates represented all K. pneumoniae isolates collected at AMUH during the study period, they were used to draw an epidemiological snapshot before and after the pandemic. The collective demographics of the two collections are presented in Table 1.The male: female ratio of patients where the isolates were obtained was very close in the two collections, average age of patients was 61.43 years in 2019 and 51.71 years in 2021.The isolates were obtained from pus (n = 9); blood (n = 10); urine (n = 21); CSF (n = 1); and mini-broncho-alveolar lavage (mini-BAL) (n = 15).The number of isolates from pus was significantly higher in the 2019 collection relative to the 2021 one. The date of patient admission was missing which prevented the determination of the type of infections (community versus nosocomial).
Table 1.
Patient demographics and sample details in the 2019 and 2021 collections
| Demographic data | 2019 (n = 19) |
2021 (n = 37) |
p-value |
|---|---|---|---|
| Gender | |||
| Male | 10 (52.6%) | 20 (54%) | 0.920 |
| Female | 9 (47.4%) | 17 (46%) | |
| Sample type | |||
| Urine | 6 (31.6%) | 15 (40.5%) | 0.512 |
| Blood | 2 (10.5%) | 8 (21.6%) | 0.467 |
| mini-BAL | 5 (26.3%) | 10 (27%) | 0.955 |
| PUS | 6 (31.6%) | 3 (8.1%) | 0.049 * |
| CSF | 0 (0%) | 1 (2.7%) | 1.000 |
| Age (years) | |||
| Min. – Max. | 21.0–85.0 | 12.0–87.0 | 0.082 |
| Mean ± SD. | 61.43 ± 23.40 | 51.71 ± 17.05 | |
| Median | 67.0 | 55.0 |
BAL: Broncho-alveolar lavage, CSF: Cerebrospinal fluid, MIN: Minimum, MAX: Maximum, SD: Standard deviation, p: p value for comparing between the two studied groups,*: Statistically significant at p ≤ 0.05
Resistance profile of the isolates
All strains were resistant to at least one antimicrobial agent tested in the study. Only ceftazidime showed a significant increase in resistance rate in 2021 relative to 2019 (Table 2). The prevalence of MDR isolates decreased from 33% in 2019 to 10% in 2021. Yet, XDR prevalence increased from 24% in 2019 to 46% in 2021 and PDR prevalence remained steady at 43%. Susceptibility data were confirmed by MIC determination, significantly higher MIC values were recorded with amikacin (p = 0.042), ampicillin/sulbactam (p = 0.001), cefepime (p = 0.041), ceftriaxone (p = 0.036)and ertapenem (p = 0.022) in 2021 relative to 2019 (Table 2 and Supplementary File 1).
Table 2.
Comparison of the resistance pattern of the isolates to tested antibiotics between 2019 and 2021
| N ( % of total ) | MIC values | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 2019 (total n = 19) | 2021 (total n = 37) | 2019 | 2021 | |||||||||
| S | I | R | SDD | S | I | R | SDD | MIC 50 | MIC 90 | MIC 50 | MIC 90 | |
| Aminoglycosides | ||||||||||||
| AK | 8(42.3) | 1(5.2) | 10(52.5) | - | 16(43.25) | 5(13.5) | 16(43.25) | - | 64 | 64 | 32 | 64 |
| GMN | 9(47.4) | 1(5.2) | 9(47.4) | - | 22(59.5) | 1(2.7) | 14(37.8) | - | 8 | 16 | 4 | 16 |
| β-lactams | ||||||||||||
| AMP | 2(10.5) | 0(0) | 17(89.5) | - | 0(0) | 0(0) | 37(100) | - | 32 | 32 | 32 | 32 |
| SAM | 1(5.2) | 0(0) | 18(94.8) | - | 0(0) | 0(0) | 37(100) | - | 512 | 2048 | 1024 | 4096 |
| TZP | 2(10.5) | - | 17(89.5) | 0(0) | 2(5.4) | - | 34(91.9) | 1(2.7) | 128 | 128 | 128 | 128 |
| AMC | 2(10.5) | 0(0) | 17(89.5) | - | 5(13.5) | 1(2.7) | 31(83.8) | - | 32 | 32 | 32 | 32 |
| CZA | 6(31.6) | 0(0) | 13(68.4) | - | 12(32.4) | 0(0) | 25(67.6) | - | 256 | 256 | 256 | 256 |
| CAZ | 3(15.8) | 0(0) | 16(84.2) | - | 0(0) | 1(2.7) | 36(97.3) | - | 64 | 64 | 64 | 64 |
| FEP | 3(15.8) | - | 14(73.7) | 2(10.5) | 2(5.4) | - | 33(89.2) | 2(5.4) | 32 | 64 | 64 | 64 |
| CRO | 0(0) | 0(0) | 19(100) | - | 0(0) | 0(0) | 37(100) | - | 16,384 | 32,768 | 16,384 | 32,768 |
| CTX | 1(5.2) | 0(0) | 18(94.8) | - | 1(2.7) | 0(0) | 36(97.3) | - | 64 | 64 | 64 | 64 |
| SCF | 0(0) | 0(0) | 19(100) | - | 0(0) | 0(0) | 37(100) | - | 1024 | 8192 | 2048 | 8192 |
| MEM | 4(21) | 0(0) | 15(79) | - | 9(24.4) | 0(0) | 28(75.6) | - | 16 | 16 | 16 | 16 |
| ETP | 2(10.5) | 0(0) | 17(89.5) | - | 8(21.6) | 0(0) | 29(78.4) | - | 4 | 8 | 8 | 8 |
| IMP | 5(26.3) | 0(0) | 14(73.7) | - | 10(27) | 0(0) | 27(73) | - | 16 | 16 | 16 | 16 |
| Fluoroquinolones | ||||||||||||
| CIP | 1(5.2) | 2(10.5) | 16(84.2) | - | 0(0) | 1(2.7) | 36(97.3) | - | 4 | 4 | 4 | 4 |
| NOR | 5(26.4) | 1(5.2) | 13(68.4) | - | 7(19) | 3(8.1) | 27(72.9) | - | 16 | 16 | 16 | 16 |
| Macrolide | ||||||||||||
| AZM | 1(5.2) | 0(0) | 18(94.8) | - | 1(2.7) | 0(0) | 36(97.3) | - | 512 | 2048 | 1024 | 4096 |
| FOS | 15(79) | 3(15.8) | 1(5.2) | - | 28(75.6) | 3(8.1) | 6(16.2) | - | 32 | 128 | 16 | 128 |
| SXT | 2(10.5) | 0(0) | 17(89.5) | - | 4(10.8) | 0(0) | 33(89.2) | - | 320 | 320 | 320 | 320 |
| DOX | 1(5.2) | 1(5.2) | 17(89.6) | - | 6(16.2) | 3(8.1) | 28(75.6) | - | 64 | 256 | 32 | 128 |
| NTF | 2(10.5) | 4(21) | 13(68.5) | - | 5(13.5) | 7(19) | 25(67.5) | - | 256 | 512 | 256 | 512 |
MIC : Minimum inhibitory concentration, SDD: Susceptible-dose dependent (which is a breakpoint category for which the susceptibility of an isolate depends on the dosing regimen used),S: Sensitive ; I : Intermediate ; R : resistant ; CIP: Ciprofloxacin; NOR: Norfloxacin; DOX: Doxycycline; NTF: Nitrofurantoin; AMP: Ampicillin ; SAM: Ampicillin/sulbactam; TZP: Piperacillin/tazobactam, AMC: Amoxicillin/clavulanic acid ; CAZ: Ceftazidime ; FEP: Cefepime ; CRO: Ceftriaxone, CTX: Cefotaxime, SCF: Cefoperazone-sulbactam ,ETP: Ertapenem; AZM: Azithromycin ; AK: Amikacin ; GMN : gentamicin ; MEM: Meropenem ; IMP :Imipenem ; CZA : Ceftazidime-avibactam ; SXT : Trimethoprim/sulfamethoxazole
Genes of resistance and their clonal distributions
The sequencing data of seven isolates from the 2019 collection and 20 from the 2021 collection passed the quality checks and were further included in the study. CheckM confirmed all sequencing was uncontaminated and all strains were confirmed as K. pneumoniae. Genomic data were used to predict the presence of antibiotic resistance genes (Table 3 and Supplementary file 2). In 58.3% of the cases, there was an increased prevalence of the genes in the 2021 collection relative to the 2019 one. This increase was significant in the case of aph(3’) – Ia, aph(3’)-VI coding for aminoglycoside resistance and mphA coding for macrolide resistance. Except for blaOXA−1 predicted in both collections, the other OXA genes (blaOXA−9, blaOXA−48) were only detected in the 2021 collection (Supplementary File2).
Table 3.
Prevalence of the most common resistance genes in the two collections
| Antibiotic class | Antibiotic resistance gene | N(% of total ) | FE p | |
|---|---|---|---|---|
| 2019 (total n = 7) |
2021 (total n = 20) |
|||
| Aminoglycosides | aph(3’) – Ia | 2 (28.6%) | 17(85%) | 0.011* |
| aac(6’) - Ib-cr7 | 5 (71.4%) | 13 (65%) | 1.000 | |
| aac(6’)-Ib7 | 1 (14.3%) | 0(0%) | 0.259 | |
| aac(3)-IIa | 0(0%) | 3(15%) | 0.545 | |
| aph(3’)-VI | 3 (42.9%) | 17 (85%) | 0.049 * | |
| aph(3’’) – Ib/ aph(6) - Id | 3 (42.9%) | 11(55%) | 0.678 | |
| aadA | 3 (42.9%) | 14 (70%) | 0.365 | |
| aadA2 | 2 (28.6%) | 5(25%) | 1.000 | |
| crcB | 1 (14.3%) | 0(0%) | 0.259 | |
| armA | 1 (14.3%) | 11(55%) | 0.091 | |
| rmtB | 0(0%) | 2(10%) | 1.000 | |
| rmtF | 2 (28.6%) | 1(5%) | 0.156 | |
| rrsB+ | 6 (85.7%) | 15(75%) | 1.000 | |
| β-lactams | bla SHV−16 | 4 (57.1%) | 13 (65%) | 1.000 |
| bla CTX−M−15 | 5 (71.4%) | 16(80%) | 0.633 | |
| bla CTX−M−2 | 0 (0%) | 8(40%) | 0.068 | |
| lap | 0 (0%) | 1(5%) | 1.000 | |
| bla LEN−1 | 4 (57.1%) | 7(35%) | 0.391 | |
|
blaOXA−20/blaOXA−24 / blaOXA−42 /porin ompC |
0 (0%) | 0 (0%) | – | |
| blaOXA−9 /blaKPC | 0 (0%) | 3(15%) | 0.545 | |
| bla OXA−48 | 0 (0%) | 5(25%) | 0.283 | |
| bla OXA−1 | 3 (42.9%) | 5(25%) | 0.633 | |
| bla NDM−1 | 3 (42.9%) | 11(55%) | 0.678 | |
| bla TEM | 4 (57.1%) | 12 (60%) | 1.000 | |
| Klebsiella pneumoniae ompK35 | 7 (100%) | 19 (95%) | 1.000 | |
|
Klebsiella pneumoniae ompK36/ ompK37 lptD / ompA |
7 (100%) | 20 (100%) | – | |
| ESBL | 3 (42.9%) | 2 (10%) | 0.091 | |
| Fosfomycin | fosA1 | 0 (0%) | 3(15%) | 0.545 |
| fosA5 | 7 (100%) | 19 (95%) | 1.000 | |
| Fluoroquinolones | qnrB | 2 (28.6%) | 6 (30%) | 1.000 |
| qnrS11 | 3 (42.9%) | 16(80%) | 0.145 | |
| oqxA | 7 (100%) | 20 (100%) | – | |
| oqxB | 7 (100%) | 19 (95%) | 1.000 | |
| acrR | 7 (100%) | 20 (100%) | – | |
| Macrolide | mphA | 2 (28.6%) | 17(85%) | 0.011 * |
| mphB | 0 (0%) | 1(5%) | 1.000 | |
| mphE | 1 (14.3%) | 12 (60%) | 0.077 | |
| msrE | 1 (14.3%) | 11(55%) | 0.091 | |
| Trimethoprim | dfrA-1 | 2 (28.6%) | 12 (60%) | 0.209 |
| dfrA12 | 2 (28.6%) | 5(25%) | 1.000 | |
| dfrA14 | 4 (57.1%) | 4 (20%) | 0.145 | |
| Sulfonamide | sul1 | 4 (57.1%) | 17 (85%) | 0.290 |
| sul2 | 4 (57.1%) | 16(80%) | 0.328 | |
| sul3 | 0(0%) | 1(5%) | 1.000 | |
| Tetracycline | tetA | 2 (28.6%) | 8(40%) | 0.678 |
| tetD | 2 (28.6%) | 0(0%) | 0.060 | |
| Multidrug |
K. pneumoniae KpnE/ KpnF/ KpnG/ Escherichia coli 23 S |
7 (100%) | 20(100%) | – |
| K. pneumoniae KpnH+ | 0(0%) | 0(0%) | – | |
| adeK / adeG | 0(0%) | 1(5%) | 1.000 | |
| ramR 1 | 1 (14.3%) | 0(0%) | 0.259 | |
| acrA | 7 (100%) | 19 (95%) | 1.000 | |
| Polymyxin | eptB / arnT | 7 (100%) | 20(100%) | – |
| Chloramphenicol | catI | 1 (14.3%) | 5(25%) | 1.000 |
| catII | 0(0%) | 5(25%) | 0.283 | |
| catB3/ floR+ | 0(0%) | 2(10%) | 1.000 | |
| catB8 | 0(0%) | 1(5%) | 1.000 | |
| Antiseptics | qacE | 4 (57.1%) | 18(90%) | 0.091 |
| Rifamycin | arr | 2 (28.6%) | 3(15%) | 0.580 |
| Bleomycin | ble MBL | 3 (42.9%) | 11(55%) | 0.678 |
FEp: p value for Fisher Exact test for comparing between 2019 and 2021
*: Statistically significant at p ≤ 0.05
As shown in supplementary file 3, mutations in existing resistance genes were detected by ResFinder database, resulting in eight groups where each group includes number of isolates sharing same pattern of mutation of ompK36,ompK37 and acrR genes. This mutation didn’t increase MIC level significantly. So, the mutations detected weren’t contributing to resistance against cephalosporins, carbapenems and fluoroquinolones.
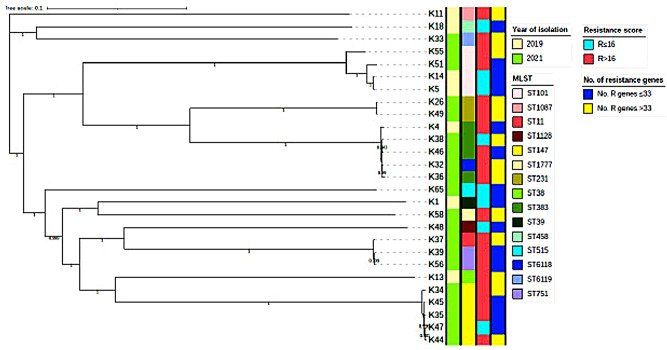
As shown in Table 4, the presence of genes responsible for resistance to different antibiotic classes was linked to the specimen type, where there was insignificant difference in their presence among different specimen types. Moreover, being resistant to a smaller number of antibiotics (smaller resistance scores) was mostly linked to carriage of a lower number of resistance genes, irrespective of the year of collection or MLST (Supplementary Files 1 and 2 and Fig. 1).
Table 4.
Prevalence of representative resistance genes relative to specimen types
| Antibiotic class | Antibiotic resistance gene | N (% of total ) | MC p | |||
|---|---|---|---|---|---|---|
| BAL (total n = 7) |
URINE (total n = 11) |
BLOOD (total n = 6 ) |
PUS (total n = 3) |
|||
| Aminoglycosides | aph(3’) – Ia | 6 (85.7%) | 5(45.5%) | 6 (100%) | 2 (66.7%) | 0.086 |
| aac(6’) - Ib-cr7 | 5(71.4%) | 7(63.6%) | 3 (50%) | 3(100%) | 0.618 | |
| aph(3’)-VI | 5(71.4%) | 5(45.5%) | 4(66.7%) | 2 (66.7%) | 0.768 | |
| aph(3’’) – Ib/ aph(6) - Id | 4(57%) | 6(54.5%) | 4(66.7%) | 0(0%) | 0.387 | |
| aac(6’)-Ib7/ crcB | 0(0%) | 1(9.1%) | 0(0%) | 0(0%) | 1.000 | |
| aac(3)-IIa | 2(28.6%) | 1(9.1%) | 0(0%) | 0(0%) | 0.602 | |
| aadA2 | 3(43%) | 3(27.3%) | 1(16.7%) | 0(0%) | 0.662 | |
| rmtB | 1(14.3%) | 1(9.1%) | 0(0%) | 0(0%) | 1.000 | |
| rmtF | 1(14.3%) | 2(18.2%) | 0(0%) | 0(0%) | 0.844 | |
| aadA | 5(71.4%) | 6(54.5%) | 4(66.7%) | 2 (66.7%) | 0.940 | |
| armA | 4(57%) | 3(27.3%) | 3 (50%) | 2 (66.7%) | 0.562 | |
| rrsB+ | 5(71.4%) | 9(81.8%) | 4(66.7%) | 3(100%) | 0.791 | |
| β-lactams | bla SHV−16 | 3(43%) | 9(81.8%) | 3 (50%) | 2 (66.7%) | 0.342 |
| bla CTX−M−15 | 5(71.4%) | 8(72.8%) | 5(83.3%) | 3(100%) | 1.000 | |
| bla CTX−M−2 | 2(28.6%) | 2(18.2%) | 4(66.7%) | 0(0%) | 0.175 | |
| bla LEN−1 | 4(57%) | 2(18.2%) | 3 (50%) | 2 (66.7%) | 0.239 | |
| lap | 0(0%) | 0(0%) | 1(16.7%) | 0(0%) | 0.345 | |
| blaOXA−20/ blaOXA−24 / blaOXA−42 | 0(0%) | 0(0%) | 0(0%) | 0(0%) | – | |
| bla OXA−9 | 1(14.3%) | 1(9.1%) | 1(16.7%) | 0(0%) | 1.000 | |
| bla OXA−48 | 1(14.3%) | 1(9.1%) | 3 (50%) | 0(0%) | 0.201 | |
| bla OXA−1 | 2(28.6%) | 4(36.4%) | 1(16.7%) | 1(33.3%) | 0.933 | |
| bla NDM−1 | 5(71.4%) | 5(45.5%) | 2(33.3%) | 2 (66.7%) | 0.565 | |
| bla TEM | 4(57%) | 6(54.5%) | 3 (50%) | 3(100%) | 0.616 | |
| bla KPC | 1(14.3%) | 1(9.1%) | 1(16.7%) | 0(0%) | 1.000 | |
| porin ompC | 0(0%) | 0(0%) | 0(0%) | 0(0%) | – | |
| Klebsiella pneumoniae ompK35 | 7(100%) | 10(91%) | 6 (100%) | 3(100%) | 1.000 | |
| Klebsiella pneumoniae ompK36/ Klebsiella pneumoniae ompK37 /lptD / ompA | 7(100%) | 11(100%) | 6 (100%) | 3(100%) | – | |
| – | ||||||
| ESBL | 1(14.3%) | 2(18.2%) | 1(16.7%) | 1(33.3%) | 1.000 | |
| Fosfomycin | fosA1 | 1(14.3%) | 1(9.1%) | 1(16.7%) | 0(0%) | 1.000 |
| fosA5 | 7(100%) | 10(91%) | 6 (100%) | 3(100%) | 1.000 | |
| Fluoroquinolones | qnrB | 4(57%) | 2(18.2%) | 2(33.3%) | 0(0%) | 0.250 |
| qnrS11 | 5(71.4%) | 7(63.6%) | 5(83.3%) | 2 (66.7%) | 0.928 | |
| oqxA | 7(100%) | 11(100%) | 6 (100%) | 3(100%) | – | |
| oqxB | 7(100%) | 11(100%) | 5(83.3%) | 3(100%) | 0.333 | |
| acrR | 7(100%) | 11(100%) | 6 (100%) | 3(100%) | – | |
| Macrolide | mphA | 7(100%) | 6(54.5%) | 5(83.3%) | 1(33.3%) | 0.154 |
| mphB | 0(0%) | 1(9.1%) | 0(0%) | 0(0%) | 1.000 | |
| mphE | 4(57%) | 4(36.4%) | 3 (50%) | 2 (66.7%) | 0.711 | |
| msrE | 4(57%) | 3(27.3%) | 3 (50%) | 2 (66.7%) | 0.572 | |
| Trimethoprim | dfrA-1 | 5(71.4%) | 3(27.3%) | 4(66.7%) | 2 (66.7%) | 0.232 |
| dfrA12 | 2(28.6%) | 4(36.4%) | 1(16.7%) | 0(0%) | 0.751 | |
| dfrA14 | 3(43%) | 4(36.4%) | 0(0%) | 1(33.3%) | 0.322 | |
| Sulfonamide | sul1 | 7(100%) | 7(63.6%) | 5(83.3%) | 2 (66.7%) | 0.318 |
| sul2 | 6 (85.7%) | 8(72.8%) | 4(66.7%) | 2 (66.7%) | 0.931 | |
| sul3 | 0(0%) | 0(0%) | 0(0%) | 1(33.3%) | 0.105 | |
| Tetracycline | tetA / tetD | 5(71.4%) | 2(18.2%) | 3 (50%) | 0(0%) | 0.076 |
| Multidrug |
K. pneumoniae KpnE/ KpnF/ KpnG/ Escherichia coli 23 S |
7(100%) | 11(100%) | 6 (100%) | 3(100%) | – |
| adeK / adeG | 1(14.3%) | 0(0%) | 0(0%) | 0(0%) | 0.588 | |
| ramR 1 | 0(0%) | 0(0%) | 0(0%) | 1(33.3%) | 0.105 | |
| acrA | 7(100%) | 11(100%) | 5(83.3%) | 3(100%) | 0.333 | |
| Polymyxin | eptB / arnT | 7(100%) | 11(100%) | 6 (100%) | 3(100%) | – |
| Chloramphenicol | catI | 3(43%) | 1(9.1%) | 2(33.3%) | 0(0%) | 0.289 |
| catII | 1(14.3%) | 3(27.3%) | 1(16.7%) | 0(0%) | 1.000 | |
| catB3 | 0(0%) | 1(9.1%) | 1(16.7%) | 0(0%) | 0.785 | |
| catB8 | 1(14.3%) | 0(0%) | 0(0%) | 0(0%) | 0.588 | |
| floR+ | 0(0%) | 0(0%) | 1(16.7%) | 1(33.3%) | 0.102 | |
| Antiseptics | qacE | 7(100%) | 8(72.8%) | 5(83.3%) | 2 (66.7%) | 0.402 |
| Rifamycin | arr- | 2(28.6%) | 2(18.2%) | 1(16.7%) | 0(0%) | 1.000 |
| Bleomycin | ble MBL | 5(71.4%) | 5(45.5%) | 2(33.3%) | 2 (66.7%) | 0.562 |
MCp: p value for Monte Carlo test for comparing between 2019 and 2021
Fig. 1.
A SNP-based phylogenetic tree annotated by year of isolation, MLST, resistance score and number of carried resistance genes (colored bands from left to right, respectively). Median resistance score and median number of resistance genes were used as cutoffs to categorize the isolates
Multilocus sequence typing and phylogeny of the isolates
The isolates belonged to 15 sequence types (STs) (Table 5 and Supplementary File 2). The STs occurring the most were ST-101 in 28.6% of the 2019 isolates and ST-147 in 25% of the 2021 isolates, with ST-383 and ST-101 being represented in both 2019 and 2021 collections. Two novel STs: ST-6118 and ST-6119, each occurring once (10%) in the 2021 collection, were identified in the study. The SNP-based phylogenetic tree (Fig. 1) largely mirrors the MLST tree (Supplementary Fig. 1) and shows that the STs are grouped in two major clades and three singletons. The phylogenetic tree confirms the clonality of the isolates belonging to each of ST101, ST383 and ST147, irrespective of the year of isolate collection.
Table 5.
Analysis of the MLST data
| Sequence type | 2019 (n = 7) N (%of total) |
2021(n = 20) N (%of total) |
|---|---|---|
| 383 | 1(14.3%) | 3(15%) |
| 101 | 2(28.6%) | 2(10%) |
| 458 | 1(14.3%) | |
| 1128 | 1(5%) | |
| 38 | 1(14.3%) | |
| 39 | 1(14.3%) | |
| 1087 | 1(14.3%) | |
| 6118* | 1(5%) | |
| 1777 | 1(5%) | |
| 231 | 2(10%) | |
| 11 | 1 (5%) | |
| 515 | 1 (5%) | |
| 751 | 2(10%) | |
| 147 | 5(25%) | |
| 6119* | 1(5%) |
*: novel MLST types identified in the current study
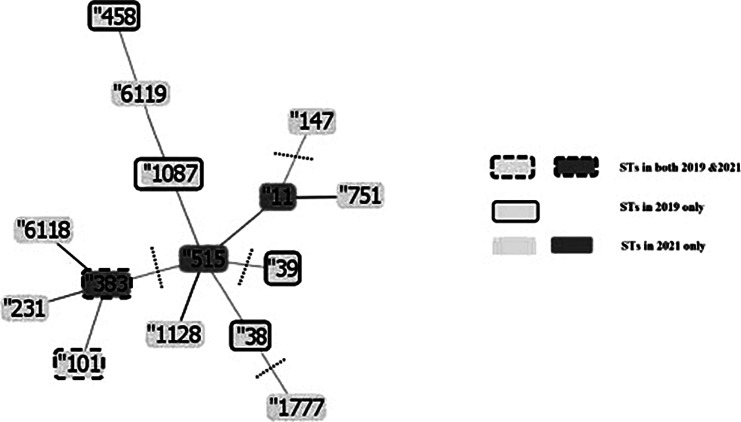
The e-BURST analysis (clustering with 3 loci matching profiles) confirmed the phylogenetic relationships and groups the STs in two clusters, showed ST-515 to likely be the ancestor ST (Fig. 2). Cluster one included nine STs: ST-515, ST-11, ST-751, ST-1128 and ST-6119 from the 2021 collection in addition to ST-1087, ST-38 and ST-458 from the 2019 collection. ST-515 was a single locus variant (SLV) of ST-1128, a double locus variant of ST-11 and a triple locus variant (TLV) of ST-751 and ST-1087. ST-11 was a SLV of ST-751 and TLV of ST-1128.ST-1128 was a TLV of ST-1087. ST-6119 was a TLV of ST-1087. All group one isolates were resistant to AZM, CTX, SCF and also had the following genes in common: ompA, ompK35, ompK36, ompK37, lptD, oqxA, kpnE, kpnF, kpnG, e.coli 23 S, acRA, eptB and arnT.
Fig. 2.
Evolutionary relationships and relatedness of MLST types using the eBURST algorithm in the PHYLOViZ software (http://www.phyloviz.net/). The identified MLST types belonged to two groups “profiles match 3 loci to any other member of the group“ of STs (group 1: ST383, ST101, ST231, ST6118 and group 2: ST515, ST11, ST751, ST1087, ST6119, ST458, ST1128, ST38) and three singleton STs (ST1777, ST147, ST39). The lengths of connecting branches between the STs are arbitrary. The dashed lines divide the diagram in groups as the STs are divided into 2 groups and 3 singletons. The darker shade of the ST indicates the ancestor STs of the tree. The strains were divided into two groups according to the relatedness to each other, whether they were within SLV, DLV or TLV of other STs
Cluster two included ST-101 and ST-383 both from 2019 to 2021 collections in addition to ST-231 and ST-6118 from the 2021 collection. ST-101 was a TLV of ST-231 and ST-383.ST-383 and ST-6118 were SLVs of each other, ST-231 and ST-383 were DLVs, and ST-231 and ST-6118 were TLVs. ST-231, ST-11, ST-751, ST-147, ST-1128, ST-1777, ST-515, ST-6118 and ST-6119 found only in the 2021 collection were significantly associated with higher MIC values relative to those found in the 2019 collection or common between the two collections.
All ST-101 isolates were positive for the aac(6’)-Ib-cR7, blaSHV−16,blaCTX−M−15, ompA, blaTEM, ompK35, ompK36, ompK37, lptD, fosA5, oqxA, oqxB, kpnE, kpnF, kpnG, E.coli 23 S, acRA, eptB, arnT genes, and were all resistant to CIP, NOR, DOX, NTF, AMP, SAM, TZP, AMC, CAZ, FEP, CRO, CTX, SCF and ETP. ST-383 isolates were positive for the aph(3’)-Ia, aph(3’)-VI, blaLEN−1, ompA, ompK35, ompK36, ompK37, lptD, fosA5, opxA, opxB, kpnE, kpnF, kpnG, eptB, arnT, blaCTX−M−15, blaCTX−M−2, blaOXA−48, aph(3’’)-Ib, aph(6)-Id genes and were all resistant to AZM, CIP, NOR, TMP/SMX, DOX, AMP, SAM, TZP, AMC, CAZ, FEP, CRO, CTX and SCF.
The 2021 unique STs were also significantly associated with the presence of aph(3’) – Ia, aph(3’)-VI and mphA. Looking at level of agreement between isolates belonging to the same ST and isolated from 2019 versus those isolated from 2021 with respect to their phenotypic resistance profile and resistance gene content, Table 6 shows that intra-collection agreement was higher than inter collection agreement for isolates belonging to the same ST. This was more evident in case of ST-101 isolates (n = 2 from each of 2019 and 2021) than ST-383 isolates (n = 1 from 2019 and n = 3 from 2021). Yet, ST-147 (n = 5 all from 2021) showed lowest level of agreement regarding their phenotypic resistance profile.These results suggest resistance evolution in isolates within same ST over years [32].
Table 6.
Resistance pattern and gene carriage inter and intra-collection agreement
| MLST | Year of isolation | Isolate number | Percentage of agreement (%) |
|---|---|---|---|
| 101 | K5 vs. K14 | ||
| 2019 | Resistance pattern | 90.9 | |
| Genes | 97.3 | ||
| 231 | 2021 | K26 vs. K49 | |
| Resistance pattern | 100 | ||
| Genes | 98 | ||
| 751 | 2021 | K39 vs. K56 | |
| Resistance pattern | 86.4 | ||
| Genes | 100 | ||
| 101 | 2021 | K51 vs. K55 | |
| Resistance pattern | 90.9 | ||
| Genes | 91.9 | ||
| 383 | 2021 | K36 vs. K38 vs. K46 | |
| Resistance pattern | 63.6 | ||
| Genes | 69.6 | ||
| 147 | 2021 | K34 vs. K35 vs. K44 vs. K45 vs. K47 | |
| Resistance pattern | 27.3 | ||
| Genes | 78.4 | ||
| 101 | 2019 + 2021 | K5 vs. K14 vs. K 51 vs. K55 | |
| Resistance pattern | 72.7 | ||
| Genes | 81.8 | ||
| 383 | 2019 + 2021 | K4 vs. K36 vs. K38 vs. K46 | |
| Resistance pattern | 63.6 | ||
| Genes | 64.2 |
Discussion
A large increase in the rates of resistance of clinical K. pneumoniae isolates to different antibacterial agents has been noted in recent years [7].Resistant strains develop because of long-term and excessive use of diverse antibiotics, where bacteria can acquire resistance genes horizontally [33].The acquisition of resistance genes under antibiotic selection pressure and further dissemination of these genes lead to the development of MDR and XDR strains [34]which challenges antimicrobial chemotherapy worldwide [35]. K. pneumoniae is becoming more recognized as an emerging pathogen, developing high levels of antibiotic resistance in Egypt [12]. After Streptococcus pneumoniae (22.5%) and Pseudomonas aeruginosa (21.7%), K. pneumoniae (16.7%) has been recognized as the third main cause of Hospital Acquired Infections (HAIs) in Egypt [36]. The current Egyptian guidelines for treating MDR K. pneumoniae include the use of the aztreonam-ceftazidime/avibactam combination or the combination of colistin or tigecycline or an aminoglycoside with a carbapenem or using ceftolozane/tazobactam combination to treat such resistant infections.
A total of 56 K. pneumoniae isolates were included in the study (n = 19 from 2019 and n = 37 from 2021). The isolates were obtained from patients with an almost equal male to female representation in both collections and with an average age above 50 years. Susceptibility to most studied antibiotics was almost equally low among the two collections. Yet on an individual isolate level, although MDR rates decreased in 2021, this was accompanied with an almost doubling in XDR rates. These high levels of XDR K. pneumoniae agree with other studies carried out in Egypt by Hassuna et al. [37]. who reported an alarming prevalence of XDR K. pneumoniae of 83.3% and Al-Baz et al. [38]. who showed that the majority (60.6%) of the isolates were XDR while 30.3% were multidrug-resistant (MDR) and only 9.2% were susceptible with CR isolates accounting for roughly a quarter. Others described resistance rates among clinical K. pneumoniae isolates to be 42.5%, 35% and 5% for MDR, XDR and PDR phenotypes, respectively [13]. This study was also comparable to another that looked at changes in drugs susceptibility patterns over a four-year period in Klebsiella pneumoniae isolates from patients in intensive care units that were mechanically ventilated in North India. As the overall rate of resistant K. pneumoniae increased from 7.5% in 2018 to 21.4% in 2022, while XDR Klebsiella pneumoniae among the mechanically ventilated ICU patients significantly increased from 62.5% in 2018 to 71% in 2022 [39].This could be explained by the massive antibiotic use during the COVID-19 pandemic. It is estimated that although < 10% of COVID-19 patients were diagnosed with a secondary bacterial infection, about 75% received an antibiotic prescription [40].
We performed successful WGS for 27 isolates (n = 7 from 2019 and n = 20 from 2021). With a few exceptions, the genomic prediction of resistance genes was almost equally high in both collections. In most cases, the carriage of a larger number of resistance genes was linked to a more resistant phenotype (higher resistance score). β-lactam resistance, including to 3rd and 4th generation cephalosporins and the carbapenems, was high in both collections. Yet, significantly higher MIC values of amikacin (p = 0.042), ampicillin/sulbactam (p = 0.001), cefepime (p = 0.041), ceftriaxone (p = 0.036)and ertapenem (p = 0.022) were recorded in 2021 versus 2019. The higher carbapenem resistance levels were confirmed by the prediction of higher carriage levels of blaKPC and blaNDM among the 2021 collection which also agreed with Bulman et al [41]. The genomic evidence supports the usefulness of MIC to assess the degree of susceptibility to various antibiotics used in the hospital for the implementation of antibiotic stewardship programs, whereupon antibiotics with MIC90 close to the breakpoint could be labelled “medications with limited access in empirical therapy”, unless absolutely needed [42].
Resistance is multifactorial, meaning that the resistance phenotype is influenced by mutations in several genes. However, this can’t be proven in our study as only mutations of only four genes were detected but weren’t affecting MIC level so switching to XDR strains can be explained by acquisition of resistance genes or mutation of other genes not detected by the database used.
In this study, K. pneumoniae isolates displayed a high genetic variability with 15 sequence types (ST), four unique to the 2019 collection and 9 to the 2021 collection (including two novel STs) and two STs common between 2019 and 2021. ST-101 and ST-147 that were most prevalent among the 2019 and 2021 collections, respectively have been previously detected as the most prevalent STs among a collection of Egyptian K. pneumoniae obtained from nine hospitals in 2018 [14].The two STs common between the 2019 and 2021 collection were ST-101 (n = 2 in each collection) and ST-383 (n = 1 in 2019 and n = 3 in 2021). Of these, only one isolate among the 2019 collection was CAZ-AVI resistant, whereas three isolates from 2021 were CAZ-AVI resistant which could reflect the fact that CAZ-AVI had not been available in clinical practice in 2019. The resistance phenotype was linked to the isolates’ carriage of the blaNDM−1 gene [43]as predicted from genomic data. Adding aztreonam to the CAZ-AVI combination is one of the recommended options to treat such resistant infections [44]. The increased resistance trend among ST-101 and ST-383 was also true for the carbapenems where resistance to meropenem and imipenem increased from 33.33% in 2019 to 83.33% in 2021. In case of ertapenem, although resistance was already at 100% among the 2019 isolates belonging to ST-101 and ST-383 and decreased to 83.33%, MIC values were higher in 2021 which shows increased resistance levels. The same pattern was also seen with trimethoprim/sulfamethoxazole and Azithromycin where there was a noticeable increase in carriage of resistance genes in the 2021 collection. For the fluoroquinolones and tetracyclines, 100% of the 2019 ST-101 and ST-383 isolates were already resistant, yet there is a distinct increase in carriage of some genes mediating resistance to these classes.
From an epidemiological point of view, it is often necessary to determine the clonality of the strains to improve the management of endemic and epidemic nosocomial outbreaks of Klebsiella infections [5]. According to the eBURST diagram, isolates within each group had shared resistance gene content and showed similar resistance phenotypes to two and three antibiotic classes, respectively. The SNP-based phylogenetic tree supported the clonal nature of the isolates within ST101, ST383 and ST147.
The only two STs common between 2019 and 2021 isolates: 101 and 383 were TLVs of each other.ST-101 isolates showed 81.8% level of agreement between the isolates concerning presence of certain resistance genes and 72.7% level of agreement concerning resistance pattern( phenotypic resistance ). De Koster et al. [45]. described high resistance levels among clinical K. pneumoniae ST-101 isolates. Yet within the ST-101 2019 collection, the intra-collection isolate agreement level was 90.9% in resistance pattern and 97.3% in presence of genes, whereas within the 2021 collection the level of the intra-collection agreement was 90.9% in resistance pattern and 91.9% in presence of genes. This shows some diversity within the same ST decreasing the overall level of agreement to 72.7% (resistance pattern) and 81.8% (resistance genes presence). That was exemplified by 2021 ST-101 isolates being resistant to meropenem, imipenem, CAZ/AVI and Fosfomycin while 2019 isolates were sensitive. A similar pattern was seen with ST-383 isolates between the 2019 and 2021 collections where 2021 isolates were resistant to Fosfomycin whereas 2019 isolates were sensitive. Guo et al. [46]. reported ST-383 to be one of two predominant clones responsible for a hospital outbreak of K. pneumoniae in a Chinese hospital.
Osman et al. [47]. described the MLSTs of 86 K. pneumoniae clinical isolates obtained from Sudan between 2016 and 2020 and found ST-101 and ST-383 to be the 4th and 7th most prevalent STs, respectively. In agreement with our findings, the Sudanese ST-101 and ST-383 isolates were MDR including carbapenem resistant with presence of one or more variant of blaCTX−M and blaNDM genes. ST-11, ST-38, ST-39, ST-147 and ST-231were other STs also detected here and among the Sudanese isolates which could be explained by geographical proximity and traffic between the two countries leading to isolate exchange. ST-101, ST-147, ST-231and ST-383 are considered emerging high-risk clones that have been identified in different parts of the world with the potential to become a global, persistent public health threat [48–51].
Conclusions
Levels of antibiotic resistance are increasing in K. pneumoniae in Egypt. Between 2019 and 2021, we detected a wide range of genotypes circulating within AMUH with ST-101 and ST-383 persisting from 2019 to 2021 which could be a risk factor for nosocomial outbreaks. This is particularly concerning because of the MDR and sometimes XDR status of some of the isolates in these two genotypes and their cargo of resistance genes. We recommend continuous surveillance of resistance levels and genotypes among K. pneumoniae isolates to better inform infection control plans, in addition to a strict implementation of antimicrobial stewardship guidelines to avoid further resistance development.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
Not applicable.
Abbreviations
- AMR
Anti-microbial resistance
- MDR
Multi-drug resistance
- XDR
Extensive drug resistance
- PDR
Pan drug resistance
- ESBL
Extended-spectrumβ-Lactamase
- KP
Klebsiella pneumoniae
- KPC
Klebsiella pneumoniae carbapenemase
- MICs
Minimum inhibitory concentrations
- MLST
Multilocus sequence typing
- STs
Sequence types
- SLV
Single locus variant
- DLV
Double locus variant
- TLV
Triple locus variant
- BAL
Broncho-alveolar lavage
- CSF
Cerebrospinal fluid
- MIN
Minimum
- MAX
Maximum
- SD
Standard deviation
- SDD
Susceptible dose dependent
- S
Sensitive
- I
Intermediate
- R
Resistant
- WGS
Whole genome sequencing
- CIP
Ciprofloxacin
- NOR
Norfloxacin
- DOX
Doxycycline
- NTF
Nitrofurantoin
- AMP
Ampicillin
- SAM
Ampicillin/sulbactam
- TZP
Piperacillin/tazobactam
- AMC
Amoxicillin/clavulanic acid
- CAZ
Ceftazidime
- FEP
Cefepime
- CRO
Ceftriaxone
- CTX
Cefotaxime
- SCF
Cefoperazone-sulbactam
- ETP
Ertapenem
- AZM
Azithromycin
- AK
Amikacin
- GMN
Gentamicin
- MEM
meropenem
- IMP
Imipenem
- CZA
Ceftazidime-avibactam
- SXT
Trimethoprim/sulfamethoxazole
- AMUH
Alexandria Main University Hospital
- ATCC
American type culture collection
Author contributions
PG: performed the experiments, analyzed the data and wrote the first draft, MA: designed the experiments and reviewed the manuscript, BE: analyzed the data and critically reviewed the manuscript, HO: designed the experiments and reviewed the manuscript, AA: conceptualized the study, analyzed the data, wrote the first draft and critically reviewed the manuscript. All authors approved of the final version of the manuscript.
Funding
The whole genome sequencing work was made possible by funding from the UEA Vice-Chancellor’s Global Challenges Research Fellowship.
Data availability
Genome assemblies and raw sequence data from SRA were deposited in NCBI’s Assembly database under BioProject accession number PRJNA1071125. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1071125.
Declarations
Ethics approval and consent to participate
All methods were carried out in compliance with the corresponding regulations and guidelines. The study was approved by the ethics committee at the Faculty of Medicine, Alexandria University (IRB No.: 00012098). The need of informed consent was waived by the ethics committee at the Faculty of Medicine, Alexandria University (IRB No.: 00012098), since all isolates were obtained as part of the routine care of the patients at the discretion of the treating physician. The isolates were included in the study anonymously and no patient intervention was decided based on the results of the study.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Reygaert WC. An overview of the antimicrobial resistance mechanisms of bacteria. AIMS Microbiol. 2018;4(3):482–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Watkins RR, Bonomo RA. Overview: global and local impact of antibiotic resistance. Infect Disease Clin. 2016;30(2):313–22. [DOI] [PubMed] [Google Scholar]
- 3.Zaman SB, et al. A review on antibiotic resistance: alarm bells are ringing. Cureus. 2017;9(6):e1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Magiorakos AP, et al. Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin Microbiol Infect. 2012;18(3):268–81. [DOI] [PubMed] [Google Scholar]
- 5.Podschun R, Ullmann U. Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin Microbiol Rev. 1998;11(4):589–603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chung PY. The emerging problems of Klebsiella pneumoniae infections: carbapenem resistance and biofilm formation. FEMS Microbiol Lett, 2016. 363(20). [DOI] [PubMed]
- 7.Bassetti M, et al. Multidrug-resistant Klebsiella pneumoniae: challenges for treatment, prevention and infection control. Expert Review of Anti-infective Therapy; 2018. p. 16. [DOI] [PubMed]
- 8.Lester CH. Fuursted K. Global spread of New Delhi metallo-beta-lactamase 1. Lancet Infect Dis. 2010;10(12):829–30. Walsh TR. [DOI] [PubMed] [Google Scholar]
- 9.Shibl A, Memish Z, Khader SA. Al-Agamy M, Memish Z, Senok A, Khader SA Assiri A. The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis 2013;17(12):e1130–3 [DOI] [PubMed]
- 10.Livermore DM. Multiresistant gram-negative bacteria: the role of high-risk clones in the dissemination of antibiotic resistance. FEMS Microbiol Rev. 2011;35(5):736–55. Turton JF. [DOI] [PubMed] [Google Scholar]
- 11.Alvarez-Uria G, et al. Global forecast of antimicrobial resistance in invasive isolates of Escherichia coli and Klebsiella pneumoniae. Int J Infect Dis. 2018;68:50–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Abdelwahab R et al. Antimicrobial Resistance and Comparative Genome Analysis of Klebsiella pneumoniae strains isolated in Egypt. Microorganisms, 2021. 9(9). [DOI] [PMC free article] [PubMed]
- 13.El-Domany RA, et al. Analysis of the correlation between antibiotic resistance patterns and virulence determinants in pathogenic Klebsiella pneumoniae isolates from Egypt. Microb Drug Resist. 2021;27(6):727–39. [DOI] [PubMed] [Google Scholar]
- 14.Sherif M, et al. Whole-genome sequencing of Egyptian multidrug-resistant Klebsiella pneumoniae isolates: a multi-center pilot study. Eur J Clin Microbiol Infect Dis. 2021;40(7):1451–60. [DOI] [PubMed] [Google Scholar]
- 15.Basic laboratory procedures in clinical bacteriology / J. Vandepitte … et al.]. 1991, World Health Organization: Geneva.
- 16.AMR NG. .H.R.U.o.G.S.o., whole-genome sequencing as part of national and international surveillance programmes for antimicrobial resistance: a roadmap. BMJ Glob Health, 2020. 5(11). [DOI] [PMC free article] [PubMed]
- 17.Howard DH. The preservation of bacteria by freezing in glycerol broth. J Bacteriol. 1956;71(5):625–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.CLSI. Performance Standards for Antimicrobial Susceptibility Testing. 31st ed. CLSI supplement M100. Clinical and Laboratory Standards Institute; 2021 [DOI] [PMC free article] [PubMed]
- 19.Ramadan AA, et al. Novel blaCTX-M variants and genotype-phenotype correlations among clinical isolates of extended spectrum beta lactamase-producing Escherichia coli. Sci Rep. 2019;9(1):4224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bankevich A, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19(5):455–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Davis JJ, et al. The PATRIC Bioinformatics Resource Center: expanding data and analysis capabilities. Nucleic Acids Res. 2020;48(D1):D606–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parks DH, et al. CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes. Genome Res. 2015;25(7):1043–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Priyam A, et al. Sequenceserver: a modern graphical user interface for Custom BLAST databases. Mol Biol Evol. 2019;36(12):2922–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Camacho C, et al. BLAST+: architecture and applications. BMC Bioinformatics. 2009;10(1):421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bortolaia V, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tatusova T, et al. NCBI prokaryotic genome annotation pipeline. Nucleic Acids Res. 2016;44(14):6614–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hunt M, et al. ARIBA: rapid antimicrobial resistance genotyping directly from sequencing reads. Microb Genom. 2017;3(10):e000131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Deng W, Nickle DC, Liu Y, Kosakovsky Pond SL. Maust BS, Nickle DC, Learn GH, Liu Y, Heath L, Kosakovsky Pond SL, Mullins JI. DIVEIN: a web server to analyze phylogenies, sequence divergence, diversity, and informative sites. BioTechniques 2010;48(5):405–8 [DOI] [PMC free article] [PubMed]
- 29.Feil EJ, Li BC, Aanensen DM, Hanage WP, Spratt BG. eBURST: inferring patterns of evolutionary descent among clusters of related bacterial genotypes from multilocus sequence typing data. J Bacteriol. 2004;186(5):1518–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kaas RS, et al. Solving the Problem of comparing whole bacterial genomes across different sequencing platforms. PLoS ONE. 2014;9(8):e104984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu M, et al. The differences and similarities between two-sample T-Test and paired T-Test. Shanghai Arch Psychiatry. 2017;29(3):184–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Abril D, et al. Within patient genetic diversity of blaKPC harboring Klebsiella pneumoniae in a Colombian hospital and identification of a new NTEKPC platform. Sci Rep. 2021;11(1):21409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maseda E, et al. Bugs, hosts and ICU environment: countering pan-resistance in nosocomial microbiota and treating bacterial infections in the critical care setting. Rev Esp Anestesiol Reanim. 2014;61(3):e1–19. [DOI] [PubMed] [Google Scholar]
- 34.Effah CY, et al. Klebsiella pneumoniae: an increasing threat to public health. Ann Clin Microbiol Antimicrob. 2020;19(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bi W, et al. Extensively drug-resistant Klebsiella pneumoniae causing nosocomial bloodstream infections in China: Molecular Investigation of Antibiotic Resistance determinants, informing therapy, and clinical outcomes. Front Microbiol. 2017;8:1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shebl E, et al. The outcome of hospital-acquired pneumonia in patients admitted for long-term care according to the antibiotic duration. Egypt J Chest Disease Tuberculosis. 2019;68:378–82. [Google Scholar]
- 37.Hassuna NA, et al. Extensively-drug resistant Klebsiella pneumoniae recovered from neonatal Sepsis cases from a major NICU in Egypt. Front Microbiol. 2020;11:1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Al-Baz AA, et al. Prevalence and Antibiotic Resistance profiles of Carbapenem-Resistant Klebsiella Pneumoniae isolated from Tertiary Care Hospital, Egypt. Egypt J Hosp Med. 2022;88(1):2883–90. [Google Scholar]
- 39.Sharma A, et al. Changing Trend in the Antibiotic Resistance Pattern of Klebsiella Pneumonia isolated from endotracheal aspirate samples of ICU patients of a Tertiary Care Hospital in North India. Cureus. 2023;15(3):e36317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Global antibiotic use during the COVID-19 pandemic: analysis of pharmaceutical sales data from 71 countries, 2020–2022 eClinicalMedicine, 2023. 57. [DOI] [PMC free article] [PubMed]
- 41.Bulman ZP, et al. Genomic Features Associated with the degree of phenotypic resistance to Carbapenems in Carbapenem-resistant Klebsiella pneumoniae. mSystems. 2021;6(5):e0019421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kowalska-Krochmal B, Dudek-Wicher R. The Minimum Inhibitory concentration of antibiotics: methods, interpretation, clinical relevance. Pathogens, 2021. 10(2). [DOI] [PMC free article] [PubMed]
- 43.Carattoli A. An ertapenem-resistant extended-spectrum-betalactamase- producing Klebsiella pneumoniae clone carries a novel OmpK36 porin variant. Antimicrob Agents Chemother. 2010;54(10):4178–84. Mancini C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mauri C et al. The Revival of Aztreonam in Combination with Avibactam against Metallo-β-Lactamase-Producing Gram-Negatives: A Systematic Review of In Vitro Studies and Clinical Cases Antibiotics (Basel), 2021. 10(8). [DOI] [PMC free article] [PubMed]
- 45.De Koster S, et al. Diversity in the characteristics of Klebsiella pneumoniae ST101 of Human, Environmental, and animal origin. Front Microbiol. 2022;13:838207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo L, et al. Nosocomial outbreak of OXA-48-Producing Klebsiella pneumoniae in a Chinese hospital: Clonal Transmission of ST147 and ST383. PLoS ONE. 2016;11(8):e0160754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osman EA et al. Klebsiella pneumonia in Sudan: Multidrug Resistance, polyclonal dissemination, and virulence. Antibiot (Basel), 2023. 12(2). [DOI] [PMC free article] [PubMed]
- 48.Roe CC et al. Diversity, virulence, and Antimicrobial Resistance in isolates from the newly emerging Klebsiella pneumoniae ST101 lineage. Front Microbiol, 2019. 10. [DOI] [PMC free article] [PubMed]
- 49.Palmieri M et al. Genomic evolution and local epidemiology of Klebsiella pneumoniae from a major hospital in Beijing, China, over a 15 year period: dissemination of known and novel high-risk clones. Microb Genom, 2019. 7(6). [DOI] [PMC free article] [PubMed]
- 50.Park J, et al. Dissemination of the high-risk cloneST147 carbapenem-resistant klebsiella pneumoniae from a local tertiary care hospital in the Republic of Korea. Ann Clin Microbiol Antimicrob. 2023;22(1):76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teo JQ, et al. Genomic surveillance of Carbapenem-resistant Klebsiella pneumoniae from a Major Public Health Hospital in Singapore. Microbiol Spectr. 2022;10(5):e0095722. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Genome assemblies and raw sequence data from SRA were deposited in NCBI’s Assembly database under BioProject accession number PRJNA1071125. https://www.ncbi.nlm.nih.gov/bioproject/PRJNA1071125.