Abstract
As an essential step in the lytic cascade, the Rta homologues of gammaherpesviruses all activate their own expression. Consistent with this biologic function, the Epstein-Barr virus (EBV) Rta protein powerfully stimulates the promoter of its own gene, Rp, in EBV-positive B cells in transient-transfection reporter-based assays. We analyzed the activity of RpCAT in response to Rta by deletional and site-directed mutagenesis. Two cognate Sp1 binding sites located at −279 and −45 relative to the transcriptional start site proved crucial for Rta-mediated activation. Previously described binding sites for the cellular transcription factor Zif268 and the viral transactivator ZEBRA were found to be dispensable for activation of RpCAT by Rta. Gel shift analysis, using extracts of B cells in latency or induced into the lytic cycle, identified Sp1 and Sp3 as the predominant cellular proteins bound to Rp near −45. During the lytic cycle, ZEBRA bound Rp near the Sp1/Sp3 site. The binding of Sp1 and Sp3 to Rp correlated with the reporter activities in the mutagenesis study, establishing a direct link between transcriptional activation of Rp by Rta and DNA binding by Sp1 and/or Sp3. The relative abundance or functional state of the cellular Sp1 and Sp3 transcription factors may be altered in response to stimuli that induce the BRLF1 promoter and thereby contribute to the activation of the viral lytic cycle.
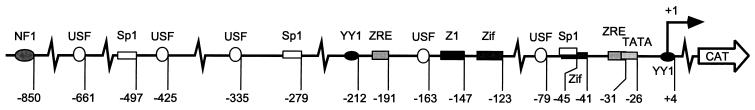
Upon induction of the lytic cycle of Epstein-Barr virus (EBV), Rp and Zp, the two promoters directing the expression of the immediate-early genes BRLF1 and BZLF1, are activated simultaneously (15, 76). Despite their similar temporal regulation, it remains unclear whether the two promoters respond to the same or different signaling cascades. Only a few known response elements for transcriptional activators are contained within Rp (Fig. 1); these include binding sites for the cellular transcription factors YY1, Zif268 (Egr1), and Sp1 and the viral transactivator ZEBRA, the product of BZLF1 (14, 70, 84–86). YY1 and Sp1 sites, as well as ZEBRA response elements (ZREs), are also present in Zp (14, 48, 58, 70, 73). ZEBRA activates Rp from the latent virus, presumably by directly binding to the ZREs (38, 42). The Zif268 sites have been implicated in phorbol ester-mediated activation of Rp, while the Sp1 sites were shown to affect the constitutive activity of Rp in cultured epithelial cells (85, 86). However, their physiological roles and contributions to autostimulation of Rp in B cells have not been assessed.
FIG. 1.
Map of RpCAT. RpCAT reporter construct used for mutagenesis analysis. Boxes and ovals represent previously documented and putative (E-boxes) transcription factor binding sites, respectively (5, 7, 22, 27–29). Numbers indicate the positions of the 3′ ends of the cis elements relative to the transcriptional start site (rightward arrow).
EBV remains predominantly latent in B lymphocytes, whereas epithelial cells are more permissive for lytic infection by the virus (5, 43, 57, 71). Consistent with this biologic observation, Rp reporters exhibit significantly higher constitutive activities in epithelial tissue culture than in B cells (18, 70, 86). We conducted an analysis of the regulation of Rp in an EBV-positive B-cell background, where EBV naturally persists in a latent state. We chose the Burkitt's lymphoma (BL) cell line Cl16 (HH514-16) for this purpose, since the EBV contained in these cells is tightly latent and yet can be induced efficiently into the lytic cycle by chemical stimuli or by transfection of ZEBRA or Rta expression vectors (30, 62, 80).
RpCAT reporters are not constitutively active in Cl16 cells and thus accurately reflect the behavior of the endogenous promoter. Moreover, in these cells Rta activates Rp both in the endogenous virus and when presented as a reporter construct (63). Since Rp lacks cognate binding sites for Rta, the mechanism of autostimulation of Rp must be indirect. Extensive mutagenesis was performed in order to identify cis elements that make Rp sensitive to activation by Rta. Two Sp1 sites contained within the −299/+58 promoter fragment, as well as sequences between +15 and +30 relative to the transcriptional start site, proved crucial for efficient autostimulation of Rp. Gel shift analysis using extracts of uninduced and Cl16 cells induced into the lytic cycle confirmed that Sp1 and Sp3 were the predominant factors binding to a promoter fragment containing one of the Sp1 sites. These data suggest that the ubiquitous housekeeping genes Sp1 and Sp3 play a role in the activation of the EBV immediate-early genes.
MATERIALS AND METHODS
Cell lines.
HH514-16 (Cl16) is a clonal derivative of the P3J-HR-1 B-cell line derived from an EBV-positive BL that is permissive for viral replication (62); Raji is a human B-cell line derived from a BL containing an EBV strain that is defective for DNA replication and late gene expression (61); and B95-8 is a marmoset B-cell line transformed with EBV (55). All cells were maintained in 5% CO2 at 37°C in RPMI 1640 supplemented with 8% fetal calf serum.
Chemical induction.
Cells were subcultured 2 to 3 days prior to drug treatment or transfection. Drug treatment consisted of the addition of 10 ng of tetradecanoylphorbol-13-acetate (TPA) per ml, 3 mM sodium butyrate, or both to the culture medium. Trichostatin A was added to 5 nM.
Expression vectors and reporter plasmids.
The ZEBRA expression vector pBXG1-genomic Z and its parent vector pBXG1, as well as the Rta expression vector RTS15 (pRTS/Rta) and the empty vector RTS15ΔHIII (pRTS), have been described previously (16, 63). The RpCAT deletion mutants were generated by PCR, based on the RpCAT (−962/+58) construct (63). The luciferase control vector pGL2 basic+HMP has been described (69). Site-directed mutations were introduced using the Quickchange mutagenesis kit (Stratagene) according to the manufacturer's instructions. All mutations were confirmed by sequencing the reporter constructs. Oligonucleotide primer sequences are available upon request.
Transfections.
Transfections were carried out using electroporation (69). A total of 1.5 × 107 cells in 0.4 ml of RPMI 1640 were exposed to 960 μF and 250 V in electroporation cuvettes with a 0.4-cm gap using a BioRad gene pulser, and 10 μg of reporter DNA plus 5 μg of expression vector pRTS/Rta or pRTS were used; 1 μg of pGL2 basic+HMP was included to control for transfection efficiency
Reporter assays.
Chloramphenicol acetyltransferase (CAT) and luciferase assays were performed as described (69). CAT and luciferase activities were determined 72 h following transfection (69). Results represent the average of at least two separate transfections.
Protein extracts and Western blots.
Cells were collected by centrifugation, washed once in phosphate-buffered saline (PBS), and resuspended in sodium dodecyl sulfate (SDS) sample buffer at 106 cells/10 μl. Prior to separation by SDS–12% polyacrylamide gel electrophoresis, samples (20 μl) were heated to 100°C for 5 min. Following electrophoresis, the proteins were transferred to nitrocellulose membranes by electroblotting and blocked in 5% nonfat dry milk overnight at 4°C. The blots were incubated with antiserum diluted in 5% nonfat dry milk at 25°C for 2 h, washed three times for 10 min in 10 mM Tris-HCl (pH 7.5)–200 mM NaCl–5% Tween 20, incubated with [125I-]protein A for 1 h, and washed again. The membranes were exposed overnight with intensifying screens to Kodak XAR-5 film at −70°C.
Cell extracts for EMSA.
Extracts were prepared for electrophoretic mobility shift assay (EMSA) as previously described (59). Cells (untreated, chemically induced, or transfected) were harvested, washed once with PBS, and collected again by centrifugation. Pellets containing 1.5 × 107 cells were flash-frozen and resuspended in 200 μl of lysis buffer (0.42 M NaCl, 20 mM HEPES [pH 7.5], 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 1 mM dithiothreitol [DTT], 1 mM phenylmethylsulfonyl fluoride, 2 μg of aprotinin per ml). Lysates were spun at 90,000 rpm at 4°C for 15 min using a TLA100 rotor (Beckmann) in a benchtop ultracentrifuge (Beckmann Optima TLX), and the supernatants were aliquoted, flash-frozen, and stored at −80°C. Protein concentrations were determined by the Bradford method (9).
EMSA.
Annealed double-stranded oligonucleotides (20 ng) were end labeled with 32P using polynucleotide kinase (Boehringer Mannheim). Binding reactions contained 7 or 10 μg of cell protein in 10 mM HEPES (pH 7.5)–50 mM NaCl–2 mM MgCl2–2.5 μM ZnSO4–0.5 mM EDTA–1 mM DTT–15% glycerol in a total volume of 20 μl. Following an incubation of 5 min at room temperature, 30,000 to 50,000 cpm of labeled oligonucleotide and 0.5 μg of poly(dI-dC) were added per reaction. Reactions with competitor DNA contained a 500-fold molar excess of unlabeled oligonucleotide in the initial reaction mix. Where indicated, antisera were added 5 min following the addition of the probe, and incubation at room temperature continued for 10 min. All antisera used, with the exception of α-Rta (63) and α-ZEBRA (36), were obtained from Santa Cruz, Calif., when available as TransCruz antiserum. The reactions were loaded onto a 4% 0.5× Tris-borate-EDTA native polyacrylamide gel and run at 200 to 280 V. Gels were dried on 3MM Whatman paper under vacuum and exposed to autoradiography film overnight.
RESULTS
Responsiveness of RpCAT to Rta is retained within the −299/+30 fragment.
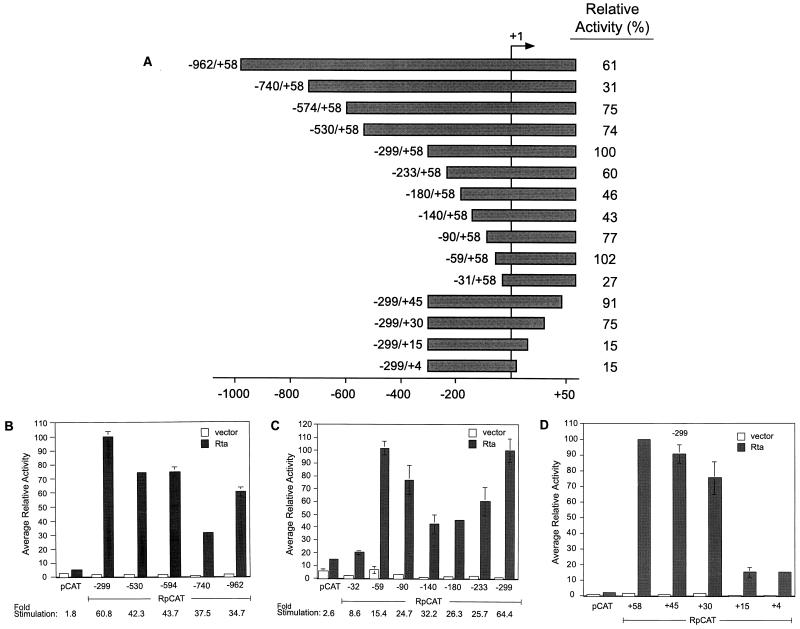
Figure 1 illustrates the described transcription factor binding sites present in Rp. To evaluate their contributions to autostimulation of the promoter, we generated a series of 5′ deletional mutants of RpCAT (Fig. 2A) and examined their activation by Rta in Cl16 cells. An initial series of large progressive 5′ deletions revealed that maximal response to Rta was maintained in a fragment of Rp containing −299 to +58 (Fig. 2B). Further analysis of constructs with shorter deletions between position −299 and the TATA box revealed a steady decrease in response to Rta with the loss of sequences down to −140, followed by an increase in activity, to the same level as the −299/+58 construct, with the −59/+58 construct (Fig. 2C). However, this increase in sensitivity of the −59/+58 fragment to Rta was accompanied by a significant increase in background activity in the absence of Rta. This resulted in stimulation of the −59/+58 reporter by Rta of only 15-fold, compared with a 64-fold stimulation of RpCAT (−299/+58). Considering that −299/+58 was activated by Rta to even higher levels than the “full-length” −962/+58 promoter while maintaining a low background, it was assumed that the −299/+58 sequence contains relevant cis elements conferring positive response to Rta.
FIG. 2.
Effect of deletions on the responsiveness of RpCAT to Rta. (A) Illustration of Rp fragments with 5′ and 3′ truncations used in deletional mutagenesis analysis. Bars represent Rp fragments linked to the CAT reporter. Designations to the left of each bar are relative to the transcriptional start site of Rp, which is indicated by +1. Relative activities of the RpCAT constructs in response to Rta are indicated on the right. (B and C) 5′ deletion analysis of RpCAT in Cl16 cells. (D) 3′ deletions of RpCAT. Average activities of deletion mutants are relative to RpCAT (−299/+58) in the presence of Rta. Cells were transfected with reporter constructs and Rta expression vector (shaded bars) or empty control vector (open bars). CAT activities are standardized to the luciferase activity of a cotransfected HMP+pGL2 construct. Error bars represent the standard error of the mean (n ≥ 2). Below the graph, the fold stimulation of the RpCAT vectors is given (reporter activity in the presence of Rta divided by the reporter activity in the absence of Rta).
A series of downstream deletions were generated in the context of Rp −299 to assess the importance of sequences 3′ of the transcriptional start site in the activation of RpCAT by Rta (Fig. 2A). The activity of the promoter construct was greatly reduced upon loss of sequences between +15 and +30 relative to the transcription start (Fig. 2D). Further 3′ deletions invading the region from +30 to +58 did not significantly affect the response of RpCAT to Rta. The data demonstrate that, in addition to the 5′ region beginning at −299 from the transcription start, 3′ sequences between +15 and +30 are needed for efficient activation of RpCAT by Rta.
Two Sp1 sites within Rp are crucial for activation by Rta.
Liu et al. reported that Rta may indirectly activate some EBV promoters lacking Rta response elements (RREs), such as the promoter of the viral DNA polymerase, via the upstream stimulating factor (USF), which binds to E-box DNA sequences of the consensus CANNTG (47). Figure 1 shows that Rp contains several of these sequences; the two most proximal to the transcriptional start site fall within the −299/+58 region at −79 and −163. However, mutation of these two E-boxes, individually or together, did not negatively affect the activation of RpCAT −299/+58 by Rta (data not shown). The E-boxes are therefore unlikely to contribute to the autostimulation of Rp.
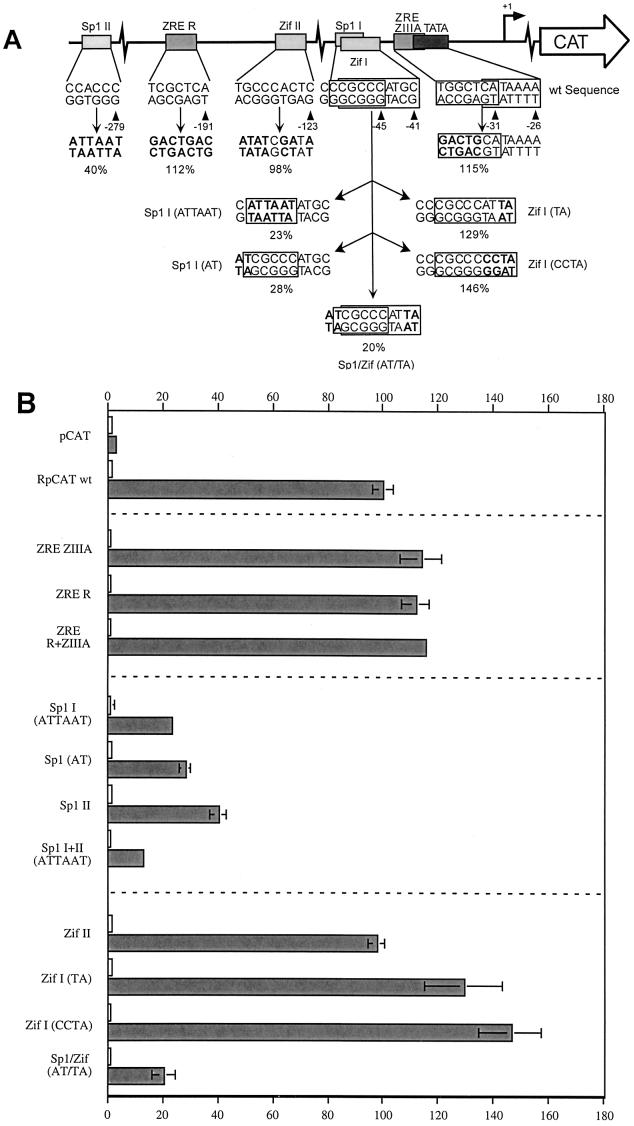
In addition to the two E-boxes, the −299/+58 fragment of Rp contains binding sites for several other transcription factors (Fig. 1). These include two Sp1 binding sites at −45 and −279, two Zif268 sites at −41 and −123, the proximal site of which overlaps one of the Sp1 sites, and two ZREs, ZIIIA at −31 and R at −191 (14, 85, 86). All of these sites were eliminated, either singly or in pairs, by site-directed mutagenesis in the context of RpCAT −299/+58. Figure 3A illustrates the individual mutations, with the altered bases indicated in bold. The proximal ZRE, a ZIIIA site, overlaps the TATA box. The introduced mutation therefore encompassed only the nucleotides at the 5′ end of the response element. Similarly, attempts were made to dissect the contributions of the overlapping Sp1 and Zif268 sites commencing at positions −45 and −41, respectively (see Fig. 3A for details).
FIG. 3.
Effect of point mutations within Sp1 sites, ZEBRA, and Zif268 response elements in Rp on activation by Rta. (A) Site-directed mutations in RpCAT (−299/+58). Boxes represent the response elements, with their names given above and their sequence in the wild-type (wt) promoter given below. The mutated sequences are shown beneath, with the changes indicated in bold. The average activity relative to wild-type −299/+58 is expressed as a percentage below the mutated sequence. The +1 above the rightward arrow above the schematic represents the transcriptional start site of Rp. (B) Average relative activity of RpCAT −299/+58 with site-directed mutations in response to Rta in Cl16 cells. Bars indicate the average activity of RpCAT reporters transfected into Cl16 cells with Rta expression vector (shaded bars) or empty control vector (open bars) and are relative to the wild-type (wt) RpCAT reporter in the presence of Rta (n ≥ 2). Constructs correspond to those illustrated in panel A.
All three reporters with mutated ZREs (R, ZIIIA, and R+ZIIIA) were stimulated slightly more than the wild-type Rp −299/+58 construct, suggesting that they do not play a role in the response of Rp to Rta (Fig. 3B).
Further results suggested that the Sp1 sites and not the Zif268 response elements within the −299/+58 Rp fragment contributed to the activation of Rp by Rta. Elimination of the distal Sp1 site located at position −279 reduced the Rp response to 40% relative to the wild-type promoter, while a 6-bp mutation (ATTAAT) within the proximal Sp1 site I reduced the response to 23% (Fig. 3B). A 2-bp mutation at the 5′ end of this Sp1 site, leaving the Zif268 response element intact, similarly reduced Rp response to 28% relative to the wild-type promoter. Removal of both Sp1 sites together led to an eightfold-lower response (12%) of Rp to Rta. Both Sp1 sites were therefore important for autostimulation of Rp by Rta.
In contrast to the distal Sp1 site, alteration of the upstream Zif268 site II had no effect on the activity of RpCAT (Fig. 3B). The analysis of the proximal Zif268 site was more complex, since it overlaps the Sp1 site. Zif268 has been reported to compete with Sp1 for DNA binding on overlapping sites (1, 6, 31, 77). A two-base change at the 3′ end of the proximal Zif268 site (at position −41) to TA, which mutates the Zif268 site but leaves the core Sp1 sequence intact, increased the promoter activity to 129% relative to wild-type Rp, consistent with a possible loss of minor inhibition by Zif268. An additional mutation within this Zif268 site, changing the 3′ end of the sequence from ATGC (wild type) to CCTA, led to a further increase in activity to 146%. This enhanced activity of the CCTA mutation may also be explained by the increased GC content surrounding the Sp1 core element, thus improving the quality of the Sp1 site.
As a final test of the relative importance of the two factors, point mutations were generated at the 5′ end of the Sp1 site and the 3′ end of the Zif268 site [Fig. 3B, Sp1/Zif (AT/TA)]. The resulting promoter activity (20%) was similar to that caused by the Sp1 (AT) mutation alone, demonstrating that the Sp1 site is dominant over the overlapping Zif268 element. Taken together, these results strongly suggest that the Sp1 sites and not the Zif268 sites contribute to the activation of Rp by Rta.
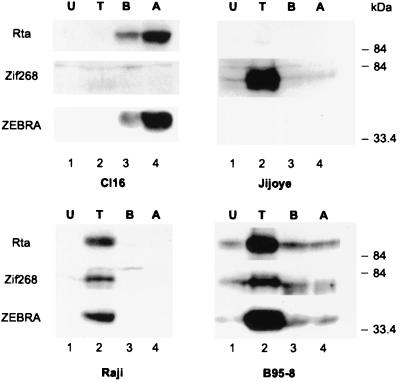
Chemical treatment does not induce Zif268 expression in Cl16 cells.
Zalani et al. reported that the phorbol ester TPA induced the expression of the transcription factor Zif268 in the B-cell line B95-8 and that this protein was capable of binding to and activating Rp (85). Since the Zif268 sites within Rp did not seem to contribute to the activation of the RpCAT reporter by Rta in Cl16 cells, we determined whether significant amounts of Zif268 were expressed in these cells. Extracts prepared from Cl16 cells treated with several inducing chemicals were analyzed by immunoblotting for the expression of Zif268 and the immediate-early lytic proteins Rta and ZEBRA. For comparison, this analysis was also performed in Jijoye cells, the parental cell line of Cl16, in Raji cells, and in the marmoset cell line B95-8. As shown in Fig. 4, with the exception of B95-8 cells, which exhibit a low level of leaky spontaneous lytic-cycle gene expression, Zif268 was not present in uninduced cells, and EBV remained tightly latent (lanes 1). TPA treatment (lanes 2) resulted in strong induction of Zif268 expression in Raji, B95-8, and Jijoye cells, but in Cl16 cells Zif268 remained undetectable. In Raji and B95-8 cells, upregulation of Zif268 by TPA was accompanied by activation of the EBV lytic cycle; both Rta and ZEBRA were expressed. EBV was not induced by TPA in Cl16 cells or in Jijoye cells. Although TPA strongly activated Zif268 in Jijoye cells, it did not do so in Cl16 cells. Two chemical agents known to induce the lytic cycle in Cl16 cells, sodium butyrate and trichostatin A, efficiently stimulated the expression of Rta and ZEBRA but did not induce Zif268 expression. Thus, the induction of Zif268 and the activation of the lytic cycle of EBV did not correlate in Jijoye and Cl16 cells.
FIG. 4.
Zif268 expression does not correlate with lytic cycle induction in Cl16 cells. Western blot analysis of total cell extracts of Cl16, Jijoye, Raji, and B95-8 cells that were either untreated (U) or treated for 48 h with either TPA (T), sodium butyrate (B), or trichostatin A (A). Immunoblots were probed with antibodies against Rta, Zif268, and ZEBRA.
Sp1 and Sp3 are the predominant cellular factors binding to Rp −64/−28.
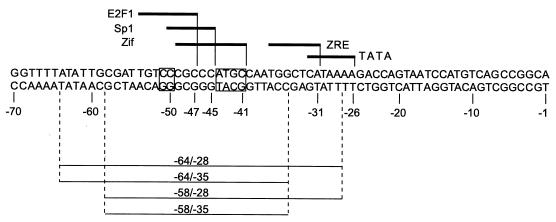
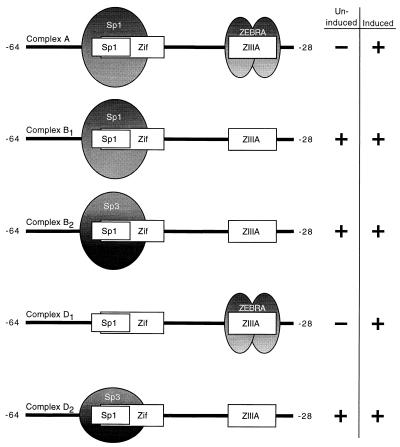
The mutagenesis analysis provided a surprising result because, as a ubiquitous housekeeping gene, Sp1 has not been considered an obvious candidate to be involved in the activation of the lytic cycle promoter Rp. Gel shift assays were performed using extracts of uninduced and induced Cl16 cells to determine which cell or viral factors were capable of binding to this promoter region in vitro (27, 28). The oligonucleotide probe, spanning Rp positions −64 to −28, was centered around the proximal Sp1 site and also included the nearby ZRE ZIIIA (Fig. 5).
FIG. 5.
Nucleotide sequence of Cl16 Rp from −70 to −1. Lines above the sequence indicate the extent of documented and putative (E2F1) cis-active response elements of DNA-binding factors. The boxes indicate bases that have been changed in mutant gel shift oligonucleotide probes: CC (−51/−50) → AT (Fig. 8A) and ATGC (−44/−41) → CCTA (Fig. 8B). Below the sequence, the extent of the oligonucleotides used in gel shifts is indicated.
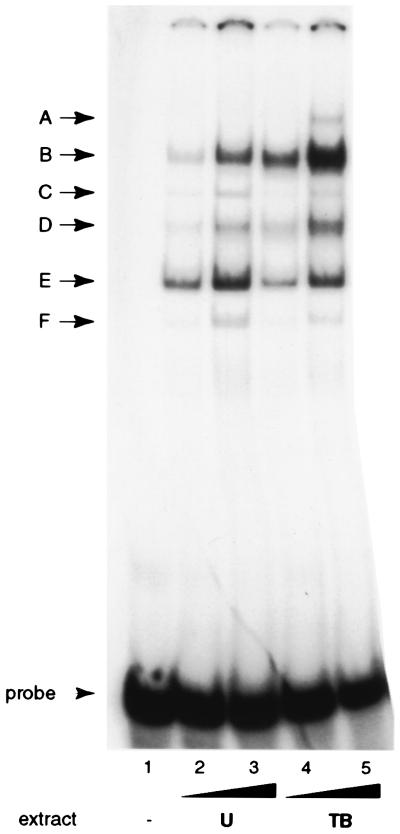
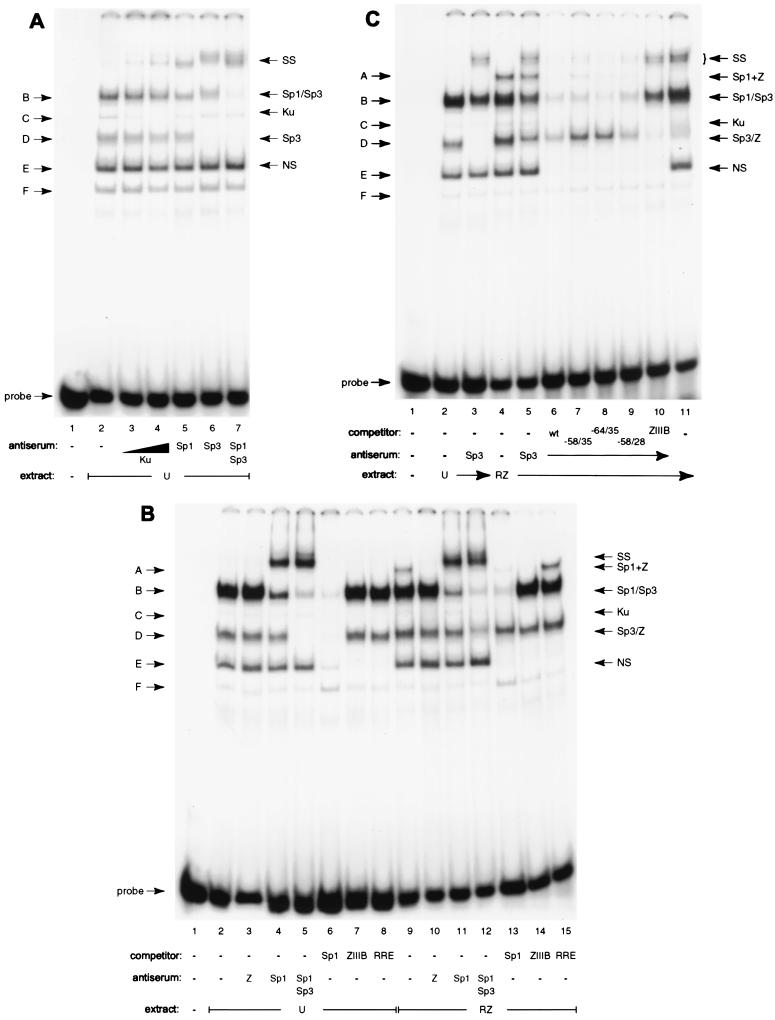
The cell extracts generated several distinct band shifts, labeled A though F (Fig. 6). The patterns produced by uninduced and chemically induced cell extracts were identical except that the slowest-mobility complex A was observed only with induced cell extract (compare lanes 2 and 3 to 4 and 5). These complexes were analyzed by supershifts using antisera against a variety of transcription factors and by means of oligonucleotide competition. These studies showed that several of the most prominent DNA-protein complexes contained Sp1 or Sp3 (Fig. 7A and data not shown). In uninduced cell extracts, α-Sp3 completely supershifted complex D (lane 6) and a lower portion of complex B (B2; lane 6), while α-Sp1 supershifted a higher portion of complex B (B1; lane 5). Addition of both antisera efficiently supershifted both components of complex B (lane 7). Sp3 exists in two major isoforms, 70 to 80 and 110 to 115 kDa in size, consistent with the two observed protein-DNA complexes B2 and D (37, 75). The only other protein factor to be identified was Ku in complex C (lanes 3 and 4). Ku binds double-stranded DNA ends irrespective of sequence (8, 20, 56, 72). Rta itself could not be detected in any of the DNA-protein complexes by antibody supershifts or competition (data not shown, and Fig. 7B, lanes 8 and 15).
FIG. 6.
Major protein-DNA complexes formed on Rp −64/−28. EMSA pattern of Cl16 extracts binding to an Rp −64/−28 oligonucleotide. Either 5 or 10 μg of total cell protein from uninduced (U, lanes 2 and 3) and chemically induced with TPA and sodium butyrate (TB, lanes 4 and 5) Cl16 cells were used in the binding reactions. Complexes are labeled with the letters A to F on the left. Probe, free probe. Lane 1, probe alone.
FIG. 7.
Identification of proteins binding to Rp −64/−28 by antibody supershifts and by oligonucleotide competition. (A) Identification of proteins in uninduced (U) Cl16 extracts binding to the Rp oligonucleotide probe. Complex designation is indicated on the left (B to F); identified proteins are on the right (SS, supershifted bands). Lane 1, probe alone; lanes 2 to 7, 7 μg of cell extract; lanes 3 to 7 contain antiserum to the indicated proteins. (B) Sp1 and ZEBRA occupy Rp −64/−28 together in complex A. Extracts of uninduced cells (U) and of Cl16 cells transfected with ZEBRA and Rta expression vectors (RZ) were incubated with the oligonucleotide probe and the indicated antisera (lanes 3 to 5 and 10 to 12) or a 500-fold excess of the indicated unlabeled competitor DNA (lanes 6 to 8 and 14 to 16). (C) Competition and supershift analysis identify ZEBRA in complex D1 in extracts of induced Cl16 cells. Uninduced (U, lanes 2 and 3) and Rta and ZEBRA-transfected (RZ, lanes 4 to 11) Cl16 extracts were incubated with the Rp probe and Sp3 (lanes 3 and 5 to 10) and ZEBRA (lane 11) antisera. Binding reactions in lanes 6 to 10 also contained a 500-fold excess of the indicated unlabeled competitor DNA (lanes 6 to 9 contained Rp −64/−28, −58/−28, −64/−35, and −58/−35, respectively; lane 10, ZRE ZIIIB).
Sp1 and ZEBRA occupy Rp −64 and −28 together in complex A.
Analysis of complex A formed by extracts of Cl16 cells that were induced into the lytic cycle revealed that complex A contained both Sp1 and ZEBRA. Several lines of evidence support this conclusion. In extracts of cells that had been transfected with ZEBRA and Rta expression vectors (Fig. 7B), complex A was eliminated by addition of α-ZEBRA serum (the signal is retained in the well; lane 10). Furthermore, an excess of unlabeled DNA containing the high-affinity ZIIIB ZEBRA recognition element efficiently competed for the binding activity in complex A (lane 14). Antibody against Sp1 also supershifted complex A (lane 11). A comparison of the Sp1 supershifts in extracts of uninduced versus induced cells (lanes 4 and 11) revealed an enlarged supershifted band with the extract of induced cells, consistent with the additional presence of a larger complex of slower mobility. Competition with an excess of unlabeled oligonucleotide containing the consensus Sp1 binding motif also eliminated most of complex A (lane 13), confirming the presence of Sp1 in the complex. It is noteworthy that although Sp3 has the same DNA-binding specificity as Sp1, it was not contained in complex A together with ZEBRA, since the α-Sp3 serum did not affect this complex (compare lanes 4, 5, and 11). On a higher percentage gel, however, a minor complex consisting of the smaller isoform of Sp3 and ZEBRA migrating between complexes A and B could be detected (data not shown). An excess of unlabeled DNA containing a binding site for Rta did not interfere with the formation of complex A (lane 15). This experiment controlled for the specificity of the competitor oligonucleotides and also showed that Rta was not present in complex A. The competition experiments also revealed that complex E was the result of nonspecific DNA-binding activity, as all three unrelated oligonucleotides competed for it with equal efficiency.
ZEBRA and Sp3 form distinct components of complex D in induced cell extracts.
The EMSA experiments also showed that complex D formed with extracts of Cl16 cells in the lytic EBV cycle contained two subcomplexes, one formed by Sp3 and the other by ZEBRA. The oligonucleotide containing the Sp1/Sp3 binding site completely competed for formation of complex D generated by extracts of latently infected cells but failed to eliminate a significant portion of complex D formed by extracts of lytically induced cells (Fig. 7B, compare lanes 6 and 13). Addition of α-Sp3, which completely supershifted complex D in extracts of uninduced cells, left a significant portion of complex D formed by extracts of induced cells (compare lanes 5 and 12). Therefore, the induced cell extract contained an additional factor binding to the Rp −64/−28 probe in complex D that was not present in uninduced Cl16 cells. Unlike the situation described for complex A, this factor is unlikely to occupy the same DNA fragment as either Sp1 or Sp3, since the Sp1 oligonucleotide did not compete for its binding. Complex D was renamed D1/2 because it is composed of at least two complexes with similar electrophoretic mobilities in induced Cl16 extracts.
The slightly slower-migrating portion of complex D (D1) was shown to contain ZEBRA by means of oligonucleotide competition and antibody supershift experiments (Fig. 7C). Addition of an excess of wild-type Rp −64/−28 37-mer unlabeled oligonucleotide completed for this binding activity (lane 6). However, a truncated version of the wild-type Rp fragment 24 nucleotides in length (−58/−35), which does not include the ZRE, competed for Sp1 and Sp3 binding in complexes A and B but did not affect the formation of complex D1 (lane 7). The competitor oligonucleotide −64/−35, which extends to the 5′ end of the 37-mer but does not contain the ZRE, similarly failed to affect formation of complex D1 (lane 8). However, the oligonucleotide −58/−28, extending to the 3′ end of the 37-mer, which encompasses the ZRE, competed for complex D1 to the same degree as the full-length wild-type −64/−28 fragment (lane 9). A ZIIIB element used as the unlabeled competitor efficiently eliminated the formation of any D1 complex (lane 10). While α-Sp3 completely supershifted complex D2 formed by extracts of uninduced cells (lane 3), the more slowly migrating D1 complex formed by induced cell extracts was not affected by antibody to Sp3 (lane 5). However, the addition of both α-Sp3 and α-ZEBRA supershifted D2 as well as D1 (lane 11).
Gel shift analysis is consistent with reporter mutagenesis data.
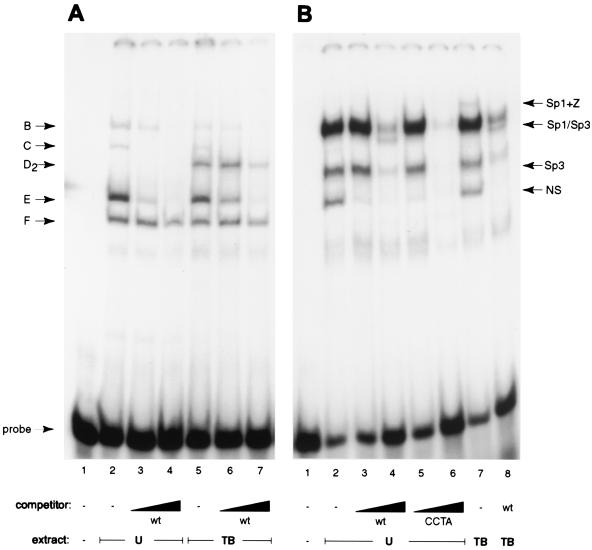
The next series of experiments demonstrated that mutations in Rp that led to decreased response to Rta (Fig. 3A) were associated with decreased binding by Sp1 and Sp3. The mutation (AT) in the 5′ end of the proximal Sp1 site of Rp that severely reduced the response of RpCAT to Rta effectively prevented binding of Sp1 and Sp3 to the Rp −64/−28 oligonucleotide (Fig. 8A). When the −64/−28 oligonucleotide containing this mutation was used as a probe, complex D2 (Sp3) was no longer observed, and only a small amount of complex B remained (Fig. 8A, lane 2). Use of the Sp1 AT mutant oligonucleotide allowed easy visualization of the D1 complex (ZEBRA) in chemically induced cell extracts (lane 5), as it was no longer masked by Sp3 in complex D2.
FIG. 8.
EMSA patterns on mutant oligonucleotides that decrease or enhance the response of Rp to Rta. Uninduced (U) and chemically induced (TB) Cl16 extracts were incubated with mutant oligonucleotide probes AT, which decreases the response to Rta (A), and CCTA, which increases the response to Rta (B). Where indicated, the reaction also included a 100- or 500-fold excess of unlabeled wild-type (wt) or mutant oligonucleotide. Complex designation is indicated on the left (B to F), identified proteins on the right (SS, supershifted bands). (A) Lane 1, probe alone; lanes 2 to 4, uninduced cell extract; lanes 5 to 7, TPA and sodium butyrate-induced cell extracts (TB). Rp −64/−28 was used as the unlabeled competitor at 100- and 500-fold excess in lanes 3, 4, 6, and 7. (B) Lane 1, probe alone; lanes 2 to 6, uninduced cell extract; lanes 7 and 8, TPA and sodium butyrate-induced cell extracts. Rp −64/−28 was used as the unlabeled competitor in lanes 3, 4, and 9; Rp −64/−28 CCTA was used in lanes 5 and 6.
An oligonucleotide probe with the CCTA mutation which eliminated the Zif268 site and resulted in enhanced response of Rp to Rta displayed a higher affinity for Sp1 and Sp3 (Fig. 8B). While this probe produced a pattern of DNA-protein complexes that was similar to that seen with the wild-type sequence, competition experiments showed that the CCTA mutant oligonucleotide competed for complexes B and D2 more efficiently than did the wild-type (−64/−28) fragment (Fig. 8B, compare lanes 3 and 4 with 5 and 6).
DISCUSSION
This paper presents several novel insights into the mechanism by which Rta protein autostimulates its promoter in EBV-positive B cells. Reporter assays demonstrated that the region between −299 and +30 relative to the transcriptional start site of Rp provides all sequences necessary for strong activation by Rta. Two Sp1 sites contained within this region proved essential for Rta to activate Rp. Other cis sequences that have been reported to be involved in the control of Rp, including two ZREs and two Zif268 sites, were dispensable for Rta-mediated autostimulation in Cl16 cells. Gel mobility shift assays determined that Sp1 and Sp3 were the principal factors binding a 37-bp Rp fragment in extracts of both uninduced and induced Cl16 cells. The binding of Sp1 and Sp3 to the probe correlated with the reporter activities in the mutagenesis study, establishing a direct link between transcriptional activation and DNA binding of Sp1 and/or Sp3. ZEBRA was also shown to bind to the Rp oligonucleotide in induced cell extracts, suggesting that it may stimulate the promoter in conjunction with Sp1.
cis-active elements mediating activation of Rp by Rta.
Mutagenesis in the region 5′ to the transcriptional start demonstrated that only the two Sp1 sites were crucial for autostimulation by Rta. Mutations eliminating either one or both of these sites reduced Rp activity between three- and eightfold relative to the wild-type promoter (Fig. 3B). While Zalani et al. had previously shown that Sp1 activates Rp in Drosophila SL2 cells and implicated the Sp1 sites in the constitutive Rp activity in epithelial cells (86), here we now assign these response elements a role in the indirect activation of Rp by Rta in B cells.
Although Zalani et al. demonstrated that Zif268 could modestly stimulate Rp in the D98/HE-R-1 fusion epithelial cell line (85), we found no evidence that Zif268 contributes positively to the regulation of Rp by Rta in Cl16 cells. Mutations of either Zif268 site did not reduce Rta-mediated Rp activity, provided that the proximal overlapping Sp1 site remained intact (Fig. 3B). There was no correlation between activation of Zif268 expression and induction of the viral lytic cycle in Cl16 cells (Fig. 4).
Mutation of the ZREs did not impair the ability of Rta to autostimulate its promoter. Thus, the indirect autostimulatory pathway at Rp is not mediated by ZEBRA's binding to the promoter. However, ZEBRA is likely to directly stimulate this promoter by binding to its cognate response elements. Therefore, Rp is stimulated by a complex mechanism involving cellular proteins, indirectly by Rta and directly by ZEBRA.
Proteins that bind the Rta-responsive region of Rp.
Figure 9 summarizes our results. The predominant factors binding to Rp −64/−28 in uninduced and induced Cl16 cell extracts were Sp1 and Sp3. ZEBRA was also found to bind the Rp fragment, either alone or in conjunction with Sp1, forming a larger complex of slow mobility. Although Sp1 and Sp3 bind to the same recognition sequences, only Sp1 and the smaller (70 to 80 kDa) isoform of Sp3 were found binding the same oligonucleotide together with ZEBRA. It is conceivable that ZEBRA interferes sterically with the binding of the large Sp3 isoform, but not Sp1. Conversely, the large (110 to 115 kDa) isoform of Sp3 may prevent ZEBRA from binding the oligonucleotide simultaneously. It is also possible that the expression of ZEBRA coincides with a disappearance of functionally competent Sp3.
FIG. 9.
Summary of protein-DNA complexes identified in EMSA experiments. The oligonucleotide probe Rp −64/−28 is illustrated with the overlapping Zif268 and Sp1 binding sites and the ZRE (ZIIIA). Ovals represent the proteins identified in complexes. Complexes A and D1 were identified only in extracts of induced cells; complexes B1/2 and D2 were found in both uninduced and induced cell extracts.
Overall, these results strongly suggest that Sp1 and/or Sp3 plays a role in the regulation of Rp. The Sp1 sites were crucial for Rta-induced promoter activity; moreover, Sp1 and Sp3 were the most prominent factors binding the proximal Sp1 site. Functional analysis by mutagenesis of cis-active sites strongly correlated with experiments examining protein-DNA interactions. A mutation causing severe reductions in promoter activity was associated with absence of Sp1 and Sp3 binding to a probe bearing the same mutation. Conversely, a gain-of-function mutation in the reporter assay also resulted in increased Sp1 and Sp3 binding in the gel shift.
Functional role of Sp1 and Sp3 in regulation of Rp.
How can the ubiquitous nature of Sp1 and Sp3 in uninduced and induced cell extracts be reconciled with their involvement in the regulation of a promoter activated only upon the switch from viral latency into the lytic cycle? Sp1 is generally considered an activator of housekeeping genes and cell cycle regulatory genes. It has also been shown to be essential for the prevention of silencing by the methylation of CpG islands (for a review, see references 10, 49, and 75). Sp1 is under posttranslational regulation by several kinases and phosphatases in response to different stimuli and signaling cascades (2, 4, 7, 32, 40, 54, 66, 68, 87). These modifications can result in altered DNA-binding affinity and transactivation potential. Consequently, despite its ubiquitous nature, Sp1 is under specific regulation and may be inhibited in its ability to activate Rp during viral latency.
While Sp1 and Sp3 are homologous proteins that have the same DNA-binding specificities, Sp3 exhibits more complex behavior than Sp1 (24, 25). Although Sp3 activates transcription under some circumstances (44, 45, 78), more often it antagonizes Sp1-mediated activation (24, 25, 50–52). Alterations in the Sp1 to Sp3 ratio may lead to activation or repression of individual promoters (3, 12, 26, 41, 75). Regulation of human papillomavirus (HPV) gene expression reveals the importance of the Sp1/Sp3 ratio in different cell backgrounds (3). While HPV may infect many different types of cells, viral gene expression is confined to epithelial cells (11, 19, 60, 65). Apt et al. demonstrated that Sp1 and Sp3 have opposing effects on HPV gene expression. Several epithelial cell lines supporting HPV gene expression exhibited higher Sp1/Sp3 ratios than other nonpermissive tissues. This observation led to the hypothesis that a high Sp1/Sp3 ratio was responsible for the cell type-specific expression of HPV genes (3).
Transcriptional regulation through the Sp1/Sp3 ratio may also be involved in control of the EBV Rp promoter. RpCAT reporters manifest higher constitutive activity in epithelial cells than in lymphocytes (18, 70, 86). This constitutive activity of RpCAT in epithelial cells has been shown to depend on the presence of the proximal Sp1 site (3, 86). These findings are consistent with the model of Apt et al. suggesting that a high Sp1/Sp3 ratio favors transcriptional activation by Sp1 in epithelial cells.
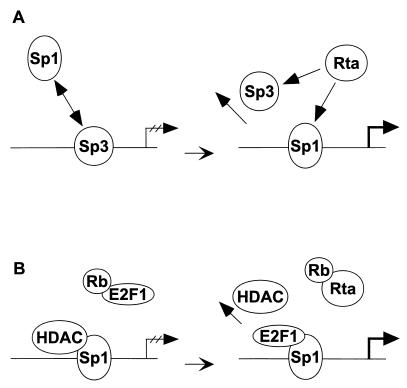
We therefore propose a model for the activation of Rp by Rta via Sp1 and Sp3 (Fig. 10). During latency, the promoter is silent due to an unfavorable functional ratio of Sp1 to Sp3 in the cell, leaving Sp3 to repress the activation of Rp. This may be achieved either by an enhanced DNA-binding affinity of Sp3 over Sp1 due to protein phosphorylation or by differences in protein concentration. An inducing stimulus for the lytic cycle of the virus may then reverse the ratio of Sp1 to Sp3, shifting it in favor of Sp1. This effect may be mimicked by expression of Rta, leading to autostimulation of Rp. There are several ways in which Rta might affect the amount of functional Sp1 or Sp3. Rta could directly activate expression of Sp1 or repress expression of Sp3. However, preliminary data from immunoblots do not suggest an alteration in Sp1 or Sp3 protein levels (data not shown). Rta could also associate with Sp3, sequestering it from the promoter. A direct interaction with Sp1 is less likely, since we cannot detect a DNA-protein complex that contains both Sp1 and Rta in our gel shift analysis. Furthermore, Rta could feed into signal transduction pathways leading to the modification of either Sp1 or Sp3.
FIG. 10.
Model for autoactivation of Rp by Rta, Sp1, and Sp3. Schematic illustration of the scenario explaining how the expression of Rta could lead to the activation of Rp through Sp1 in a permissive B-cell background such as Cl16 cells. (A) Rp is shown in its silent form during latency on the left and in its activated form on the right. Ovals represent the protein factors Sp1, Sp3, and Rta involved in the repression or activation processes. (B) Potential HDAC repression of Rp. See text for details.
The gel shift analysis did not reveal a change in abundance of Sp1 or Sp3 following expression of Rta. Their complex intensities were similar regardless of whether the binding extracts were prepared from uninduced or induced Cl16 cells. However, any subtle change in complex formation using induced Cl16 cell extracts is likely to be masked by an abundance of uninduced cells in the population. Chemical treatment induces about 40% of the cells into the lytic cycle, while transfection of Rta or ZEBRA expression vectors has an efficiency of approximately 1%. Determination of changes in complex abundance thus awaits an experimental strategy allowing 100% of the cells to be induced into the lytic cycle.
In a recent report, Doetzlhofer et al. proposed that inhibition of activation by Sp1 could result from an association with members of the histone deacetylase (HDAC) family (13). Not only could HDAC1 prevent Sp1 from engaging in other protein-protein interactions necessary for transcriptional activation, but the presence of HDAC1 could also stabilize the repressive effect of higher-order chromatin structures. Trichostatin A, an HDAC inhibitor, was reported to relieve this repression by HDAC1 (13). Other groups have also reported that trichostatin A enhances Sp1-mediated activity and that this process requires the concomitant expression of CBP/p300, a histone acetyltransferase (81). These events would be fully consistent with the activation of the EBV lytic cycle that is observed upon treatment of Cl16 cells with trichostatin A (Fig. 4).
The cell cycle-regulated transcription factor E2F1 may also be involved in the autostimulation of Rp by Rta. E2F1 might contribute to autoactivation in one of two ways, by binding the DNA or by binding to Sp1. It had previously been suggested that Rta activation of the EBV pol gene was mediated by an E2F-like protein binding to the DNA (47). Although Rp contains a potential binding site for E2F1, partially overlapping the proximal Sp1 site, we were unable to demonstrate direct binding of E2F1 to Rp in our gel shift assays. All members of the Sp1 protein family have been shown to be capable of interacting with E2F1 (67), and genes may be activated synergistically by the combination of Sp1 and E2F1 (34, 46, 67). In fact, E2F1 and HDAC1 interact with the same surface of Sp1. In their competition for binding to Sp1, E2F1 displaced HDAC1 and led to activation of transcription by the two factors (13). Either mechanism of action of E2F1 might be the result of the capacity of the Rta protein to interact with Rb and thereby cause the release of E2F1 (83).
The indirect transactivation mechanism of Rta appears to differ from that of other viral transcription factors, such as the herpes simplex virus type 1 VP16 protein and the EBV latency protein EBNA2. Neither of these proteins binds to its cognate DNA sequence alone, but requires cellular factors for recruitment to DNA. RBP-Jκ is responsible for targeting EBNA2 to DNA (29, 33, 79), while in the case of VP16, two proteins, Oct-1 and host cell factor, are required (17, 35, 39, 53, 74, 82). However, both EBNA2 and VP16 eventually come into contact with the DNA as the result of tethering by cell proteins. Rta is already capable of binding to DNA sequence specifically without the assistance of cofactors in vitro (21–23). This property presumably forms the basis of the direct activation of some viral genes, such as BaRF1, by Rta (64). The indirect mechanism of action of Rta, on the other hand, may not involve DNA binding to the viral promoter, since we and others have been unable to detect Rta binding to Rp probes in EMSA (86). Therefore, the ability of Rta to activate promoters lacking RREs may be the consequence of manipulation of cellular signaling pathways. This capacity may or may not involve Rta binding to the promoters of cellular genes or their protein products. We propose that this indirect function of Rta may evoke a change in the effective Sp1 to Sp3 ratio, leading to Sp1-mediated gene activation. The Rta protein thus has at its disposal at least two mechanisms by which it contributes to activation of the lytic cascade.
ACKNOWLEDGMENTS
This work was supported by NIH grants CA16038 and CA12055 to G.M.
We thank T. Serio for helpful discussions and critical reading of the manuscript and S. Ghosh for generous use of laboratory equipment.
REFERENCES
- 1.Ackerman S L, Minden A G, Williams G T, Bobonis C, Yeung C Y. Functional significance of an overlapping consensus binding motif for Sp1 and Zif268 in the murine adenosine deaminase gene promoter. Proc Natl Acad Sci USA. 1991;88:7523–7527. doi: 10.1073/pnas.88.17.7523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alroy I, Soussan L, Seger R, Yarden Y. Neu differentiation factor stimulates phosphorylation and activation of the Sp1 transcription factor. Mol Cell Biol. 1999;19:1961–1972. doi: 10.1128/mcb.19.3.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Apt D, Watts R M, Suske G, Bernard H U. High Sp1/Sp3 ratios in epithelial cells during epithelial differentiation and cellular transformation correlate with the activation of the HPV-16 promoter. Virology. 1996;224:281–291. doi: 10.1006/viro.1996.0530. [DOI] [PubMed] [Google Scholar]
- 4.Armstrong S A, Barry D A, Leggett R W, Mueller C R. Casein kinase II-mediated phosphorylation of the C terminus of Sp1 decreases its DNA binding activity. J Biol Chem. 1997;272:13489–13495. doi: 10.1074/jbc.272.21.13489. [DOI] [PubMed] [Google Scholar]
- 5.Babcock G J, Decker L L, Volk M, Thorley-Lawson D A. EBV persistence in memory B cells in vivo. Immunity. 1998;9:395–404. doi: 10.1016/s1074-7613(00)80622-6. [DOI] [PubMed] [Google Scholar]
- 6.Barroso I, Santisteban P. Insulin-induced early growth response gene (Egr-1) mediates a short term repression of rat malic enzyme gene transcription. J Biol Chem. 1999;274:17997–18004. doi: 10.1074/jbc.274.25.17997. [DOI] [PubMed] [Google Scholar]
- 7.Black A R, Jensen D, Lin S Y, Azizkhan J C. Growth/cell cycle regulation of Sp1 phosphorylation. J Biol Chem. 1999;274:1207–1215. doi: 10.1074/jbc.274.3.1207. [DOI] [PubMed] [Google Scholar]
- 8.Blier P R, Griffith A J, Craft J, Hardin J A. Binding of Ku protein to DNA: measurement of affinity for ends and demonstration of binding to nicks. J Biol Chem. 1993;268:7594–7601. [PubMed] [Google Scholar]
- 9.Bradford M M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 10.Brandeis M, Frank D, Keshet I, Siegfried Z, Mendelsohn M, Nemes A, Temper V, Razin A, Cedar H. Sp1 elements protect a CpG island from de novo methylation. Nature. 1994;371:435–438. doi: 10.1038/371435a0. [DOI] [PubMed] [Google Scholar]
- 11.Cripe T P, Haugen T H, Turk J P, Tabatabai F, Schmid P G d, Durst M, Gissmann L, Roman A, Turek L P. Transcriptional regulation of the human papillomavirus-16 E6–E7 promoter by a keratinocyte-dependent enhancer, and by viral E2 trans- activator and repressor gene products: implications for cervical carcinogenesis. EMBO J. 1987;6:3745–3753. doi: 10.1002/j.1460-2075.1987.tb02709.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Discher D J, Bishopric N H, Wu X, Peterson C A, Webster K A. Hypoxia regulates beta-enolase and pyruvate kinase-M promoters by modulating Sp1/Sp3 binding to a conserved GC element. J Biol Chem. 1998;273:26087–26093. doi: 10.1074/jbc.273.40.26087. [DOI] [PubMed] [Google Scholar]
- 13.Doetzlhofer A, Rotheneder H, Lagger G, Koranda M, Kurtev V, Brosch G, Wintersberger E, Seiser C. Histone deacetylase 1 can repress transcription by binding to Sp1. Mol Cell Biol. 1999;19:5504–5511. doi: 10.1128/mcb.19.8.5504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Flemington E, Speck S H. Autoregulation of Epstein-Barr virus putative lytic switch gene BZLF1. J Virol. 1990;64:1227–1232. doi: 10.1128/jvi.64.3.1227-1232.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flemington E K, Goldfeld A E, Speck S H. Efficient transcription of the Epstein-Barr Virus immediate-early BZLF1 and BRLF1 genes requires protein synthesis. J Virol. 1991;65:7073–7077. doi: 10.1128/jvi.65.12.7073-7077.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Francis A L, Gradoville L, Miller G. Alteration of a single serine in the basic domain of the Epstein-Barr virus ZEBRA protein separates its functions of transcriptional activation and disruption of latency. J Virol. 1997;71:3054–3061. doi: 10.1128/jvi.71.4.3054-3061.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gerster T, Roeder R G. A herpesvirus trans-activating protein interacts with transcription factor OTF-1 and other cellular proteins. Proc Natl Acad Sci USA. 1988;85:6347–6351. doi: 10.1073/pnas.85.17.6347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Glaser G, Vogel M, Wolf H, Niller H H. Regulation of the Epstein-Barr viral immediate early BRLF1 promoter through a distal NF1 site. Arch Virol. 1998;143:1967–1983. doi: 10.1007/s007050050433. [DOI] [PubMed] [Google Scholar]
- 19.Gloss B, Bernard H U, Seedorf K, Klock G. The upstream regulatory region of the human papilloma virus-16 contains an E2 protein-independent enhancer which is specific for cervical carcinoma cells and regulated by glucocorticoid hormones. EMBO J. 1987;6:3735–3743. doi: 10.1002/j.1460-2075.1987.tb02708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gottlieb T M, Jackson S P. The DNA-dependent protein kinase: requirement for DNA ends and association with Ku antigen. Cell. 1993;72:131–142. doi: 10.1016/0092-8674(93)90057-w. [DOI] [PubMed] [Google Scholar]
- 21.Gruffat H, Duran N, Buisson M, Wild F, Buckland R, Sergeant A. Characterization of an R-binding site mediating the R-induced activation of the Epstein-Barr virus BMLF1 promoter. J Virol. 1992;66:46–52. doi: 10.1128/jvi.66.1.46-52.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gruffat H, Manet E, Rigolet A, Sergeant A. The enhancer factor R of Epstein-Barr virus (EBV) is a sequence-specific DNA binding protein. Nucleic Acids Res. 1990;18:6835–6843. doi: 10.1093/nar/18.23.6835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gruffat H, Sergeant A. Characterization of the DNA-binding site repertoire for the Epstein-Barr virus transcription factor R. Nucleic Acids Res. 1994;22:1172–1178. doi: 10.1093/nar/22.7.1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hagen G, Dennig J, Preiss A, Beato M, Suske G. Functional analyses of the transcription factor Sp4 reveal properties distinct from Sp1 and Sp3. J Biol Chem. 1995;270:24989–24994. doi: 10.1074/jbc.270.42.24989. [DOI] [PubMed] [Google Scholar]
- 25.Hagen G, Muller S, Beato M, Suske G. Sp1-mediated transcriptional activation is repressed by Sp3. EMBO J. 1994;13:3843–3851. doi: 10.1002/j.1460-2075.1994.tb06695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hata Y, Duh E, Zhang K, Robinson G S, Aiello L P. Transcription factors Sp1 and Sp3 alter vascular endothelial growth factor receptor expression through a novel recognition sequence. J Biol Chem. 1998;273:19294–19303. doi: 10.1074/jbc.273.30.19294. [DOI] [PubMed] [Google Scholar]
- 27.Heinemeyer T, Chen X, Karas H, Kel A E, Kel O V, Liebich I, Meinhardt T, Reuter I, Schacherer F, Wingender E. Expanding the TRANSFAC database towards an expert system of regulatory molecular mechanisms. Nucleic Acids Res. 1999;27:318–322. doi: 10.1093/nar/27.1.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Heinemeyer T, Wingender E, Reuter I, Hermjakob H, Kel A E, Kel O V, Ignatieva E V, Ananko E A, Podkolodnaya O A, Kolpakov F A, Podkolodny N L, Kolchanov N A. Databases on transcriptional regulation: TRANSFAC, TRRD and COMPEL. Nucleic Acids Res. 1998;26:362–367. doi: 10.1093/nar/26.1.362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Henkel T, Ling P D, Hayward S D, Peterson M G. Mediation of Epstein-Barr virus EBNA2 transactivation by recombination signal-binding protein J kappa. Science. 1994;265:92–95. doi: 10.1126/science.8016657. [DOI] [PubMed] [Google Scholar]
- 30.Heston L, Rabson M, Brown N, Miller G. New Epstein-Barr virus variants from cellular subclones of P3J-HR-1 Burkitt lymphoma. Nature. 1982;295:160–163. doi: 10.1038/295160a0. [DOI] [PubMed] [Google Scholar]
- 31.Huang R P, Fan Y, Ni Z, Mercola D, Adamson E D. Reciprocal modulation between Sp1 and Egr-1. J Cell Biochem. 1997;66:489–499. [PubMed] [Google Scholar]
- 32.Jackson S P, MacDonald J J, Lees-Miller S, Tjian R. GC box binding induces phosphorylation of Sp1 by a DNA-dependent protein kinase. Cell. 1990;63:155–165. doi: 10.1016/0092-8674(90)90296-q. [DOI] [PubMed] [Google Scholar]
- 33.Johannsen E, Koh E, Mosialos G, Tong X, Kieff E, Grossman S R. Epstein-Barr virus nuclear protein 2 transactivation of the latent membrane protein 1 promoter is mediated by J kappa and PU.1. J Virol. 1995;69:253–262. doi: 10.1128/jvi.69.1.253-262.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Karlseder J, Rotheneder H, Wintersberger E. Interaction of Sp1 with the growth- and cell cycle-regulated transcription factor E2F. Mol Cell Biol. 1996;16:1659–1667. doi: 10.1128/mcb.16.4.1659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Katan M, Haigh A, Verrijzer C P, van der Vliet P C, O'Hare P. Characterization of a cellular factor which interacts functionally with Oct-1 in the assembly of a multicomponent transcription complex. Nucleic Acids Res. 1990;18:6871–6880. doi: 10.1093/nar/18.23.6871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Katz D A, Baumann R P, Sun R, Kolman J L, Taylor N, Miller G. Viral proteins associated with the Epstein-Barr virus transactivator, ZEBRA. Proc Natl Acad Sci USA. 1992;89:378–382. doi: 10.1073/pnas.89.1.378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kennett S B, Udvadia A J, Horowitz J M. Sp3 encodes multiple proteins that differ in their capacity to stimulate or repress transcription. Nucleic Acids Res. 1997;25:3110–3117. doi: 10.1093/nar/25.15.3110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kolman J L, Taylor N, Gradoville L, Countryman J, Miller G. Comparing transcriptional activation and autostimulation by ZEBRA and ZEBRA/c-Fos chimeras. J Virol. 1996;70:1493–1504. doi: 10.1128/jvi.70.3.1493-1504.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kristie T M, Sharp P A. Interactions of the Oct-1 POU subdomains with specific DNA sequences and with the HSV alpha-trans-activator protein. Genes Dev. 1990;4:2383–2396. doi: 10.1101/gad.4.12b.2383. [DOI] [PubMed] [Google Scholar]
- 40.Kumar A P, Butler A P. Serum responsive gene expression mediated by Sp1. Biochem Biophys Res Commun. 1998;252:517–523. doi: 10.1006/bbrc.1998.9676. [DOI] [PubMed] [Google Scholar]
- 41.Kumar A P, Butler A P. Transcription factor Sp3 antagonizes activation of the ornithine decarboxylase promoter by Sp1. Nucleic Acids Res. 1997;25:2012–2019. doi: 10.1093/nar/25.10.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Le Roux F, Sergeant A, Corbo L. Epstein-Barr virus (EBV) EB1/Zta protein provided in trans and competent for the activation of productive cycle genes does not activate the BZLF1 gene in the EBV genome. J Gen Virol. 1996;77:501–509. doi: 10.1099/0022-1317-77-3-501. [DOI] [PubMed] [Google Scholar]
- 43.Li Q X, Young L S, Niedobitek G, Dawson C W, Birkenbach M, Wang F, Rickinson A B. Epstein-Barr virus infection and replication in a human epithelial cell system. Nature. 1992;356:347–350. doi: 10.1038/356347a0. [DOI] [PubMed] [Google Scholar]
- 44.Liang Y, Robinson D F, Dennig J, Suske G, Fahl W E. Transcriptional regulation of the SIS/PDGF-B gene in human osteosarcoma cells by the Sp family of transcription factors. J Biol Chem. 1996;271:11792–11797. doi: 10.1074/jbc.271.20.11792. [DOI] [PubMed] [Google Scholar]
- 45.Liang Y, Robinson D F, Kujoth G C, Fahl W E. Functional analysis of the SIS proximal element and its activating factors: regulated transcription of the c-SIS/PDGF-B gene in human erythroleukemia cells. Oncogene. 1996;13:863–871. [PubMed] [Google Scholar]
- 46.Lin S Y, Black A R, Kostic D, Pajovic S, Hoover C N, Azizkhan J C. Cell cycle-regulated association of E2F1 and Sp1 is related to their functional interaction. Mol Cell Biol. 1996;16:1668–1675. doi: 10.1128/mcb.16.4.1668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu C, Sista N D, Pagano J S. Activation of the Epstein-Barr virus DNA polymerase promoter by the BRLF1 immediate-early protein is mediated through USF and E2F. J Virol. 1996;70:2545–2555. doi: 10.1128/jvi.70.4.2545-2555.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu S, Borras A M, Liu P, Suske G, Speck S H. Binding of the ubiquitous cellular transcription factors Sp1 and Sp3 to the ZI domains in the Epstein-Barr virus lytic switch BZLF1 gene promoter. Virology. 1997;228:11–18. doi: 10.1006/viro.1996.8371. [DOI] [PubMed] [Google Scholar]
- 49.Macleod D, Charlton J, Mullins J, Bird A P. Sp1 sites in the mouse aprt gene promoter are required to prevent methylation of the CpG island. Genes Dev. 1994;8:2282–2292. doi: 10.1101/gad.8.19.2282. [DOI] [PubMed] [Google Scholar]
- 50.Majello B, De Luca P, Hagen G, Suske G, Lania L. Different members of the Sp1 multigene family exert opposite transcriptional regulation of the long terminal repeat of HIV-1. Nucleic Acids Res. 1994;22:4914–4921. doi: 10.1093/nar/22.23.4914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Majello B, De Luca P, Lania L. Sp3 is a bifunctional transcription regulator with modular independent activation and repression domains. J Biol Chem. 1997;272:4021–4026. doi: 10.1074/jbc.272.7.4021. [DOI] [PubMed] [Google Scholar]
- 52.Majello B, De Luca P, Suske G, Lania L. Differential transcriptional regulation of c-myc promoter through the same DNA binding sites targeted by Sp1-like proteins. Oncogene. 1995;10:1841–1848. [PubMed] [Google Scholar]
- 53.McKnight J L, Kristie T M, Roizman B. Binding of the virion protein mediating alpha gene induction in herpes simplex virus 1-infected cells to its cis site requires cellular proteins. Proc Natl Acad Sci USA. 1987;84:7061–7065. doi: 10.1073/pnas.84.20.7061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Merchant J L, Du M, Todisco A. Sp1 phosphorylation by Erk 2 stimulates DNA binding. Biochem Biophys Res Commun. 1999;254:454–461. doi: 10.1006/bbrc.1998.9964. [DOI] [PubMed] [Google Scholar]
- 55.Miller G, Lipman M. Release of infectious Epstein-Barr virus by transformed marmoset leukocytes. Proc Natl Acad Sci USA. 1973;70:190–194. doi: 10.1073/pnas.70.1.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mimori T, Hardin J A. Mechanism of interaction between Ku protein and DNA. J Biol Chem. 1986;261:10375–10379. [PubMed] [Google Scholar]
- 57.Miyashita E M, Yang B, Lam K M, Crawford D H, Thorley-Lawson D A. A novel form of Epstein-Barr virus latency in normal B cells in vivo. Cell. 1995;80:593–601. doi: 10.1016/0092-8674(95)90513-8. [DOI] [PubMed] [Google Scholar]
- 58.Montalvo E A, Cottam M, Hill S, Wang Y J. YY1 binds to and regulates cis-acting negative elements in the Epstein- Barr virus BZLF1 promoter. J Virol. 1995;69:4158–4165. doi: 10.1128/jvi.69.7.4158-4165.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Mosser D D, Theodorakis N G, Morimoto R I. Coordinate changes in heat shock element-binding activity and HSP70 gene transcription rates in human cells. Mol Cell Biol. 1988;8:4736–4744. doi: 10.1128/mcb.8.11.4736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Muller M, Gissmann L, Cristiano R J, Sun X Y, Frazer I H, Jenson A B, Alonso A, Zentgraf H, Zhou J. Papillomavirus capsid binding and uptake by cells from different tissues and species. J Virol. 1995;69:948–954. doi: 10.1128/jvi.69.2.948-954.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pulvertaft R J V. Cytology of Burkitt's tumor (African lymphoma) Lancet. 1964;i:238–240. doi: 10.1016/s0140-6736(64)92345-1. [DOI] [PubMed] [Google Scholar]
- 62.Rabson M, Heston L, Miller G. Identification of a rare Epstein-Barr virus variant that enhances early antigen expression in Raji cells. Proc Natl Acad Sci USA. 1983;80:2762–2766. doi: 10.1073/pnas.80.9.2762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ragoczy T, Heston L, Miller G. The Epstein-Barr virus Rta protein activates lytic cycle genes and can disrupt latency in B lymphocytes. J Virol. 1998;72:7978–7984. doi: 10.1128/jvi.72.10.7978-7984.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ragoczy T, Miller G. Role of the Epstein-Barr virus Rta protein in activation of distinct classes of viral lytic cycle genes. J Virol. 1999;73:9858–9866. doi: 10.1128/jvi.73.12.9858-9866.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Roden R B, Kirnbauer R, Jenson A B, Lowy D R, Schiller J T. Interaction of papillomaviruses with the cell surface. J Virol. 1994;68:7260–7266. doi: 10.1128/jvi.68.11.7260-7266.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rohlff C, Ahmad S, Borellini F, Lei J, Glazer R I. Modulation of transcription factor Sp1 by cAMP-dependent protein kinase. J Biol Chem. 1997;272:21137–21141. doi: 10.1074/jbc.272.34.21137. [DOI] [PubMed] [Google Scholar]
- 67.Rotheneder H, Geymayer S, Haidweger E. Transcription factors of the Sp1 family: interaction with E2F and regulation of the murine thymidine kinase promoter. J Mol Biol. 1999;293:1005–1015. doi: 10.1006/jmbi.1999.3213. [DOI] [PubMed] [Google Scholar]
- 68.Schafer D, Hamm-Kunzelmann B, Brand K. Glucose regulates the promoter activity of aldolase A and pyruvate kinase M2 via dephosphorylation of Sp1. FEBS Lett. 1997;417:325–328. doi: 10.1016/s0014-5793(97)01314-8. [DOI] [PubMed] [Google Scholar]
- 69.Serio T R, Kolman J L, Miller G. Late gene expression from the Epstein-Barr virus BcLF1 and BFRF3 promoters does not require DNA replication in cis. J Virol. 1997;71:8726–8734. doi: 10.1128/jvi.71.11.8726-8734.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sinclair A J, Brimmell M, Shanahan F, Farrell P J. Pathways of activation of the Epstein-Barr virus productive cycle. J Virol. 1991;65:2237–2244. doi: 10.1128/jvi.65.5.2237-2244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sixbey J W, Nedrud J G, Raab-Traub N, Hanes R A, Pagano J S. Epstein-Barr virus replication in oropharyngeal epithelial cells. N Engl J Med. 1984;310:1225–1230. doi: 10.1056/NEJM198405103101905. [DOI] [PubMed] [Google Scholar]
- 72.Spain T A, Sun R, Miller G. The locus of Epstein-Barr virus terminal repeat processing is bound with enhanced affinity by Sp1 and Sp3. Virology. 1997;237:137–147. doi: 10.1006/viro.1997.8770. [DOI] [PubMed] [Google Scholar]
- 73.Speck S H, Chatila T, Flemington E. Reactivation of Epstein-Barr virus: regulation and function of the BZLF1 gene. Trends Microbiol. 1997;5:399–405. doi: 10.1016/S0966-842X(97)01129-3. [DOI] [PubMed] [Google Scholar]
- 74.Stern S, Tanaka M, Herr W. The Oct-1 homeodomain directs formation of a multiprotein-DNA complex with the HSV transactivator VP16. Nature. 1989;341:624–630. doi: 10.1038/341624a0. [DOI] [PubMed] [Google Scholar]
- 75.Suske G. The Sp-family of transcription factors. Gene. 1999;238:291–300. doi: 10.1016/s0378-1119(99)00357-1. [DOI] [PubMed] [Google Scholar]
- 76.Takada K, Ono Y. Synchronous and sequential activation of latently infected Epstein-Barr virus genomes. J Virol. 1989;63:445–449. doi: 10.1128/jvi.63.1.445-449.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Thottassery J V, Sun D, Zambetti G P, Troutman A, Sukhatme V P, Schuetz E G, Schuetz J D. Sp1 and egr-1 have opposing effects on the regulation of the rat Pgp2/mdr1b gene. J Biol Chem. 1999;274:3199–3206. doi: 10.1074/jbc.274.5.3199. [DOI] [PubMed] [Google Scholar]
- 78.Udvadia A J, Templeton D J, Horowitz J M. Functional interactions between the retinoblastoma (Rb) protein and Sp- family members: superactivation by Rb requires amino acids necessary for growth suppression. Proc Natl Acad Sci USA. 1995;92:3953–3957. doi: 10.1073/pnas.92.9.3953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Waltzer L, Bourillot P Y, Sergeant A, Manet E. RBP-J kappa repression activity is mediated by a co-repressor and antagonized by the Epstein-Barr virus transcription factor EBNA2. Nucleic Acids Res. 1995;23:4939–4945. doi: 10.1093/nar/23.24.4939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Weigel R, Miller G. Major EB virus-specific cytoplasmic transcripts in a cellular clone of the HR-1 Burkitt lymphoma line during latency and after induction of viral replicative cycle by phorbol esters. Virology. 1983;125:287–298. doi: 10.1016/0042-6822(83)90202-7. [DOI] [PubMed] [Google Scholar]
- 81.Xiao H, Hasegawa T, Isobe K. p300 collaborates with Sp1 and Sp3 in p21(waf1/cip1) promoter activation induced by histone deacetylase inhibitor. J Biol Chem. 2000;275:1371–1376. doi: 10.1074/jbc.275.2.1371. [DOI] [PubMed] [Google Scholar]
- 82.Xiao P, Capone J P. A cellular factor binds to the herpes simplex virus type 1 transactivator Vmw65 and is required for Vmw65-dependent protein-DNA complex assembly with Oct-1. Mol Cell Biol. 1990;10:4974–4977. doi: 10.1128/mcb.10.9.4974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Zacny V L, Wilson J, Pagano J S. The Epstein-Barr virus immediate-early gene product BRLF1 interacts with the retinoblastoma protein during the viral lytic cycle. J Virol. 1998;72:8043–8051. doi: 10.1128/jvi.72.10.8043-8051.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Zalani S, Coppage A, Holley-Guthrie E, Kenney S. The cellular YY1 transcription factor binds a cis-acting, negatively regulating element in the Epstein-Barr virus BRLF1 promoter. J Virol. 1997;71:3268–3274. doi: 10.1128/jvi.71.4.3268-3274.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Zalani S, Holley-Guthrie E, Kenney S. The Zif268 cellular transcription factor activates expression of the Epstein-Barr virus immediate-early BRLF1 promoter. J Virol. 1995;69:3816–3823. doi: 10.1128/jvi.69.6.3816-3823.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zalani S, Holley-Guthrie E A, Gutsch D E, Kenney S C. The Epstein-Barr virus immediate-early promoter BRLF1 can be activated by the cellular Sp1 transcription factor. J Virol. 1992;66:7282–7292. doi: 10.1128/jvi.66.12.7282-7292.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zhang S, Kim K H. Protein kinase CK2 down-regulates glucose-activated expression of the acetyl-CoA carboxylase gene. Arch Biochem Biophys. 1997;338:227–232. doi: 10.1006/abbi.1996.9809. [DOI] [PubMed] [Google Scholar]