Abstract
As advances in analytical separation techniques, mass spectrometry instrumentation, and data processing platforms continue to spur growth in the lipidomics field, more structurally unique lipid species are detected and annotated. The lipidomics community is in need of benchmark reference values to assess the validity of various lipidomics workflows in providing accurate quantitative measurements across the diverse lipidome. LipidQC addresses the harmonization challenge in lipid quantitation by providing a semiautomated process, independent of analytical platform, for visual comparison of experimental results of National Institute of Standards and Technology Standard Reference Material (SRM) 1950, “Metabolites in Frozen Human Plasma”, against benchmark consensus mean concentrations derived from the NIST Lipidomics Interlaboratory Comparison Exercise.
Graphical Abstract
Lipids are a diverse group of biomolecules that play an important role in membrane structure, energy storage, and cell signaling/trafficking that have been used to assess human health and disease.1–3 With recent advancements in analyte separation, mass spectrometry instrumentation, and data processing platforms, many structurally unique lipid species have been detected and annotated.1,4–8 Despite this progress, standardized workflows for sample preparation, analytical platforms, data processing, and lipid annotation are not fully characterized and a comparison of lipidomics results between analytical platforms and laboratories remains a challenge.9,10 As an example, no two lipidomics workflows were the same among the 31 participating laboratories in the National Institute of Standards and Technology (NIST) Lipidomic Interlaboratory Comparison Exercise (NIST-ILCE).11
Quality control samples are commonly incorporated in routine sample analysis to assess data quality and account for variance introduced from sample preparation, systematic noise, and post-processing steps such as batch or drift correction.12 In addition, quality control measures can be used to assess the reproducibility and quality of a dataset, since resulting assignments affect the biochemical interpretation.13,14 The NIST Standard Reference Material, SRM 1950 “Metabolites in Frozen Plasma”, is often extracted alongside test plasma samples to serve as a matrix-matched quality control for metabolomics and lipidomics applications.15,16 However, certified values for SRM 1950 are only available for selected metabolites (e.g., amino acids, vitamins, carotenoids, etc.), fatty acids, and total cholesterol. These certified values are metrologically traceable to the International System of Units (SI) and are suitable for measurement standardization. Additional values of interest (labeled as “reference” or “information” values17) are also provided that are only defined by specialized analytical methods and can useful when assessing measurements from similar systems, since no metrological traceability chain has been established. The lipidomics field needs such values that better reflect the diversity of the lipidome for interand intralaboratory harmonization, which is the first step toward standardization.
In 2011, LIPID Metabolites and Pathways Strategy (LIPID MAPS) quantified 588 of the more abundant lipid species present in SRM 1950, using a targeted triple quadrupole mass spectrometry platform.18 These values were obtained by a single laboratory and are limited to targeted lipidomics workflows; therefore, these (1) may not be comparable to results from other analytical platforms or (2) may not reflect the diversity present in the results from untargeted lipidomics studies. To efficiently harmonize results across laboratories, the lipidomics community needs more robust benchmark values that are independent of the analytical platform or the biomarker discovery workflow design.
The NIST-ILCE11 provided robustly measured consensus mean value estimates for SRM 1950, using submissions from 31 diverse national and international laboratories, each employing a different lipidomics workflow. The interlaboratory is composed of both global and targeted lipidomic methodologies spanning across academia, industry, and core facilities. Consensus mean value estimates were reported for the following lipid categories: fatty acyls (FA), glycerolipids (GL), glycerophospholipids (GP), sphingolipids (SP), and sterols (ST). While the consensus values generated from the NIST-ILCE are not metrologically traceable to the SI, these values were measured by a cross-section of the lipidomics community and are useful for harmonization purposes.
LipidQC is visualization tool that provides a means—independent of sample preparation methods, MS instruments, and lipid adduct—to rapidly compare lipid concentration measurements (nmol/mL) with the available consensus mean value estimates for SRM 1950. Users can quickly compare experimental concentrations (nmol/mL) of lipid species quantified for the SRM material against the consensus estimates and corresponding uncertainties obtained from the NIST-ILCE.11 Lipid classes supported in LipidQC, as shown in Table S1 in the Supporting Information, include the following: free fatty acids (FFA), eicosanoids, diacylglycerols (DAG), triacylglycerols (TAG), lysophosphatidylcholines (LPC), phosphatidylcholines (PC), phosphatidylcholines (PC), lysophosphatidylethanolamines (LPE), phosphatidylethanolamines (PE), phosphatidylglycerols (PG), phosphatidylinositols (PI), phosphatidylserines (PS), ceramides (CER), dihydroceramides (CerOH), hexosylceramides (HexCer), lactosylceramides (LacCer), sphingomyelin (SM), sphingosine-1-phosphate (S1P), sphinganine-1-phosphate (dhS1P), cholesteryl esters (CE), free cholesterol/cholesterol derivates (FC/CHOL), and bile acids (BA).
EXPERIMENTAL SECTION
Sample Preparation.
To highlight the applicability of LipidQC for multiple analytical platforms (e.g., direct infusion versus chromatography and low-resolution vs high-resolution), lipids were extracted from SRM 1950 using both a Bligh–Dyer19 lipid extraction with 30 μL of plasma (Method A) and a modified Bligh–Dyer lipid extraction protocol with 25 μL of plasma (Method B). Only generic information regarding the method and instrument platform is given, to provide anonymity of the data and prevent the endorsement of any method/analytical platform. The data obtained from these methods were not used to generate the consensus mean values and corresponding uncertainties in the NIST-ILCE.
Instrument Platforms.
The lipid extracts from Method A were analyzed using a high-resolution orbitrap mass spectrometer coupled to an ultrahigh-performance liquid chromatography (UHPLC) system. A Waters Acquity C18 BEH column (2.1 mm × 100 mm, 1.7 μm particle size, Waters, Milford, MA) maintained at 60 °C was used for all lipid extracts. The injection volume was 5 and 10 μL in positive and negative ion mode, respectively, with a mobile phase flow rate of 450 μL/min. The gradient program consisted of mobile phase C [60:40, v/v, acetonitrile/water] and mobile phase D [90:8:2, v/v/v, isopropanol/acetonitrile/water], each containing 10 mmol/L ammonium formate and 0.1% formic acid. Full scan and data-dependent MS/MS (top10-ddMS2) were collected at m/z 150–2000. The data were collected and analyzed using an untargeted lipidomics approach.
The lipid extracts from Method B were analyzed using a triple quadrupole mass spectrometer coupled to a LC system for direct flow injection. Two injections of 50 μL each was used for analysis. MRM transitions were used for the analysis of FFA, TAG, DAG, CE, PC, LPC, PE, LPE, SM, and CER lipid classes.
NIST Lipidomics Interlaboratory Comparison.
Thirty-one (31) laboratories participated in the NIST-ILCE, providing submissions for lipid species quantified in NIST SRM 1950. SRM 1950 was developed in 2006 as a “normal” human plasma reference material that was constructed from 100 fasting individuals, ages 40–50, who represented the average U.S. population, as defined by race, sex, and health.
Consensus means were calculated in the NIST-ILCE, using the Median of Means (MEDM) estimation method,20 for lipid species measured by five or more participating laboratories with coefficient of dispersion (COD) values of ≤40%.21 To expand the lipidome coverage, additional consensus means were also calculated, using the DerSimonian Laird (DSL) estimation method22 for lipid species measured by three or four laboratories with CODs of ≤40% and a ≤ 20% percent difference between the MEDM and DSL estimations. The DSL estimation method was employed for these tentative values, because the uncertainty calculation for the MEDM method cannot be applied to instances where values are reported by less than five laboratories. The uncertainty calculations used for the MEDM and DSL estimation methods are described elsewhere.23 All consensus mean estimates, uncertainties, and calculations associated with the interlaboratory exercise for SRM 1950 are provided in the NIST Internal Report.24
Supported Nomenclature.
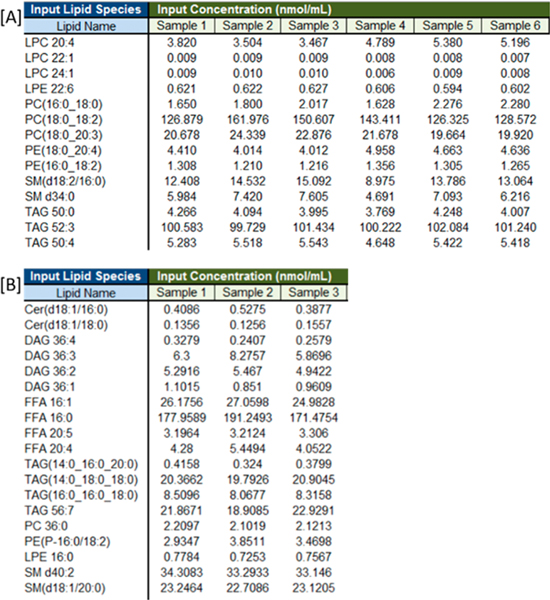
The NIST-ILCE consensus mean values were reported using sum composition annotations (e.g., PC 34:2). LipidQC supports inputs at the sum composition [PC 36:0], fatty acid position level with known fatty acyl position [PC(16:0/18:1)], and fatty acid level with unknown fatty acyl position [PC (16:0_18:1)]. Lipid species names provided by the user, as shown in Figure 1, are parsed to determine the following: (1) lipid class, (2) sum composition of each lipid species using the methodology employed in LipidPioneer,25 and (3) sum concentration of isobaric lipid species of the same lipid class. Figure 1B is an example where the experimental means of three isomeric TAG lipid species [TAG(14:0_16:0_20:0), TAG(14:0_18:0_18:0), and TAG(16:0_16:0_18:0)] were summed. The sum is reported as a mean for TAG 50:0 in Figure S1-B in the Supporting Information.
Figure 1.
[A] Screen capture of the input concentration table for example lipid species quantified (nmol/mL) in Bligh–Dyer lipid extracts analyzed by ultrahigh-performance liquid chromatography–high-resolution mass spectroscopy (UHPLC-HRMS). [B] Screen capture of input concentration table for example lipid species quantified (nmol/mL) in modified Bligh–Dyer lipid extracts analyzed by DI-MS/MS. Figures S1 and S2 in the Supporting Information show the summary table and accuracy assessment, respectively, for the DI-MS/MS input data.
The NIST-ILCE also combined isomeric ether-linked lipid species for the PE and PC lipid classes at the sum composition level (e.g., PC O-40:5 and PC P-40:4). Therefore, if LipidQC recognizes separate entries for isobaric species within the same class that were reported as a single value in the NIST Interlaboratory consensus mean report (e.g., PC O-40:5, PC P-40:4, and PC 39:5), those isobaric entries are then combined in a similar manner. A note is provided in the final summary table, indicating which lipids were combined for comparison from the input concentration table (see Figure S1-B).
Table 1 lists the included eicosanoids and bile acids, along with the supported abbreviation. Lipid name entries should begin with the abbreviation for the lipid class (Table S1 in the Supporting Information) followed by the tail information. For ether-linked lipid species, the first acyl chain should have a prefix of “P-” for plasmenyl ethers and “O-” for plasmanyl ether-linked lipid species. LipidQC only supports individual entries for lipid species (i.e., PC O-40:7 and PC P-40:6 are separate inputs, instead of a submission as PC O-40:7/PC P-40:6), as annotations for isobaric species are not supported. Detailed instructions for the annotation style or lipid nomenclature supported in LipidQC can be found on the tool’s “Instructions” tab.
Table 1.
Supported Eicosanoid and Bile Acids
| eicosanoids | bile acids (BA) | ||
|---|---|---|---|
| 5-HETE | 5-HEPE | CDCA | TDCA |
| 8-HETE | 9-HEPE | CA | TLCA |
| 9-HETE | 18-HEPE | DCA | UDCA |
| 12-HETE | 12-HHTrE | GCDCA | TLCA-S |
| 15-HETE | 13-HOTrE | GDCA | MCA |
| 20-HETE | 14-HDoHE | GLCA | LCA |
| 11,12-DiHETErE | 17-HDoHE | GUDCA | TCDCA |
| 11-HDoHE | 5,6-EET | GCA | TCA |
| 9,10-DiHOME | 8-HETrE | ||
| 12,13-DiHOME | 9-HODE | ||
| 12,13-EpOME | 9-OxoODE | ||
| PGE2 | |||
Computation.
LipidQC is a Microsoft Excel-based system that uses Visual Basic for Applications scripting and runs on Windows 2010 and later platforms.
RESULTS AND DISCUSSION
Comparison to NIST-ILCE Consensus Values.
LipidQC compares input experimental data to benchmark consensus mean concentrations (nmol/mL). Currently, consensus values are available only for SRM 1950; however, a dropdown box is provided to enable the selection of different reference materials when data become available.
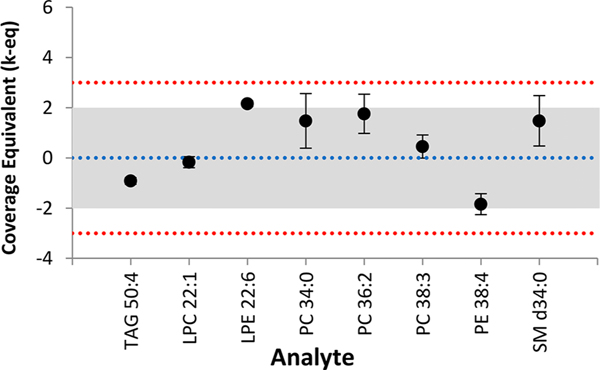
Along with the mean, the standard deviation is also calculated for experimentally obtained concentrations of lipids quantified. LipidQC produces a result table (see Figure S1) and landing strip style control plot (see Figure 2) that compares experimental results to the consensus values. The user should determine the appropriate number of significant figures for the reported mean and standard deviation based on the analytical platform employed. The result table indicates the number of laboratories that contributed to the reported consensus mean value (see Figure S1-A). The control plot (Figure S2 in the Supporting Information) shows the coverage equivalent for each lipid species.
Figure 2.
Example control plot. Results are presented as normalized coverage equivalents (k-eq) at the mean (data points) and standard deviation (error bars). The blue dotted line represents the consensus mean. The middle shaded region represents the 95% expanded uncertainty interval for the NIST-ILCE consensus mean estimate; the red dotted lines bound the 99% expanded uncertainty.
LipidQC also displays ∼95% and ∼99% expanded uncertainty regions on the consensus mean. If the experimental mean and/or error bars for a lipid species fall within the 95% (middle shaded) region, the measurement is largely consistent with the lipidomics community. If the experimental mean falls outside the 99% (red dotted line) region, the measurement is likely inconsistent, although the inconsistency may be due to the summation of lipids. The consensus means were generated using the summation of all reported isobaric lipid species within the same class. A list of all summed lipid species used to generate the consensus mean values for SRM 1950 can be found in the NIST-ILCE summary report.24
In summary, LipidQC currently provides comparisons against 254 values (n ≥ 5 laboratories reporting) and 62 values (n = 3 or 4 laboratories reporting) for SRM 1950. Table S2 in the Supporting Information details the all consensus mean values included in LipidQC per lipid class.
Applicability of LipidQC to Multiple Analytical Platforms.
LipidQC opens on the “Instructions” page and prompts the user to save a new copy of the program. The user inputs measured concentrations on the input table by selecting “Enter Values”. The user should indicate, on the input table, the appropriate number of headers for replicates, because this field is initially left blank (e.g., Sample 1, Sample 2, Sample, 3, etc.). After values have been entered in the input table, the user selects the “Click Here When Finished” button in the bottom left corner to start the visualization tool comparison. Values can be edited in the input table by selecting the “Click Here to Edit Value” button in the bottom left corner.
LipidQC generates the sum composition for all lipid species included in the input concentration table. Isobaric lipid species within the same class are summed and compared to the corresponding consensus mean value. For example, Method 1 incorporated six replicates (Figure 1A) and Method 2 incorporated three replicates (Figure 1B). It is recommended that at least three replicates are used for each material.
Figure S2-A shows agreement between the UHPLC-HRMS/MS data and the consensus mean values. However, lipid species that fall out of the 95% and 99% uncertainty regions are easily identified (e.g., PE 34:2, PE 38:4, LPC 20:4). The control plot allows for class-dependent trends to quickly be identified, because this may imply that the lipid class was not measured appropriately with the user’s current methodology. For example, in the DI-MS/MS data (Figure S2-B), two out of the three FFA lipid species and both chosen SM lipid species fell outside of the 95% and 99% uncertainty regions.
CONCLUSION
Various sample handling/preparation methodologies, internal standards, sample introduction and chromatographic methods, mass spectrometer platforms, tandem MS scanning approaches, and data processing software are employed within the lipidomics community. As the lipidomics field advances, however, it becomes increasingly important to address harmonization limitations.
Literature has shown the analysis of SRM 1950 to be reproducible and transferable across multiple LC/MS platforms for metabolomics applications.15 LipidQC allows lipidomics users to visually compare SRM 1950 data from not only LC platforms, but also direct infusion platforms and various mass spectrometers (high resolution and low resolution).
LipidQC provides an easy, routine, and automated tool for comparing experimental data consensus estimates for well-characterized reference materials. SRM 1950 is the first such material, but was not designed for lipidomic harmonization and is not necessarily optimal for this purpose. We anticipate that new reference materials will be developed specifically for and by the lipidomic community. We hope that these materials, interlaboratory studies such as NIST-ILCE, and the LipidQC tool will help the community harmonize their measurement processes.
Supplementary Material
ACKNOWLEDGMENTS
The work performed and the consensus mean value estimates obtained in this study would not be possible without the funding support (either partial or full) from each contributing laboratory in the NIST Lipidomics Interlaboratory Comparison exercise. This research was done in collaboration between Core 1 and Core 3 of the Southeast Center for Metabolomics (SECIM) ( http://secim.ufl.edu/) (NIH Grant No. U24 DK097209).
Footnotes
ASSOCIATED CONTENT
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acs.analchem.7b04042.
Reference validation tool for lipids observed in SRM 1950 by participating laboratories in the NIST Lipidomics Interlaboratory Comparison Exercise. (LipidQC v.1.0.xlsm, Microsoft Excel Macro-Enabled Workbook). LipidQC is freely available for download at http://secim.ufl.edu/secim-tools/ (ZIP)
Example summary table for experimental results compared against NIST consensus mean values for SRM 1950 (Figure S-1); example control plot for experimental results compared against NIST consensus mean values for SRM 1950 (Figure S-2); LipidQC-supported lipid classes and their corresponding abbreviations (Table S-1); breakdown of LipidQC lipid species included with NIST consensus mean values (Table S-2) (PDF)
Certain commercial equipment, instruments, or materials are identified in this paper to specify adequately the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology; nor does it imply that the materials or equipment identified are necessarily the best for the purpose.
The authors declare no competing financial interest.
REFERENCES
- (1).Yetukuri L; Ekroos K; Vidal-Puig A; Oresǐ M. Mol. BioSyst 2008, 4, 121–127. [DOI] [PubMed] [Google Scholar]
- (2).Kolovou G; Kolovou V; Mavrogeni S. Vasc. Health Risk Manage 2015, 11, 333–342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Loizides-Mangold U. FEBS J. 2013, 280, 2817–2829. [DOI] [PubMed] [Google Scholar]
- (4).Li M; Zhou Z; Nie H; Bai Y; Liu H. Anal. Bioanal. Chem 2011, 399, 243–249. [DOI] [PubMed] [Google Scholar]
- (5).Narvaez-Rivas M; Zhang QJ Chromatogr. A 2016, 1440, 123–134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (6).Yamada T; Bamba T. In Lipidomics; Wood, Ed.; Springer: New York, 2017; pp 109–131. [Google Scholar]
- (7).Paglia G; Astarita G. Nat. Protoc 2017, 12, 797–813. [DOI] [PubMed] [Google Scholar]
- (8).Hines KM; Herron J; Xu LJ Lipid Res. 2017, 58 (4), 809–819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Cajka T; Fiehn O. TrAC, Trends Anal. Chem 2014, 61, 192–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (10).Yang L; Li M; Shan Y; Shen S; Bai Y; Liu HJ Sep. Sci 2016, 39, 38–50. [DOI] [PubMed] [Google Scholar]
- (11).Bowden JA; Heckert A; Ulmer CZ; Jones CM; Koelmel JP; Abdullah L; Ahonen L; Alnouti Y; Armando A; Asara JM; Bamba T; Barr JR; Bergquist J; Borchers CH; Brandsma J; Breitkopf SB; Cajka T; Cazenave-Gassiot A; Checa A; Cinel MA, et al. J. Lipid Res 2017, in press; (DOI: 10.1194/jlr.M07901210.1194/jlr.M079012). [DOI] [Google Scholar]
- (12).Luan H; Meng N; Liu P; Fu J; Chen X; Rao W; Jiang H; Xu X; Cai Z; Wang J. GigaScience 2015, 4, 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Trabado S; Al-Salameh A; Croixmarie V; Masson P; Corruble E; Fève B; Colle R; Ripoll L; Walther B; Boursier-Neyret C; Werner E; Becquemont L; Chanson P. PLoS One 2017, 12, e0173615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Sales S; Graessler J; Ciucci S; Al-Atrib R; Vihervaara T; Schuhmann K; Kauhanen D; Sysi-Aho M; Bornstein SR; Bickle M; Cannistraci CV; Ekroos K; Shevchenko A. Sci. Rep 2016, 6, 27710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Simon-Manso Y; Lowenthal MS; Kilpatrick LE; Sampson ML; Telu KH; Rudnick PA; Mallard WG; Bearden DW; Schock TB; Tchekhovskoi DV; Blonder N; Yan X; Liang Y; Zheng Y; Wallace WE; Neta P; Phinney KW; Remaley AT; Stein SE Anal. Chem 2013, 85, 11725–11731. [DOI] [PubMed] [Google Scholar]
- (16).Telu KH; Yan X; Wallace WE; Stein SE; Simón-Manso Y. Rapid Commun. Mass Spectrom 2016, 30, 581–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).May W; Parris R; Beck C; Fassett J; Greenberg R; Guenther F; Kramer G; Wise S; Gills T; Colbert J; Gettings R; MacDonald B. NIST Spec. Publ 2000, 260–136, 1–18. [Google Scholar]
- (18).Quehenberger O; Armando AM; Brown AH; Milne SB; Myers DS; Merrill AH; Bandyopadhyay S; Jones KN; Kelly S; Shaner RL; Sullards CM; Wang E; Murphy RC; Barkley RM; Leiker TJ; Raetz CR; Guan Z; Laird GM; Six DA; Russell DW; et al. J. Lipid Res 2010, 51, 3299–3305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Bligh EG; Dyer WJ Can. J. Biochem. Physiol 1959, 37, 911–917. [DOI] [PubMed] [Google Scholar]
- (20).Harris P; Cox M; Forbes A; Matthews C. Estimation of a consensus KCRV and associated Degrees of Equivalence, Consultative Committee for Amount of Substance (CCQM). CCQM Guidance Note; Bureau International des Poids et Mesures (BIPM): Sèvres, Paris, 2013, 10, 1–34. https://www.bipm.org/cc/CCQM/Allowed/19/CCQM13-22_Consensus_KCRV_v10.pdf [Google Scholar]
- (21).Bonett DG; Seier E. Biom. J 2006, 48, 144–148. [DOI] [PubMed] [Google Scholar]
- (22).DerSimonian R; Laird N. Controlled Clin. Trials 1986, 7, 177–188. [DOI] [PubMed] [Google Scholar]
- (23).Horn SD; Horn RA; Duncan DB J. Am. Stat. Assoc 1975, 70, 380–385. [Google Scholar]
- (24).Bowden JA; Heckert A; Ulmer CZ; Jones CM NIST Interagency/Internal Report, No. (NISTIR)-8185, 2017; pp 1–451. [Google Scholar]
- (25).Ulmer CZ; Koelmel JP; Ragland JM; Garrett TJ; Bowden JA J. Am. Soc. Mass Spectrom 2017, 28, 562–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.