Abstract
A CD4-independent version of the X4 human immunodeficiency virus type 1 (HIV-1) HXBc2 envelope (Env) protein, termed 8x, mediates infection of CD4-negative, CXCR4-positive cells, binds directly to CXCR4 in the absence of CD4 due to constitutive exposure of a conserved coreceptor binding site in the gp120 subunit, and is more sensitive to antibody-mediated neutralization. To study the relationships between CD4 independence, neutralization sensitivity, and exposure of CD4-induced epitopes associated with the coreceptor binding site, we generated a large panel of Env mutants and chimeras between 8x and its CD4-dependent parent, HXBc2. We found that a frameshift mutation just proximal to the gp41 cytoplasmic domain in 8x Env was necessary but not sufficient for CD4 independence and led to increased exposure of the coreceptor binding site. In the presence of this altered cytoplasmic domain, single amino acid changes in either the 8x V3 (V320I) or V4/C4 (N386K) regions imparted CD4 independence, with other changes playing a modulatory role. The N386K mutation resulted in loss of an N-linked glycosylation site, but additional mutagenesis showed that it was the presence of a lysine rather than loss of the glycosylation site that contributed to CD4 independence. However, loss of the glycosylation site alone was sufficient to render Env neutralization sensitive, providing additional evidence that carbohydrate structures shield important neutralization determinants. Exposure of the CD4-induced epitope recognized by monoclonal antibody 17b and which overlaps the coreceptor binding site was highly sensitive to an R298K mutation at the base of the V3 loop and was often but not always associated with CD4 independence. Finally, while not all neutralization-sensitive Envs were CD4 independent, all CD4-independent Envs exhibited enhanced sensitivity to neutralization by HIV-1-positive human sera, indicating that the humoral immune response can exert strong selective pressure against the CD4-independent phenotype in vivo. Whether this can be used to advantage in designing more effective immunogens remains to be seen.
The entry of human immunodeficiency virus type 1 (HIV-1) into cells requires that a membrane fusion reaction occur between the viral and cellular membranes. As for other enveloped viruses, this function is mediated by a virally encoded type 1 membrane protein (14). In the case of HIV-1, receptor binding and fusion are mediated by the Env protein, a trimeric protein in which each monomer consists of a surface subunit (gp120) noncovalently associated with the gp41 transmembrane subunit (41). Binding to CD4 triggers conformational changes in the gp120 subunit that enable it to efficiently interact with a viral coreceptor (22, 36, 40), most often the chemokine receptors CCR5 and CXCR4 (6). Coreceptor binding is thought to lead to the final conformational changes in Env needed for the membrane fusion reaction (7).
Primate lentiviruses that short-circuit the normal entry pathway by interacting directly with the coreceptors have been described (8, 10–12, 19, 29). As a result, these viruses can infect CD4-negative cells provided that they express the appropriate coreceptor, thereby broadening viral tropism in vitro and perhaps in vivo as well. CD4 independence on CCR5 is a particularly common feature of primary simian immunodeficiency virus (SIV) and HIV-2 strains (10, 11, 28), suggesting that CCR5 may have served as the primordial receptor for the primate lentiviruses. While all primary HIV-1 strains studied to date require CD4 to infect cells efficiently, HIV-1 can be rendered CD4 independent through in vitro passaging. Three CD4-independent HIV-1 strains have been identified to date, often as a result of relatively subtle mutations, indicating that the structure of HIV-1 Env can be altered so as to overcome the CD4 requirement (8, 16, 19, 21). Why CD4-independent, primary strains of HIV-1 have not been identified to date remains an open question.
Previous studies described the generation of a CD4-independent variant of HIV-1 HXBc2 termed 8x (16, 21). This CD4-independent Env mediates infection of CD4-negative, CXCR4-positive cells. It was found that mutations in 8x that rendered it CD4 independent resulted in the stable, constitutive exposure of a chemokine receptor binding site in gp120, enabling it to bind directly to CXCR4 (16). In addition, the 8x virus was considerably more sensitive to neutralization by HIV-1-positive human sera. In the present study, we have more fully mapped the determinants in 8x that render it CD4 independent and have investigated the relationship between CD4 independence, neutralization sensitivity, and the exposure of CD4-induced antigenic epitopes that overlap the coreceptor-binding site in gp120. We identified specific residues in both the V3 and V4/C4 regions of 8x gp120 that contribute to the CD4-independent phenotype. In addition, a frameshift (FS) mutation in the cytoplasmic domain of 8x gp41 and a conservative Arg to Lys mutation at the base of the V3 loop contributed to CD4 independence and influenced exposure of CD4-induced determinants in gp120. HXBc2 Envs made CD4 independent by the introduction of regions or mutations from 8x were invariably more sensitive to neutralization by HIV- 1-positive human sera, suggesting that the humoral immune response may provide strong selective pressure against the CD4-independent phenotype in vivo. The study of CD4-independent viruses provides a means to dissect the steps leading to Env-mediated membrane fusion by genetically identifying residues in Env that presumably subserve the role of CD4 in triggering conformational changes. In addition, CD4-independent viruses provide a way to study the evolution of receptor use by primate lentiviruses in vivo and, through the identification of neutralization-sensitive Env proteins, suggest possible ways to modify Env so as to generate more effective immunogens.
MATERIALS AND METHODS
Plasmids and viruses.
Parental 8x and HXBc2 Envs were expressed in pSP73 (Promega) as described previously (16). HXBc2-based chimeras containing the entire 8x gp120 or the 8x V1/V2, V3, or V4/C4 domains either alone or with the 8x gp41 were constructed using KpnI, DraIII, StuI, Bsu36I, BsaBI and BamHI sites as described previously (21). Site-directed mutations in V3, V4/C4, and gp41 were made using the Quickchange site-directed mutagenesis kit (Stratagene) by using conditions recommended by the manufacturer. Human CXCR4 and CD4 were expressed in pCDNA3 (Invitrogen), while the luciferase gene was expressed in pGEM2 (Promega) under control of the T7 promoter. Chimeras between 8x and JRFL were made by using the conserved BsaBI site at nucleotide 7673 of the HXBc2 sequence. Finally, the HIV-1 ADA Env protein was made CD4 independent through the introduction of two previously described amino acid changes, R190S and S197N (19).
Cell-cell fusion assay.
This assay has been described in more detail elsewhere (32). Briefly, effector quail QT6 cells were infected with recombinant vaccinia virus vTF1.1 expressing T7 polymerase (1) and were transfected with Env constructs via CaPO4. Target QT6 cells were transfected with CXCR4 and/or CD4 plasmids under control of the cytomegalovirus promoter and the luciferase gene under control of the T7 promoter. The next day, effector cells were added to target cells and allowed to fuse for at least 7 h. Fusion was measured by quantification of luciferase in cell lysates. For neutralization experiments, sera from HIV-1-positive individuals or seronegative controls were incubated with effector cells at the indicated dilutions for 1 h prior to addition to target cells. Serum was present at the same dilution during the cell-cell fusion assay, and neutralization was scored as a percent reduction in luciferase activity.
Surface expression and 17b binding assays.
293T cells were transfected with a plasmid containing the Env of interest and then were incubated overnight at 37°C. The next day, Env-bearing cells were washed once with phosphate-buffered saline. Cells were resuspended in binding buffer (50 mM HEPES [pH 7.4], 2 mM magnesium chloride, 2 mM calcium chloride, 0.5% bovine serum albumin) and were incubated with either 1 μg of 17b/106 cells or with HIV-positive human sera (1:100 dilution incubated with 106 cells) for 20 min at room temperature. Cells were washed once with phosphate-buffered saline and were resuspended in 50 μl of binding buffer. Iodinated anti-human immunoglobulin G (100,000 cpm in 50 μl of binding buffer) was added to the cells and was incubated for 1 h at room temperature. Cells were collected onto Brandel grade GF/B filters with wash buffer (the same as binding buffer plus 150 mM sodium chloride and without bovine serum albumin) using a cell harvester. Filters were counted using a Wallac Wizard 1470 automatic gamma counter. Percent binding was determined by dividing the counts from the filters by the input radioactivity.
RESULTS
Regions of gp120 responsible for CD4 independence.
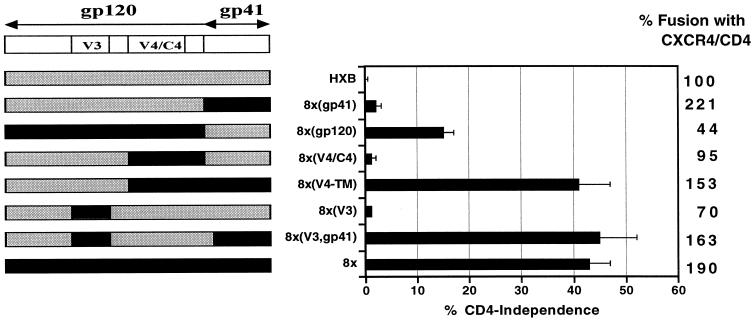
A CD4-independent variant of the prototype X4 HIV-1 HXBc2 Env protein termed 8x that mediates infection and cell-cell fusion on several CXCR4-positive, CD4-negative cell types has been described previously (16, 21). In addition, 8x Env was neutralization sensitive and exhibited constitutive exposure of CD4-induced epitopes recognized by monoclonal antibodies 17b and 48d (16). Using chimeras between HXBc2 and 8x, it was found that the V4/C4 and gp41 regions of 8x contributed to the CD4-independent phenotype (21). Thus, CD4 independence mapped, in part, outside regions of Env previously implicated in determining coreceptor specificity (i.e., V1/V2 and V3) and potentially within a highly conserved area of the gp120 core thought to comprise a chemokine receptor binding site (31). Previous experiments showing the stable exposure of the 17b epitope in 8x gp120, which overlaps this region, are consistent with the idea that CD4-independent Envs exist in a partially triggered conformation.
To identify regions of the 8x Env that conferred CD4 independence, neutralization sensitivity, and exposure of the 17b epitope, we introduced additional portions of the 8x Env into the CD4-dependent HXBc2 protein and tested the abilities of the chimeras to mediate fusion with cells expressing CXCR4 alone or in combination with CD4. The chimeras are specified by the region(s) of 8x introduced into an HXBc2 background. Thus, 8x(V3) refers to a chimera in which the V3 region of 8x is introduced into an HXBc2 background, 8x(gp41) refers to a chimera containing the HXBc2 gp120 and 8x gp41 subunits, while 8x(V3,gp41) refers to a chimera containing both the 8x V3 loop and gp41 subunits. Cell-cell fusion assays were employed rather than virus infection assays because we have been unable to efficiently pseudotype the 8x Env protein onto virus particles.
As previously shown, neither the 8x gp41 nor the V1/V2, V3, or V4/C4 regions alone conferred CD4 independence on HXBc2 (Fig. 1; 21). A chimera containing the entire 8x gp120 subunit with the HXBc2 gp41 mediated a modest degree of CD4 independence, though the fusion activity of this chimera in the presence of CD4 was reduced. The other chimeric Env proteins efficiently elicited membrane fusion when CD4 was present, indicating that they were functional (Fig. 1). However, the combination of the V3 region of 8x along with the 8x gp41 subunit [8x(V3,gp41)] conferred CD4 independence on HXBc2 that was comparable to that seen with 8x Env. Similarly, as reported previously (21), the V4/C4 region of 8x in conjunction with the 8x gp41 domain conferred CD4 independence equally well (Fig. 1). The 8x V1/V2 region did not impart CD4 independence, either with or without the 8x gp41 (data not shown and reference 21). These results indicated that both the V3 and V4/C4 domains of the 8x protein contained important determinants for CD4 independence but that these functioned only in the presence of the 8x gp41 subunit.
FIG. 1.
Regions in 8x gp120 that confer CD4 independence. Cell-cell fusion assays were performed to identify regions required for CD4 independence as described in Materials and Methods. (Left panel) Env chimeras containing regions from the CD4-independent 8x (shown in black) and the parental CD4-dependent HXBc2 (shown in gray) were generated. (Right panel) Fusion efficiency relative to wild-type HXBc2 for each construct in the presence of CD4 is shown by the column of numbers on the right side of the figure. Shown is the extent of CD4-independent fusion graphed as a percentage of maximal fusion seen with CD4 for each Env chimera ± the standard error of the mean.
The contribution of gp41 to CD4 independence.
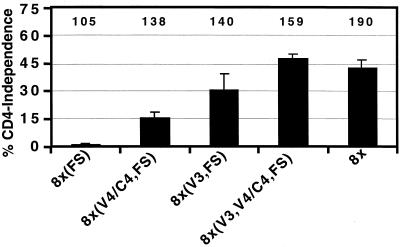
There are six mutations in the 8x gp41 ectodomain, two of which have been observed in other Env clones derived from the HIV-1 IIIB family. In addition, the 8x gp41 has a single nucleotide deletion that results in an FS at position 706 and a truncated cytoplasmic domain of only 27 amino acids. While truncations in the cytoplasmic domain of SIV Env proteins derived from viruses grown in human cells are relatively common, truncations of the HIV-1 Env cytoplasmic domain are unusual. In addition, the 8x gp41 also has two point mutations distal to the FS in the “nonsense” region of the tail that changes a Glu to Lys at position 706 and a Pro to His at position 723 (21). To determine if the 8x FS mutation contributed to CD4 independence, we introduced the identical mutation in HXBc2 Env [8x(FS)]. The resulting 8x(FS) protein was strictly CD4 dependent (Fig. 2). We then introduced the 8x V3 region [8x(V3,FS)], the 8x V4/C4 region [8x(V4/C4,FS)], or both domains [8x(V3,V4/C4,FS)] into the HXBc2 Env containing the gp41 FS. All three of these chimeras were CD4 independent, with the 8x(V3,V4/C4,FS) chimera giving maximal fusion activity in the absence of CD4 (Fig. 2). All Envs were competent for fusion in the presence of CD4 (Fig. 2). The FS mutation did not affect surface expression of the proteins (data not shown) but rather influenced exposure of the 17b epitope as discussed below. Finally, we introduced a stop codon in gp41 at the site of the FS mutation. We found that constructs bearing this premature stop codon functioned identically to those with the FS mutation (data not shown). Thus, truncation of the gp41 cytoplasmic domain either by a premature stop codon or by an FS mutation in combination with either the V3 or V4 regions of 8x gp120 was sufficient to confer the CD4-independent phenotype on HXBc2.
FIG. 2.
The transmembrane region of 8x is required for CD4 independence. Cell-cell fusion assays were performed to determine the contribution of the FS mutation present in 8x gp41 to CD4 independence. On the left, percent CD4 independence refers to the amount of fusion observed in the absence of CD4 relative to the amount of fusion seen in its presence ± the standard error of the mean. The numbers at the top of the graph indicate the amount of fusion observed in the presence of CD4 and CXCR4 relative to that seen with the HXBc2 Env protein.
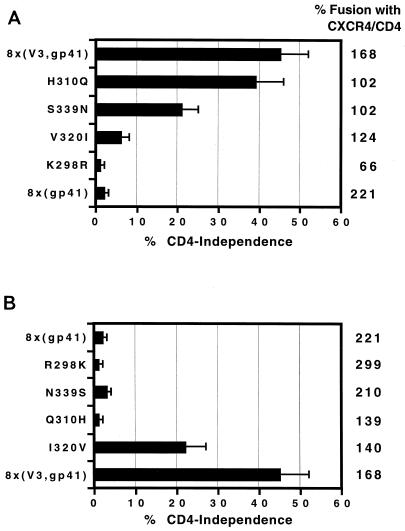
Determinants in the V3 region important for CD4 independence.
The V3 loop region of 8x contains four mutations (R298K, Q310H, I320V, and N339S) relative to the parental HXBc2 (21). To determine which mutations were necessary for CD4 independence, we used the 8x(V3,gp41) chimera as a template. Each residue was changed individually back to the wild-type sequence, and the effects on cell-cell fusion were determined (Fig. 3A). Then, to determine which residues were sufficient for CD4 independence, we introduced each mutation individually into a protein containing the HXBc2 gp120 subunit and the 8x gp41 domain [8x(gp41)] (Fig. 3B).
FIG. 3.
Determinants in the V3 region important for CD4 independence. (A) To identify residues required for CD4 independence, the 8x(V3,gp41) chimera was used. Each of the four V3 loop mutations found in 8x Env was changed individually back to the wild-type HXBc2 sequence, and their effects on fusion in the presence of CD4 and CXCR4 (far right column of numbers) or in the absence of CD4 (graphed as percent CD4 independence) are shown. The percent CD4 independence represents the extent of fusion observed in the absence of CD4 divided by the extent of fusion in its presence for each individual construct ± the standard error of the mean. (B) To identify residues in the V3 region sufficient for CD4-independent membrane fusion, individual 8x mutations were introduced into the 8x(gp41) chimera and the extent of fusion in the presence and absence of CD4 was determined as in panel A.
When 8x mutations were removed individually from the 8x(V3,gp41) chimera, K298R eliminated CD4-independent fusion, V320I almost completely eliminated CD4 independence, S339N had a moderate effect, and H310Q had a negligible effect on CD4-independent membrane fusion (Fig. 3A). When 8x mutations were introduced into the 8x(gp41) chimera, I320V was the only mutation that was sufficient to confer CD4 independence, reaching a level approximately half that of the 8x Env protein (Fig. 3B). Therefore, the R298K and I320V mutations appear to be the major contributors to the CD4-independent phenotype in the 8x V3 region.
Determinants in the V4/C4 region important for CD4 independence.
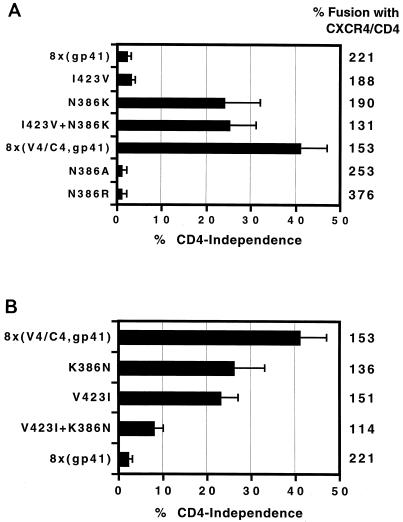
The V4/C4 region of the 8x Env contains four point mutations (N386K, E403K, I423V, and K429E) and a five-amino-acid deletion (WFNST at position 395 to 399) compared to the parental HXBc2. Among these, E403K, K429E, and the deletion are present in other members of the IIIB virus family, making it less likely that they substantially impact CD4 independence (21). Therefore, we examined the effects of the two novel mutations, N386K and I423V, on CD4-independent fusion. We confirmed that the N386K mutation resulted in the loss of a carbohydrate structure, as this mutation resulted in a 2-to-3-kDa shift in apparent molecular mass as judged by sodium dodecyl sulfate-polyacrylamide gel electrophoresis and Western blotting (data not shown), consistent with an earlier study demonstrating that all potential N-linked carbohydrate addition sites in gp120 are in fact utilized (24). To determine if either mutation was sufficient for CD4-independent fusion activity, we introduced I423V or N386K into the 8x(gp41) chimera (Fig. 4A). The N386K mutation alone was sufficient to confer nearly full CD4 independence on the 8x(gp41) chimera, while the I423V mutation had little effect either alone or in combination with N386K. To determine if either mutation was necessary for CD4 independence, we changed each residue back to the HXBc2 sequence in the 8x(V4/C4,gp41) chimera, which contained these two mutations (Fig. 4B). Both the K386N and V423I mutations reduced CD4 independence about a third, while changing both of these residues back to the HXBc2 sequence further reduced CD4-independent fusion. Therefore, the N386K mutation is the most important determinant for CD4 independence in the V4/C4 region, although the I423V mutation also contributes to this phenotype.
FIG. 4.
Determinants in the V4 region important for CD4 independence. (A) To identify residues in the V4 region that were sufficient for CD4 independence, the two novel mutations (I423V and N386K) found in 8x were introduced individually or together into the 8x(gp41) chimera. To determine if it was the loss of a glycosylation site at position 386 or the presence of a Lys residue that contributed to CD4 independence, additional substitutions at this site were made, either with or without the I423V mutation. The extent of fusion in the presence of CD4 is shown (relative to HXBc2 Env) by the column of numbers on the far right, while the panel depicts the extent of fusion observed in the absence of CD4 relative to the amount of fusion observed in its presence for each construct ± the standard error of the mean. (B) To identify residues in the V4 region required for CD4-independent membrane fusion, the 8x(V4,gp41) chimera was used. Residues 386 and 423 were changed back to the wild-type HXBc2 sequence alone or in combination, and the extent of fusion was observed in the presence or absence of CD4, determined as in panel A.
Because the N386K mutation resulted in the removal of a highly conserved N-linked glycosylation site in the vicinity of the chemokine receptor binding site in Env (31), we substituted additional residues at this position to determine whether it was the loss of glycosylation or the presence of a lysine that was responsible for the CD4-independent phenotype (Fig. 4A). Introduction of an alanine at position 386 (N386A) failed to confer CD4 independence on 8x(gp41), indicating that loss of the glycosylation site at this position did not account for CD4 independence. Interestingly, introduction of a different, positively charged amino acid at this position (N386R) also failed to confer CD4-independent fusion. Thus it appears that the specific acquisition of a lysine at this position is responsible for the CD4-independent phenotype.
Regions in 8x responsible for neutralization sensitivity.
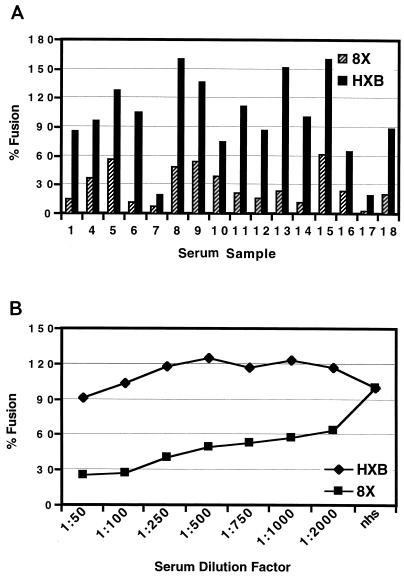
In addition to being CD4 independent, the 8x virus is approximately one log more sensitive to neutralization by HIV-positive human sera (16). We have found that a number of CD4-independent SIV strains are also neutralization sensitive relative to closely related, CD4-dependent virus strains (B. Puffer and R. W. Doms, unpublished data). To determine if CD4 independence and neutralization sensitivity are linked, we examined the Env proteins used to map determinants for CD4 independence for their sensitivity to neutralization. Because our previous study looked at neutralization by only two human serum samples, we first determined if the neutralization-sensitive phenotype of 8x would be manifest using a larger panel of HIV immune sera by the cell-cell fusion assay. We found that the 8x Env protein was more neutralization sensitive to every serum tested in this assay (Fig. 5A). A representative serum sample (sample 15 in Fig. 5A) was then tested for the ability to neutralize fusion mediated by the 8x and HXBc2 Env proteins over a broad concentration range. The 8x Env was more sensitive to neutralization than HXBc2 Env at all serum concentrations tested (Fig. 5B). In subsequent experiments, we used a 1:100 dilution of this serum to determine if the various chimeric and mutant Envs were neutralization sensitive.
FIG. 5.
Neutralization sensitivity of 8x Env. (A) The ability of 16 different HIV-positive human sera to inhibit HXBc2 or 8x Env-mediated cell-cell fusion was determined at a 1:100 dilution. The graph is a representative experiment, with 100% fusion being the activity observed in the presence of normal human serum. (B) Serum sample no. 15 was tested for its ability to block 8x and HXBc2-mediated membrane fusion at various concentrations. The percent fusion is based on the ability of each Env to elicit fusion in the presence of HIV-negative human serum at the same concentration. The graph is a representative experiment.
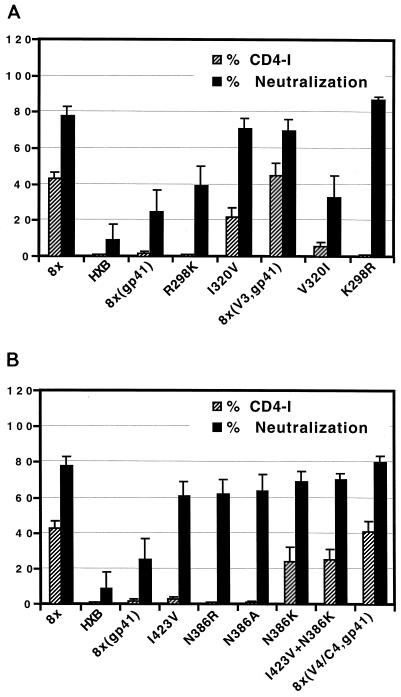
We found that CD4-independent chimeric Env proteins containing the 8x gp41 and either the V3 [8x(V3,gp41)] or V4/C4 [8x(V4/C4,gp41)] regions of 8x Env were as neutralization sensitive as the full-length 8x Env protein (Fig. 6). Thus, changes in both the V3 and V4/C4 regions influence neutralization sensitivity as well as CD4 independence in the context of the 8x gp41. We next examined the neutralization sensitivity of the various point mutants used to map determinants for CD4 independence by introducing them into the 8x(gp41) chimera (Fig. 6A). The R298K mutation in the V3 region of 8x Env did not confer CD4 independence and only slightly increased the neutralization sensitivity of this Env. By contrast, the I320V mutation, which conferred partial CD4 independence, had a more profound effect, increasing neutralization sensitivity to a level comparable with that of 8x. The reciprocal mutants were also examined using the 8x(V3,gp41) chimera, which is relatively CD4 independent and neutralization sensitive (Fig. 6A). Changing Val 320 back to the Ile in the HXBc2 sequence markedly reduced CD4 independence and made the Env protein more resistant to neutralization. However, changing Lys 298 back to the Arg in the HXBc2 sequence, which ablated CD4 independence, resulted in an Env that remained neutralization sensitive. Therefore, within the V3 region of 8x the I320V mutation is largely responsible for increased sensitivity to neutralization by HIV-positive human sera.
FIG. 6.
Relationship between CD4 independence and neutralization sensitivity. Fusion assays with the indicated Env proteins were performed in the presence of a 1:100 dilution of either normal human serum or HIV-positive human serum. The efficiency with which each Env mediated CD4-independent membrane fusion (derived from Fig. 2 to 4) is shown for convenience. Chimeras containing the indicated 8x domains and the V3 region point mutations described in Fig. 3 are shown in panel A, while the V4 point mutations described in Fig. 4 are shown in panel B.
Mutations in the V4/C4 region of 8x were also examined for their ability to modulate neutralization sensitivity (Fig. 6B). When introduced into the 8x(gp41) chimera, all mutations at positions 423 and 386 increased sensitivity to neutralization and did so to similar degrees. However, as noted previously, only the N386K mutation was capable of conferring CD4 independence. Mutations N386R and N386A did not impart CD4 independence but did render the Env more sensitive to neutralization (Fig. 6B). Thus, loss of an N-linked glycosylation site at position 386 results in enhanced neutralization sensitivity but not necessarily in CD4 independence.
Exposure of the 17b determinant.
A region known as the bridging sheet, which connects the inner and outer domains of gp120, has been shown to play a critical role in CCR5 binding. Due to its highly conserved nature, this domain likely interacts with CXCR4 as well (31). The 17b monoclonal antibody (MAb) recognizes a CD4-induced epitope that largely overlaps this putative coreceptor binding site (20, 35). It has been argued that detection of this epitope can be used as a surrogate for exposure of the chemokine receptor binding site (16, 31). As we have shown, 17b binds well to soluble 8x gp120 in the absence of CD4 whereas HXBc2 does not (16). To determine to what extent 17b reactivity correlated with CD4 independence and neutralization sensitivity, we developed a cell-surface binding assay in which 17b was incubated with cells expressing the desired Env protein for 20 min, after which bound 17b was detected with an iodinated secondary antibody. This assay was used because, once bound, 17b exhibits a relatively fast off rate from 8x relative to HXBc2 gp120, most likely due to a mutation involving a contact site for this antibody (16). We found that very brief wash steps could be employed with this cell-surface binding assay, whereas the additional wash steps required for fluorescence-activated cell sorter analysis resulted in a considerable loss of signal. Even with the cell-surface binding assay, however, we are probably underestimating the amount of 17b bound to the 8x protein.
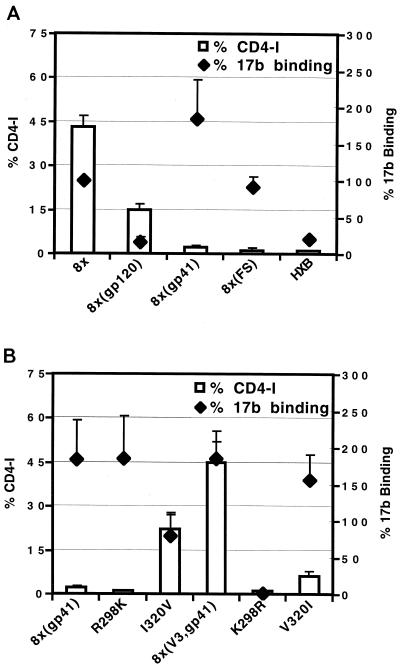
We found that cells expressing the 8x Env bound 17b approximately fivefold better than cells expressing HXBc2 Env, consistent with our earlier findings using soluble gp120 (Fig. 7A). Surprisingly, exposure of the 17b epitope was found to be largely influenced by changes in gp41. As shown in Fig. 7A, 17b bound well to 8x(gp41) but poorly to the 8x(gp120) chimera. Since the FS in the 8x gp41 cytoplasmic tail contributes to CD4 independence (Fig. 2), we measured 17b binding to an HXBc2 Env that contained only this mutation, 8x(FS). Although this protein remained CD4 dependent, it bound 17b as well as the 8x Env (Fig. 7A). Importantly, when HIV-positive human sera were used to assess surface expression, all Env proteins were shown to be expressed at similar levels (data not shown). Thus, although the 8x FS mutation alone was not sufficient to induce CD4 independence, it did cause an apparent conformational change in gp120 that increased exposure of the 17b epitope.
FIG. 7.
Relationship between CD4 independence and exposure of a CD4-induced epitope. The CD4-induced MAb, 17b, was used in a cell-surface binding assay as described in Materials and Methods. The Env proteins indicated in panels A and B were expressed in 293T cells, and the extent of 17b binding relative to 8x Env, ± the standard error of the mean, was determined. The efficiency with which each Env mediated CD4-independent membrane fusion (derived from Fig. 2 and 3) is shown for convenience.
We also evaluated 17b epitope exposure in chimeras containing the 8x(V3) or 8x(V4/C4) domains. In general, Envs containing the 8x gp41 domain exhibited enhanced binding to 17b relative to the HXB Env regardless of whether they were CD4 independent or not (Fig. 7B and data not shown). Thus, exposure of the coreceptor binding site as judged by 17b binding does not strictly correlate with the CD4-independent phenotype. Interestingly, we did find that residue 298 in the V3 loop could affect 17b binding. The 8x(V3,gp41) chimera bound 17b well and was CD4 independent (Fig. 7B). However, if residue 298 was then changed from a Lys to an Arg (as is found in the HXBc2 sequence), both CD4 independence and 17b binding were lost (Fig. 7B). Since the 17b epitope does not include any portion of the V3 loop, we speculate that mutation of residue 298, located at the base of the V3 loop, modulates the orientation of the V3 loop. In the native HXBc2 Env, the V3 loop may help shield the 17b epitope, while in the 8x Env the R298K mutation appears to alter the orientation of the V3 loop in such a way that the 17b determinant is more accessible. In contrast, the I320V mutation in the 8x V3 loop, also shown to affect CD4 independence, had the opposite effect. Introduction of I320V into the 8x(gp41) Env allowed CD4-independent fusion but actually reduced 17b reactivity.
Introduction of CD4-independent determinants into heterologous Env proteins.
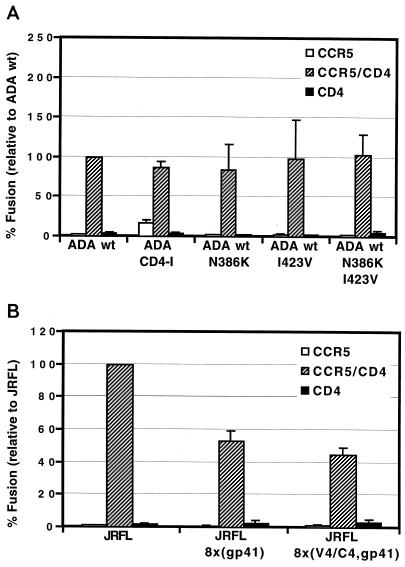
Finally, we determined whether the N386K and I423V mutations could render a heterologous Env protein CD4 independent. These residues were chosen because of their conserved nature and their ability to render HXBc2 CD4 independent provided the 8x gp41 subunit was present. We used the R5 virus strain ADA, and we also used a CD4-independent variant of ADA described by Kolchinsky et al. (ADA-CD4i) containing two amino acid changes in the V1/V2 region (R190S and S197N) as a positive control (19). Fusion elicited by ADA-CD4i in the absence of CD4 was observed at a level only 20% that of this Env with CD4 (Fig. 8A). Introducing N386K and I423V into ADA either alone or in combination on the CD4-dependent ADA Env failed to confer CD4 independence on ADA. Unfortunately, an ADA gp120/8x gp41 chimera was nonfunctional (data not shown). The N386K and I423V mutations also failed to render the R5-tropic Env, JRFL, CD4 independent on CCR5-expressing target cells when the 8x V4/C4 domain was transferred to JRFL alone or in combination with the 8x gp41 (Fig. 8B). Thus, the ability of the N386K and I423V mutations in conjunction with changes in gp41 to confer CD4 independence is context dependent.
FIG. 8.
Regions in 8x important for CD4 independence are not transferable to heterologous envelopes. Cell-cell fusion assays were used to determine if residues in 8x gp120 important for CD4 independence can make heterologous Envs CD4 independent. The indicated mutations or 8x domains were introduced into the R5 Envs ADA (A) and JRFL (B). The percentage of fusion relative to the wild-type (wt) Env is shown ± the standard error of the mean.
DISCUSSION
Sequential binding of the HIV-1 Env protein to CD4 and a coreceptor provides the signals necessary to activate Env's membrane fusion potential (7). Since CXCR4 is expressed in many CD4-negative cell types, the requirement for CD4 binding would appear to greatly restrict the tropism of X4 and R5X4 virus strains. Nonetheless, naturally occurring CD4-independent HIV-1 strains have not been identified. This stands in stark contrast to SIV, as many primary SIV strains have been identified that can infect CCR5-positive cells independently of CD4, suggesting that CCR5 was the primordial receptor for the primate lentiviruses (2, 9–11, 17, 25, 28). While CD4-independent primary HIV-1 strains have not been identified, viruses that can utilize either CCR5 or CXCR4 in the absence of CD4 are relatively easy to generate in vitro by passaging virus on CD4-negative, coreceptor-positive cells (8, 16, 19, 21). In all cases, CD4 independence is the consequence of a small number of mutations in Env. Given the replication rate of HIV-1 and the mutability of this virus in vivo, one would anticipate that CD4-independent viruses would arise in vivo unless there was selective pressure that prevented their development.
Our study shows that one consequence of CD4 independence exhibited by 8x is markedly increased sensitivity to antibody-mediated neutralization. Every CD4-independent version of HIV-1 HXBc2 Env we examined was considerably more sensitive to neutralization by HIV-positive human sera than was the parental HXBc2 Env. In addition, we found that CD4-independent SIV strains are universally more sensitive to neutralization by SIV-positive rhesus macaque sera than are closely related CD4-dependent SIV strains (Puffer and Doms, unpublished). This suggests that the humoral immune response applies significant selective pressure against CD4 independence in vivo. In the case of SIV, this selective pressure may be counteracted by the fact that macrophages from rhesus macaques have very low levels of CD4, providing selective pressure for CD4 independence (27). By contrast, human macrophages typically express relatively high levels of CD4 per cell, though there is considerable donor variability (23). In addition, most CD4-independent SIV isolates have been obtained from rhesus macaques. It will be important to determine if CD4-independent SIV strains are common in their natural hosts. As noted previously, the majority of primary HIV-2 isolates have been shown to be CD4 independent on either CCR5 or CXCR4 to some degree (28). In the case of HIV-1, it will be interesting to determine if CD4-independent HIV-1 strains evolve in vivo at late stages of disease, in individuals with poor humoral responses, or in antibody-privileged sites such as the central nervous system.
While all CD4-independent Envs identified in this study were neutralization sensitive, we have not fully identified the determinants responsible for this phenotype. In an earlier study, it was shown that a V3 loop antibody neutralized CD4-dependent and -independent versions of HXBc2 equally well, while the CD4-induced antibodies 17b and 48d were far more effective at neutralizing 8x (16). Whether the CD4-induced determinants recognized by these antibodies are responsible for the enhanced neutralization activity exhibited by HIV-positive human sera for CD4-independent Envs remains to be determined. However, we found a good but imperfect correlation between exposure of the 17b epitope and neutralization sensitivity. While most Envs that bound 17b in the absence of CD4 were neutralization sensitive, there were exceptions. For example, the Env chimera containing the HXBc2 gp120 and 8x gp41 subunits bound 17b efficiently but was CD4 dependent and neutralization resistant. This makes it likely that the mechanisms responsible for neutralization sensitivity are complex, involving multiple determinants. Indeed, several studies have shown that deletions in the V1/V2 region of Env result in enhanced sensitivity to neutralization and that this involves determinants associated with the V3 loop and the CD4-binding site, as well as conserved regions in gp120 that have not yet been identified (3, 5, 34). The conserved bridging sheet region in gp120, already shown to play an important role in coreceptor binding and containing a portion of the 17b epitope, is an obvious target for such antibodies. It will be useful to probe closely related CD4-dependent and -independent Envs with a panel of MAbs to identify which regions of Env are responsible for neutralization sensitivity and whether this is due simply to increased antibody binding or to other reasons.
The failure of Env immunogens tested thus far to elicit broadly cross-reactive neutralizing antibodies has spurred interest in generating modified forms of Env in the hopes that antibodies can be elicited to functionally important regions that are either not immunogenic or are poorly accessible in the native Env trimer. Structural studies have identified highly conserved regions in Env responsible for receptor binding and membrane fusion that are real or potential targets of small molecule inhibitors or neutralizing antibodies (4, 7, 20, 39). Generally, these regions are exposed transiently as a consequence of the conformational changes initiated by receptor binding. Such structural intermediates of the fusion process can be targets for small molecule inhibitors. The peptide T20 binds to the triple-stranded coiled-coil structure of gp41, preventing it from forming the six-helix bundle that is the proximal cause of membrane fusion (18, 26). Whether antibodies will have access to these or other conserved determinants and so be able to inhibit membrane fusion is not clear. The fact that our CD4-independent Envs exhibit enhanced sensitivity to neutralization by all HIV-positive human sera we have tested indicates that the epitopes responsible for this phenotype are targeted in the majority of patients. Nevertheless, there is no evidence to suggest that this has a significant effect upon virus load. Rather, this suggests that these regions are simply not accessible in CD4-dependent Env proteins either in the native state or during the course of virus entry, arguing that while antibodies to these determinants can be generated, they are not effective at neutralizing CD4-dependent, neutralization-resistant virus strains. Such antibodies could, however, preclude the development of CD4-independent viruses in vivo.
While 8x gp120 proved to be incapable of eliciting antibodies competent to neutralize CD4-dependent, primary isolates, there are potentially many ways to genetically modify Env that could result in a more effective immunogen. Perhaps the best evidence that genetic modification of Env can generate a more potent immune response comes from a study by Reitter and colleagues (30). In this study, elimination of two N-linked glycosylation sites in SIVmac239 resulted in a fully replication-competent virus that was attenuated in vivo, with the low virus loads being associated with high levels of neutralizing antibodies. Interestingly, these changes also made the virus largely CD4 independent (Puffer and Doms, unpublished). Importantly, sera from animals infected with the partially deglycosylated viruses were more effective in neutralizing the parental, fully glycosylated SIVmac239, a virus that is notoriously difficult to neutralize. In our study, we used a highly laboratory-adapted virus strain; perhaps modification of primary Env proteins would produce a more effective immunogen. Recent advances in understanding the structure of Env coupled with the identification of the receptors needed to trigger conformational changes should make it possible to more systematically modify Env and to assess the effects of these modifications on immunogenicity and antibody neutralization mechanisms.
Our study identified specific changes that can render the HIV-1 HXBc2 Env CD4 independent. These changes were context dependent in that they did not render heterologous HIV-1 Envs CD4 independent. The most surprising finding was that the FS mutation in gp41 was necessary for CD4 independence. Biochemical studies have shown that truncations in the cytoplasmic domain of gp41 can affect the conformation of the gp41 ectodomain, although the mechanism for this effect and its consequences are unknown (33). Interestingly, we showed that the 8x FS mutation, which leads to a truncated cytoplasmic domain of gp41, affected the conformation of gp120. Thus, HXBc2 Env with the 8x FS mutation exhibited markedly enhanced binding to 17b in the absence of CD4. Provided that the FS mutation was present, a single amino acid change in either the V3 loop (I320V) or V4 region (N386K) could independently confer CD4 independence. Remarkably, loss of the 8x R298K mutation in the 8x(V3,gp41) chimera resulted in complete loss of CD4 independence and 17b exposure, indicating that this conservative change at the base of the V3 loop significantly impacts the structure and function of Env. An Arg at this position is highly conserved among HIVs and SIVs and has been implicated as playing an important role in chemokine receptor interactions (37, 38).
Changes in V4/C4 contributed to CD4 independence to a lesser extent. The mutation at position 386 resulted in the loss of an N-linked glycosylation site, but it was the specific substitution of a Lys at this position that was required for CD4 independence. Since this residue is either a part of or close to the conserved coreceptor binding region, the introduction of a positively charged residue could enhance binding of gp120 to CXCR4, which is highly negatively charged. However, we have found that 8x gp120 binds to CXCR4 with an affinity that is very close to that of the parental HXBc2 gp120 (15). A plausible mechanism by which the conservative I320V mutation contributes to CD4 independence is not readily apparent.
In a previous study, it was observed that 8x gp120 bound directly to CXCR4 and readily bound MAb 17b (16). Thus, it was concluded that CD4 independence was associated with stable and constitutive exposure of the coreceptor binding site. The results from the present study indicate that this view is overly simplistic and that mere exposure of the coreceptor binding site alone is not sufficient for CD4 independence. Likewise, CD4 independence was not always associated with exposure of the 17b epitope. As shown in Fig. 6A and B, constructs such as 8x(gp41) with or without the R298K mutation exhibited 17b reactivity but were completely CD4 dependent. It is possible that differences between the 17b epitope and the coreceptor binding site could explain these findings. Full understanding of how an Env protein can function independently of CD4 will require a better understanding of the roles CD4 and coreceptor play in triggering the conformational changes in Env that ultimately lead to membrane fusion. At present, it is known that binding to CD4 either is required for subsequent coreceptor binding or makes coreceptor binding far more efficient. At some point, conformational changes are triggered in the gp41 subunit, involving the formation of a triple-stranded coiled coil composed of one N-terminal helical domain contributed by each gp41 subunit in the Env trimer. Subsequent to this, C-terminal helices pack into grooves present on the outside of the triple-stranded coiled coil, forming a six-helix bundle (4, 39). The formation of the six-helix bundle is the proximal cause of membrane fusion. A recent study by Melikyan et al. (26) coupled with an earlier study by Furuta et al. (13) argue that CD4 binding alone is sufficient to result in formation of the triple-stranded coiled coil. If so, then coreceptor binding alone must subserve this function in CD4-independent Env proteins. As a result, the changes we and others have identified as being important for CD4 independence could exert their influence not only in modulating how Env binds to coreceptors but in how the conformational changes are triggered.
ACKNOWLEDGMENTS
We thank James Robinson (Tulane University) for providing MAb 17b.
This work was supported by National Institutes of Health grants NIH R21 AI44308 to C.C.L., NIH R01 45378 to J.A.H., and NIH R01 35383 and -40880 to R.W.D. This work was also supported by a Burroughs Wellcome Fund Translational Research Award to R.W.D. R.W.D. is a recipient of an Elizabeth Glaser Scientist Award from the Pediatric AIDS Foundation. S.W. and F.B. (grant number 823A-61172) were supported by fellowships from the Swiss National Science Foundation.
REFERENCES
- 1.Alexander W A, Moss B, Fuerst T R. Regulated expression of foreign genes in vaccinia virus under the control of bacteriophage T7 RNA polymerase and the Escherichia coli lac repressor. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borsetti A, Parolin C, Ridolfi B, Sernicola L, Geraci A, Ensoli B, Titti F. CD4-independent infection of two CD4−/CD5−/CXCR4+ pre-T-cell lines by human and simian immunodeficiency viruses. J Virol. 2000;74:6689–6694. doi: 10.1128/jvi.74.14.6689-6694.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chan D C, Fass D, Berger J M, Kim P S. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89:263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 5.Cherpelis S, Shrivastava I, Gettie A, Jin X, Ho D D, Barnett S W, Stamatatos L. DNA vaccination with the human immunodeficiency virus type 1 SF162V2 envelope elicits immune responses that offer partial protection from simian/human immunodeficiency virus infection to CD8+ T-cell-depleted rhesus macaques. J Virol. 2001;75:1547–1550. doi: 10.1128/JVI.75.3.1547-1550.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Doms R, Edinger A, Moore J. Coreceptor use by primate lentiviruses. In: Korber B, Foley B, Leitner T, Myers G, Hahn B, McCutchan F, Mellors J, Kuiken C, editors. Human retroviruses and AIDS. Los Alamos National Laboratory: Theoretical Biology and Biophysics. N.Mex: Los Alamos; 1999. [Google Scholar]
- 7.Doms R W, Moore J P. HIV-1 membrane fusion: targets of opportunity. J Cell Biol. 2000;151:F9–F13. doi: 10.1083/jcb.151.2.f9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Edinger A, Clements J, Doms R. Chemokine and orphan receptors in HIV-2 and SIV tropism and pathogenesis. Virology. 1999;260:211–221. doi: 10.1006/viro.1999.9819. [DOI] [PubMed] [Google Scholar]
- 10.Edinger A, Mankowski J L, Doranz B J, Margulies B J, Lee B, Rucker J, Sharron M, Hoffman T L, Berson J F, Zink C, Hirsch V M, Clements J E, Doms R W. CD4-independent, CCR5-dependent infection of brain capillary endothelial cells by neurovirulent SIV. Proc Natl Acad Sci USA. 1997;94:14742–14747. doi: 10.1073/pnas.94.26.14742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Edinger A L, Blanpain C, Kunstman K J, Wolinsky S M, Parmentier M, Doms R W. Functional dissection of CCR5 coreceptor function through the use of CD4-independent simian immunodeficiency virus strains. J Virol. 1999;73:4062–4073. doi: 10.1128/jvi.73.5.4062-4073.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Endres M J, Clapham P R, Marsh M, Ahuja M, Turner J D, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 13.Furuta R A, Wild C T, Weng Y, Weiss C D. Capture of an early fusion-active conformation of gp41. Nat Struct Biol. 1998;5:276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 14.Hernandez L D, Hoffman L R, Wolfsberg T G, White J M. Virus-cell and cell-cell fusion. Annu Rev Cell Dev Biol. 1996;12:627–661. doi: 10.1146/annurev.cellbio.12.1.627. [DOI] [PubMed] [Google Scholar]
- 15.Hoffman T L, Canziani G, Jia L, Rucker J, Doms R W. A biosensor assay for studying ligand-membrane receptor interactions: binding of antibodies and HIV-1 env to chemokine receptors. Proc Natl Acad Sci USA. 2000;97:11215–11220. doi: 10.1073/pnas.190274097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hoffman T L, LaBranche C, Zhang W, Canziani G, Robinson J, Chaiken I, Hoxie J A, Doms R W. Stable exposure of the coreceptor binding site in a CD4-independent HIV-1 envelope protein. Proc Natl Acad Sci USA. 1999;96:6359–6364. doi: 10.1073/pnas.96.11.6359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Iyengar S, Schwartz D H, Clements J E, Hildreth J E K. CD4-independent, CCR5-dependent simian immunodeficiency virus infection and chemotaxis of human cells. J Virol. 2000;74:6720–6724. doi: 10.1128/jvi.74.15.6720-6724.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kilby J M, Hopkins S, Venetta T M, DiMassimo B, Cloud G A, Lee J Y, Alldredge L, Hunter E, Lambert D, Bolognesi D, Matthews T, Johnson M R, Nowak M A, Shaw G M, Saag M S. Potent suppression of HIV-1 replication in humans by T-20, a peptide inhibitor of gp41-mediated virus entry. Nat Med. 1998;4:1302–1307. doi: 10.1038/3293. [DOI] [PubMed] [Google Scholar]
- 19.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L, Choe H, Sodroski J. Adaptation of a CCR5-using, primary HIV-1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwong P D, Wyatt R, Robinson J, Sweet R W, Sodroski J, Hendrickson W A. Structure of an HIV gp120 envelope glycoprotein in complex with the CD4 receptor and a neutralizing human antibody. Nature. 1998;393:648–659. doi: 10.1038/31405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.LaBranche C, Hoffman T L, Romano J, Haggarty B S, Edwards T, Matthews T J, Doms R W, Hoxie J A. Determinants of CD4-independence for HIV-1/IIIBx map outside regions required for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Evidence for cell-surface association between fusin and the CD4-gp120 complex in human cell lines. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 23.Lee B, Sharron M, Montaner L J, Weissman D, Doms R W. Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages. Proc Nat Acad Sci USA. 1999;96:5215–5220. doi: 10.1073/pnas.96.9.5215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leonard C K, Spellman M W, Riddle L, Harris R J, Thomas J N, Gregory T J. Assignment of intrachain disulfide bonds and characterization of potential glycosylation sites of the type 1 recombinant human immunodeficiency virus envelope glycoprotein (gp120) expressed in Chinese hamster ovary cells. J Biol Chem. 1990;265:10373–10382. [PubMed] [Google Scholar]
- 25.Martin K A, Wyatt R, Farzan M, Choe H, Marcon L, Desjardins E, Robinson J, Sodroski J, Gerard C, Gerard N P. CD4-independent binding of SIV gp120 to rhesus CCR5. Science. 1997;278:1470–1473. doi: 10.1126/science.278.5342.1470. [DOI] [PubMed] [Google Scholar]
- 26.Melikyan G B, Markosyan R M, Hemmati H, Delmedico M K, Lambert D M, Cohen F S. Evidence that the transition of HIV-1 gp41 into a six-helix bundle, not the bundle configuration, induces membrane fusion. J Cell Biol. 2000;151:413–423. doi: 10.1083/jcb.151.2.413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mori K, Rosenzweig M, Desrosiers R C. Mechanisms for adaption of simian immunodeficiency virus to replication in alveolar macrophages. J Virol. 2000;74:10852–10859. doi: 10.1128/jvi.74.22.10852-10859.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Reeves J, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira J, Moniz-Pereira J, Clapham P. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 30.Reitter J N, Means R E, Desrosiers R C. A role for carbohydrates in immune evasion in AIDS. Nat Med. 1998;4:679–684. doi: 10.1038/nm0698-679. [DOI] [PubMed] [Google Scholar]
- 31.Rizzuto C D, Wyatt R, Hernandez-Ramos N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 32.Rucker J, Doranz B J, Edinger A E, Long D, Berson J F, Doms R W. Use of a cell-cell fusion assay to study the role of chemokine receptors in human immunodeficiency virus type 1 (HIV-1) entry. Methods Enzymol. 1997;288:118–133. doi: 10.1016/s0076-6879(97)88011-1. [DOI] [PubMed] [Google Scholar]
- 33.Spies C P, Compans R W. Effects of cytoplasmic domain length on cell surface expression and syncytium-forming capacity of the simian immunodeficiency virus envelope glycoprotein. Virology. 1994;203:8–19. doi: 10.1006/viro.1994.1449. [DOI] [PubMed] [Google Scholar]
- 34.Stamatatos L, Cheng-Mayer C. An envelope modification that renders a primary, neutralization-resistant clade B human immunodeficiency virus type 1 isolate highly susceptible to neutralization by sera from other clades. J Virol. 1998;72:7840–7845. doi: 10.1128/jvi.72.10.7840-7845.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thali M, Moore J, Furman C, Charles M, Ho D, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. J Virol. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Trkola A, Drajic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. CD4-dependent, antibody-sensitive interactions between HIV-1 and its coreceptor CCR-5. Nature. 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 37.Wang W-K, Dudek T, Zhao Y-J, Brublay H G, Essex M, Lee T-H. CCR5 coreceptor utilization involves a highly conserved arginine residue of HIV type 1 gp120. Proc Natl Acad Sci USA. 1998;95:5740–5745. doi: 10.1073/pnas.95.10.5740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W K, Dudek T, Essex M, Lee T H. Hypervariable region 3 residues of HIV type 1 gp120 involved in CCR5 coreceptor utilization: therapeutic and prophylactic implications. Proc Natl Acad Sci USA. 1999;96:4558–4562. doi: 10.1073/pnas.96.8.4558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Weissenhorn W, Dessen A, Harrison S C, Skehel J J, Wiley D C. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 40.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature. 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 41.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]