Abstract
Interpreting the phenotypes of blaSHV alleles in Klebsiella pneumoniae genomes is complex. Whilst all strains are expected to carry a chromosomal copy conferring resistance to ampicillin, they may also carry mutations in chromosomal blaSHV alleles or additional plasmid-borne blaSHV alleles that have extended-spectrum β-lactamase (ESBL) activity and/or β-lactamase inhibitor (BLI) resistance activity. In addition, the role of individual mutations/a changes is not completely documented or understood. This has led to confusion in the literature and in antimicrobial resistance (AMR) gene databases [e.g. the National Center for Biotechnology Information (NCBI) Reference Gene Catalog and the β-lactamase database (BLDB)] over the specific functionality of individual sulfhydryl variable (SHV) protein variants. Therefore, the identification of ESBL-producing strains from K. pneumoniae genome data is complicated. Here, we reviewed the experimental evidence for the expansion of SHV enzyme function associated with specific aa substitutions. We then systematically assigned SHV alleles to functional classes (WT, ESBL and BLI resistant) based on the presence of these mutations. This resulted in the re-classification of 37 SHV alleles compared with the current assignments in the NCBI’s Reference Gene Catalog and/or BLDB (21 to WT, 12 to ESBL and 4 to BLI resistant). Phylogenetic and comparative genomic analyses support that (i) SHV-1 (encoded by blaSHV-1) is the ancestral chromosomal variant, (ii) ESBL- and BLI-resistant variants have evolved multiple times through parallel substitution mutations, (iii) ESBL variants are mostly mobilized to plasmids and (iv) BLI-resistant variants mostly result from mutations in chromosomal blaSHV. We used matched genome–phenotype data from the KlebNET-GSP AMR Genotype-Phenotype Group to identify 3999 K. pneumoniae isolates carrying one or more blaSHV alleles but no other acquired β-lactamases to assess genotype–phenotype relationships for blaSHV. This collection includes human, animal and environmental isolates collected between 2001 and 2021 from 24 countries. Our analysis supports that mutations at Ambler sites 238 and 179 confer ESBL activity, whilst most omega-loop substitutions do not. Our data also provide support for the WT assignment of 67 protein variants, including 8 that were noted in public databases as ESBL. These eight variants were reclassified as WT because they lack ESBL-associated mutations, and our phenotype data support susceptibility to third-generation cephalosporins (SHV-27, SHV-38, SHV-40, SHV-41, SHV-42, SHV-65, SHV-164 and SHV-187). The approach and results outlined here have been implemented in Kleborate v2.4.1 (a software tool for genotyping K. pneumoniae), whereby known and novel blaSHV alleles are classified based on causative mutations. Kleborate v2.4.1 was updated to include ten novel protein variants from the KlebNET-GSP dataset and all alleles in public databases as of November 2023. This study demonstrates the power of sharing AMR phenotypes alongside genome data to improve the understanding of resistance mechanisms.
Keywords: AMR, β-lactamase, BLI resistance, extended-spectrum β-lactamase (ESBL), genotype, K. pneumoniae, prediction, SHV
Impact Statement
Since every K. pneumoniae genome has an intrinsic SHV β-lactamase and may also carry additional mobile forms, the correct interpretation of blaSHV genes detected in genome data can be challenging and can lead to K. pneumoniae being misclassified as ESBL-producing. Here, we use matched K. pneumoniae genome and drug susceptibility data contributed from dozens of studies, together with a systematic literature review of experimental evidence, to improve our understanding of blaSHV allele variation and mapping of genotype to phenotype. This study shows the value of coordinated data sharing, in this case via the KlebNET-GSP AMR Genotype-Phenotype Group, to improve our understanding of the evolutionary history and functionality of blaSHV genes. The results are captured in an open-source AMR dictionary utilized by the Kleborate genotyping tool, which could easily be incorporated into or used to update other tools and AMR gene databases. This work is part of the wider efforts of the KlebNET-GSP group to develop and support a unified platform tailored for the analysis and interpretation of K. pneumoniae genomes by a wide range of stakeholders.
Data Summary
BlaSHV allele sequences and class assignments are distributed with Kleborate v2.4.1 (DOI: 10.5281/zenodo.10469001). The following supplementary tables are included in Supplementary Material 1. Table S1 provides a summary of blaSHV alleles, including primary accessions, class-modifying mutations and supporting evidence for class assignments that differ from the NCBI’s Reference Gene Catalog or BLDB. The whole-genome sequence data are publicly available as reads and/or assemblies, and individual accessions are given in Table S2; corresponding genotypes and antibiotic susceptibility phenotypes and measurements are available in Tables S3 and S4, respectively; genomic location of SHVs is summarised in Table S5.
Introduction
Klebsiella pneumoniae are typically resistant to ampicillin due to the production of a chromosomally encoded Ambler class A β-lactamase enzyme, sulfhydryl variable (SHV). Indeed, the European Committee on Antimicrobial Susceptibility Testing (EUCAST) terms this ‘expected resistance’ (formerly ‘intrinsic resistance’) and recommends against phenotypic testing of ampicillin resistance in K. pneumoniae, as a susceptible result is likely to be incorrect.
Whilst blaSHV is a core chromosomal gene in K. pneumoniae, it has been mobilized out of the K. pneumoniae chromosome at least twice via IS26 transposition [1], into multiple plasmid backbones [2] that, in turn, have spread between bacterial species. Chromosomal and plasmid forms of SHV have undergone allelic diversification to generate variants with differing functional activity, including extended-spectrum β-lactamases (ESBLs) conferring resistance to third-generation cephalosporins (3GCs) and alleles conferring resistance to β-lactamase inhibitors (BLIs).
The ESBL phenotype is facilitated through the overexpression of IS26 [3] (e.g. SHV-2 and SHV-12). Consequently, blaSHV genes identified in species other than K. pneumoniae are typically mobile and confer ESBL activity due to IS26, leading to a general conflation of SHV enzymes with ESBL. The existence of blaSHV alleles with different activity profiles, including the potential for both chromosomal- and plasmid-encoded genes with differing functions in a single isolate, has created confusion and can lead to an incorrect interpretation of the phenotypic impact of the molecular detection of blaSHV genes in K. pneumoniae.
It is not surprising that confusion exists around the interpretation of SHV variants, given the circuitous routes through which current understanding of the origins and evolution of the enzyme has emerged. First described in 1972 as a plasmid-encoded protein of Escherichia coli str. 453 conferring resistance to ampicillin [4], the enzyme was later given the name SHV-1 and was reported in multiple Klebsiella, E. coli and Proteus mirabilis isolates [5]. A 1979 study reported the blaSHV-1 gene as being chromosomally located in several Klebsiella, with a second plasmid-borne copy in one strain [6]. That study also demonstrated the transposition of the blaSHV-1 gene into different plasmid backbones and reported the detection of blaSHV-1 on naturally occurring plasmids of diverse types [6]. A naturally occurring plasmid-encoded variant, designated SHV-2 [7], displayed ESBL activity and conferred resistance to 3GCs, was reported in K. pneumoniae, K. ozaenae (now known as K. pneumoniae subsp. ozaenae) and Serratia marcescens in 1983 [8]. SHV-2 differs from SHV-1 by a single substitution (Gly to Ser) at Ambler codon 238 (aa 213 of the mature protein), which is sufficient to change its spectrum of activity [9]. BlaSHV-2 and blaSHV-12 are well known to be plasmid borne, following the transposition from the K. pneumoniae chromosome by IS26 [1], and found outside K. pneumoniae [10,11]. By 1997, 12 protein variants of SHV had been reported, most of them ESBL and mostly in K. pneumoniae [2]. These were designated consecutive numbers (SHV-3, -4, etc.), with the exception of the ESBL variant SHV-2a, so named due to its similar kinetic properties to SHV-2, although it is not derived from SHV-2 [12].
Besides codon 238S, aa substitutions identified as conferring ESBL activity mostly affect the omega-loop of SHV, including substitutions at Ambler position 179 (SHV-8 [13] and SHV-24 [14]) or 169 (SHV-57 [15]) and an insertion at 163 (SHV-16 [16]). The first variant displaying resistance to BLI (clavulanate and tazobactam) was SHV-10, which owes its unique phenotype to a substitution at codon 130 [17]. Other reported BLI-resistant substitutions are located at Ambler codons 69 (SHV-49 [18]), 234 (SHV-56 [19] and SHV-72 [20]) and 235 (SHV-107 [21]).
In the sequencing era, new blaSHV alleles are frequently reported and now number in the hundreds (as of November 2023, up to blaSHV-232 have been assigned by the NCBI’s Reference Gene Catalog [22], https://www.ncbi.nlm.nih.gov/pathogens/refgene/). Most of these alleles are also catalogued in the β-lactamase database (BLDB) [23] and the Comprehensive Antibiotic Resistance Database (CARD) [11]. Additions to these databases are based on novel aa sequences, without a requirement for biochemical characterization of enzyme function [10,24]. Database curators attempt to assign β-lactamase alleles to the functional groups according to their reported spectrum of activity: narrow spectrum, extended-spectrum (ESBL) and/or BLI resistant. Unfortunately, the primary literature used to support the functional classifications varies widely in terms of experimental design. In some cases, the presence of a blaSHV allele in a K. pneumoniae isolate displaying ESBL activity has been used to ascribe ESBL functionality to the SHV enzyme, without ruling out the presence of other ESBL enzymes. This has led to the assignment of chromosomal variants with WT activity being reported in the literature as ESBL variants [25] (e.g. SHV-27 and SHV-41), where the error propagated to multiple antimicrobial resistance (AMR) gene databases (in November 2023, SHV-27 and SHV-41 were still recorded as ESBL in BLDB and CARD, despite the error being reported in a 2006 publication [25]). Other examples are summarized in Table S1 (see column ‘Evidence’ for a discussion of discrepancies between databases).
Neubauer et al. [26] recently sought to clarify the role of specific substitutions in SHV functionality by systematically reconstructing isogenic mutants carrying individual substitutions (identified in naturally occurring variants with modified activity). Mutations were introduced into the blaSHV-1 background, and the spectrum of enzyme activity was assessed in an E. coli strain lacking any other β-lactamase [26]. This confirmed the role of some substitutions in conferring ESBL activity (at Ambler site 238) or BLI resistance (at Ambler sites 69, 234 and 240); however, some mutants could not be generated. In 2021, we used these findings, together with a review of experimental evidence from the literature, to systematically assign blaSHV alleles to functional classes. We also incorporated the resulting blaSHV database and list of functionally relevant mutations in the Kleborate tool (v2.0) for genotyping of K. pneumoniae genomes [27].
Here, we aimed to systematically assess the evidence for enzyme activity by exploring genotype–phenotype relationships for naturally occurring blaSHV alleles using a diverse set of 3999 K. pneumoniae isolates with matched genome–phenotype data, which lack non-blaSHV alleles. We also explored the evolutionary relationships and genetic context of blaSHV alleles, with the aim of further clarifying the emergence and spread of SHV variants.
Methods
SHV reference database
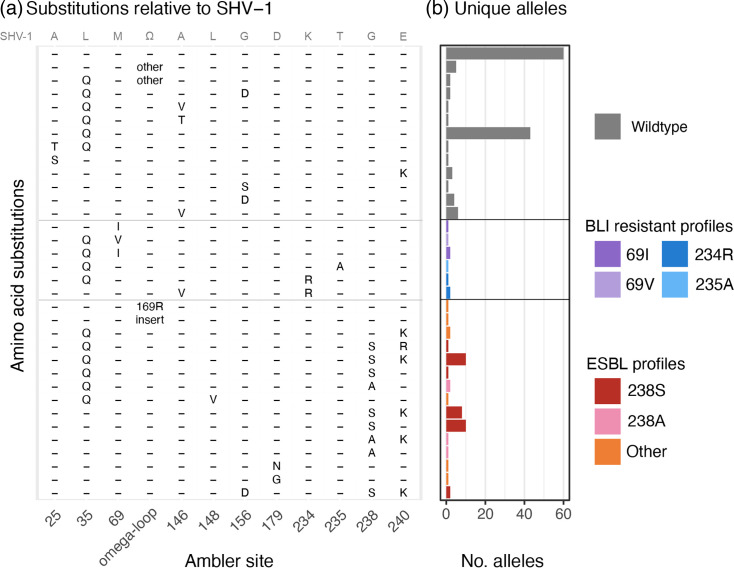
We curated an updated set of blaSHV nt alleles for Kleborate based on a comparison of CARD (v3.2.8) [11], the NCBI’s Reference Gene Catalog [22] and BLDB (as of November 2023, up to allele number blaSHV-228). BlaSHV-6 and blaSHV-10 were excluded as the published nt sequence for blaSHV-6 is incomplete, and there is no nt sequence for blaSHV-10 (only an aa sequence for SHV-10, which yields no exact matches to any six-frame translations of nt sequences in the NCBI using tblastn). BlaSHV-11, blaSHV-28 and blaSHV-31 were represented by two nt sequences each (labelled .v1 and .v2) and the rest by a single nt sequence. In addition to reporting matches to known alleles and the corresponding functional class, Kleborate specifically checks for and reports mutations of known functional relevance (listed in Fig. 1; plus Ambler position 130, which was found in two novel alleles and was reported as responsible for BLI resistance in SHV-10 [17]).
Fig. 1. Amino acid substitution profiles associated with n = 181 blaSHV alleles. (a) Positions studied in the article by Neubauer et al. and tracked in Kleborate v2 are shown as columns; these include sites where substitutions have a clear association with ESBL activity [148, 179, 238 and 240, omega-loop (positions 164–178)] or BLI resistance activity (69, 234 and 235), plus some sites (25, 35, 146 and 156) that are associated with increased MIC to ceftaroline but not ceftriaxone or inhibitor resistance. Position 179 is also a part of the omega-loop and is specifically separated to show its association with ESBL profiles. Position 130 is not included, as it is found only in SHV-10 (BLI resistant) for which there is no nt sequence available. Each row indicates a unique combination of aas across these variable sites. The aas present in SHV-1 are indicated in grey at the top of the panel, where the omega-loop sequence (Ω) is RWETELNEALPGDARD. (b) The number of unique nt alleles associated with each aa profile (row) is shown as a barplot. Colours indicate the functional class assigned to these alleles, on the basis of the mutations shown here and supporting literature (cited in the text and in Table S1).
We used the rules previously established for Kleborate (v2.0), as described in Supplementary Note 3 of Lam et al. [27], to assign functional classes (WT, ESBL or BLI resistant). This relied primarily on the presence of well-supported SHV substitutions as noted above (ESBL: mutation at Ambler sites 238, 179 and 169; BLI resistance: mutation at sites 69, 130 and 234), or direct experimental evidence for individual blaSHV alleles (detailed in Results). The curated set of allele sequences and their assignment to functional classes is included in a new release of Kleborate (v2.4.1, DOI: 10.5281/zenodo.10469001) and Table S1. Table S1 also includes information on each allele extracted from the NCBI’s Reference Gene Catalog (including PubMed ID, subclass, protein and nt accessions), BLDB (including phenotype, sequence accessions and alternative names) and CARD (Antibiotic Resistance Ontology identifiers and sequence accessions).
Our curated class assignments were compared with those of BLDB and the NCBI’s Reference Gene Catalog, which were interpreted as follows: WT (phenotype ‘2b’ in BLDB and subclass ‘BETA-LACTAM’ in the NCBI), ESBL (phenotype ‘2be’ in BLDB and subclass ‘CEPHALOSPORIN’ in the NCBI) and BLI resistant (‘2br’ in BLDB and subclass ‘BETA-LACTAM’ in the NCBI but with ‘product name’ typically including the prefix ‘inhibitor resistant’). Discrepancies between our class assignments and those of BLDB or the NCBI’s Reference Gene Catalog are reported in Table S1, which includes a summary of evidence based on the literature review and the phenotype data presented in this study.
Novel alleles, each encoding for unique aa sequences, identified in our sequence data (described below) were submitted to the NCBI’s Reference Gene Catalog to obtain allele numbers in November 2023. These blaSHV alleles (encoding SHV-233 to 237 and SHV-239 to 243), together with 12 additional blaSHV alleles present in the NCBI in November 2023 but not yet in our database (encoding SHV-115, SHV-116, SHV-132, SHV-146, SHV-171, SHV-190, SHV-191, SHV-202 and SHV-229 to 232), were also added to Kleborate v2.4.1. We also updated the database to include a single representative sequence per allele in Kleborate v2.4.1. For blaSHV-11, we selected GenBank accession AY293069 (as per BLDB, labelled .v1 in earlier versions of Kleborate), as this sequence is the correct length, and we confirmed exact matches in >1000 of our K. pneumoniae genomes in >300 multi-locus sequence types (STs), whilst .v2 was not detected. In the NCBI’s Reference Gene Catalog and CARD, blaSHV-11 is represented by a different sequence (GenBank accession X98101.1) that has additional nts at the start and end, differs from AY293069 at two synonymous mutations and had no exact matches in K. pneumoniae whole genomes. For blaSHV-28, we used GenBank accession AF299299.1 (as per CARD and BLDB; formerly .v2 in earlier versions of Kleborate), and for blaSHV-31, we used GenBank accession AY277255.2 (as per the NCBI’s Reference Gene Catalog, CARD and BLDB; formerly .v2 in Kleborate).
Matched genome and phenotype data for K. pneumoniae species complex isolates
A global collection of K. pneumoniae species complex genomes with matched antimicrobial susceptibility testing (AST) data was aggregated by the KlebNET-GSP AMR Genotype-Phenotype project group (summarized in Tables S2–S4). This collection includes human, animal and environmental isolates collected between 2001 and 2021 from 24 countries across six continents. Genomes were assembled from Illumina reads using Unicycler (v0.4.8) (accessions and assembly metrics in Table S2) and analysed using Kleborate (v2.2.0) (results in Table S3), which identifies the presence of acquired resistance genes/alleles including SHV, SHV protein variants and porin defects associated with AMR [28] (i.e. loss of OmpK35 or OmpK36 and insertions in loop 3 of OmpK36). All assemblies met the pre-agreed KlebNET-GSP criteria of <5% contamination (assessed using KmerFinder [29] (v3.2)): Kleborate-designated species match of ‘strong’, ≤500 contigs; genome size 4 969 898–6 132 846 bp; and G+C content in the range 56.35–57.98%. Contig metrics across the dataset were as follows: contig count, mean 131.5 and sd 83.1; N50, mean 376 272.4 bp and sd 557 147.6 bp; genome size, mean 5 418 671.9 bp and sd 163 789.2 bp; and G+C content, mean 57.3% and sd 0.2%. We included only K. pneumoniae genomes with an exact aa sequence match to one or more known blaSHV alleles and excluded those in which other (i.e. non-SHV) β-lactamases were detected (total n = 3999 isolates for analysis).
The available AST data were determined by the contributing laboratories using a range of methods, including disc diffusion, agar dilution, broth microdilution and semi-automated methods (Vitek 2 or Phoenix), that were performed based on Clinical and Laboratory Standards Institute (CLSI) or EUCAST guidelines. AST data were shared in the form of disc diffusion zone sizes or minimum inhibitory concentrations (MICs). We interpreted as ‘susceptible’ (S), ‘intermediate/susceptible, increased exposure’ (I) or ‘resistant’ (R) using the EUCAST (v13.0) or CLSI (M100 33rd edition) breakpoints, as appropriate to the assay used. As there were very few isolates categorized as I, we grouped I/R (i.e. non-WT) together and refer to this group as ‘resistant’ (data in Table S4).
Phylogenetic analysis
BlaSHV nt sequences were aligned using MAFFT [30] (v6.861). Pairwise distances between aligned nt sequences were calculated using the ‘dist.dna’ function in the ‘ape’ package (v5.7-1) for R (v4.2.3), and a phylogeny was inferred using the BioNJ algorithm in the same package. The minimum spanning tree was inferred using GrapeTree [31] (v1.5.0) using MSTreeV2. R packages ggplot2 (v3.4.4) and ggtree [32] (v3.6.2) were used for data visualization.
Genomic context of SHV alleles
The genomic location (chromosome or mobile) of blaSHV allelic variants was determined using a combination of literature review; the CARD Prevalence, Resistomes and Variants database (v3.0.9); and blastn (100% nt identity and coverage) searches of blaSHV allelic variants in publicly available complete genome sequences and assembly graphs of K. pneumoniae. At the time of the search, CARD Prevalence, Resistomes and Variants [11] (v3.0.9, accessed October 2021) included 874 and 5466 K. pneumoniae chromosomes and plasmids, respectively, from NCBI genomes. The presence of blaSHV alleles amongst these chromosomes and plasmids was extracted from the CARD database. Plasmer (v0.1) [33] was used to predict the genomic locations of blaSHV in the KlebNET-GSP collection. In addition, complete K. pneumoniae genomes (n = 1296 chromosomes and n = 4217 plasmids) were downloaded from the NCBI using ncbi-genome-download (v0.3.1) to directly examine the genomic context of blaSHV alleles. We used Kleborate [27] (v2.2.0) to search for SHV variants across complete K. pneumoniae genomes from the NCBI and the KlebNET-GSP collection.
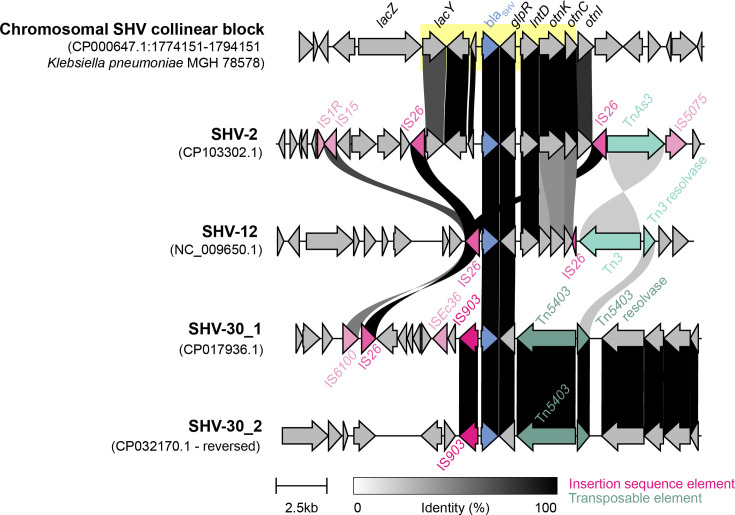
To investigate the genetic context of mobile blaSHV variants (represented in this analysis by blaSHV-2, blaSHV-12 and blaSHV-30), we used a subset of the publicly available complete genome sequences of K. pneumoniae from the NCBI. We selected 30 random genomes from different STs [determined by Kleborate (v.2.2.0)] and ran Mauve [34] (v2015-02-25) to identify the collinear block containing blaSHV. We then used blastn to search for this collinear block in all publicly available complete chromosome sequences of K. pneumoniae from the NCBI, to confirm its broader conservation. To visualize the genetic context of mobile variants of blaSHV compared with the typical chromosomal context from which they were presumably mobilized, we extracted 10 kbp of sequence upstream and downstream of the gene from genomes CP103302.1 (blaSHV-2), NC_009650.1 (blaSHV-12), NZ_CP017936.1 (blaSHV-30) and NZ_CP032170.1 (blaSHV-30). We then used Prokka [35] (v1.14.6) and clinker [36] (v0.0.24) to annotate and visually compare the extracted genomic regions with a representative sequence of the chromosomal collinear block extracted from the chromosome of K. pneumoniae strain MGH 78578 (accession CP000647.1). In addition, we used flankophile [37] (v0.2.10) to extract blaSHV flanking regions (5 kbp upstream and downstream) across the previously used NCBI’s publicly available complete chromosome K. pneumoniae sequences to capture genetic variation in blaSHV flanking regions. We then used CD-HIT-EST [38] (v4.8.1) to cluster the flanking regions with ≥90% nt sequence similarity. We used the same visualization methods as described above. We also investigated the presence of insertion elements 10 kbp upstream of WT blaSHV in the genomes of phenotypically 3GC resistant isolates using the blastn and the ISfinder [39] database.
For the set of complete genomes, the blaSHV copy number was calculated based on the number of unique non-overlapping blastn hits. For the matched genotype–phenotype dataset, the copy number of blaSHV in draft genomes was estimated by analysing Illumina read sets, calculating the ratio of read depth for blaSHV vs the mean read depth of the seven K. pneumoniae loci used for multi-locus sequence typing and using SRST2 [40] (v0.2.0) to perform the mapping and depth calculations. The copy number estimates are included along with other genotype information in Table S3.
Results
Distribution of activity-modifying mutations
In Kleborate v2.2.0 database, there are a total of 181 unique blaSHV alleles, corresponding to 178 unique protein sequences or variants (Table S1). Figure 1 illustrates the distribution of key aa substitutions (hereafter the term mutations is used for both nt and aa variation for convenience, even though aa changes are a consequence of the actual mutations) across these alleles. Amongst the blaSHV alleles identified, 38 encoded mutations relative to SHV-1 at Ambler positions 238 (n = 36) and 179 (n = 2). These specific mutations have been observed by Neubauer et al. [26] to confer 3GC resistance, classifying these variants as ESBL. Five additional protein variants were assigned as ESBL based on primary literature reports (Table S1): SHV-16 [16] (omega-loop insertion between Ambler sites 167 and 168), SHV-57 [15] (omega-loop substitution 169R), SHV-31 (encoded by divergent alleles SHV-31.v1 and SHV-31.v2 [41], each carrying mutations 35Q and 240K) and SHV-70 [42] (mutation 148V). Eight alleles harboured a substitution at a site associated with BLI resistance [26] and were classified accordingly: Ambler site 69 (SHV-49 [18], SHV-52, SHV-92 and SHV-203) or 234 (SHV-56 [19], SHV-72 [20] and SHV-73). SHV-107 (harbouring mutation 235A) was also classified as BLI resistant based on primary literature [21]. The remaining 130 alleles were assigned as WT.
Evolutionary relationships and genomic context
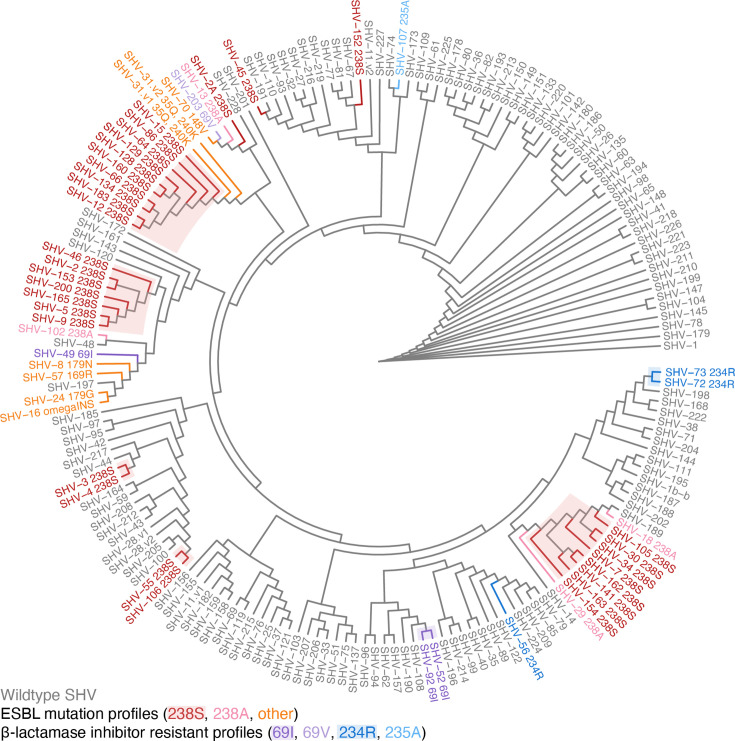
To understand the evolutionary relationships between blaSHV alleles, we inferred a cladogram and minimum spanning tree from the nt sequence alignment (Figs 2 and S1). Pairwise genetic distances between allele sequences support blaSHV-1 as the ancestral form, as it has the smallest distance to all other variants (mean 5.1 substitutions and total distance 918; compared with mean 5.4 and total 969 for blaSHV-11, which had the next lowest values). We therefore rooted the phylogeny at blaSHV-1. We identified a chromosomal blaSHV collinear block (7585 bp) conserved in 90.5% (n = 1236/1366) of complete K. pneumoniae genomes with >90% nt identity and >90% coverage (Fig. 3). In general, there is a low genetic variation of the chromosomal blaSHV flanking regions (5 kbp upstream and downstream), where 95.1% (n = 1229/1292) of complete genomes had ≥90% nt sequence similarity in their flanking regions. Comparing the chromosomal blaSHV collinear block with the genomic context of plasmid-borne and IS26-mediated blaSHV-2 and blaSHV-12, the chromosomal blaSHV collinear block is conserved in the genomic context of blaSHV-2 and flanked by IS26. Similarly, the chromosomal blaSHV collinear block is partially conserved (59% coverage and 99% identity of 7585 bp) in the blaSHV-12 genomic context. The gene directly downstream of blaSHV, glpR, encodes a glycerol-3-phosphate regulon repressor, which is conserved across the genomic contexts of chromosomal and plasmid-borne blaSHVs.
Fig. 2. Cladogram for n = 181 blaSHV alleles. The cladogram was inferred from a pairwise genetic distance matrix calculated from nt sequences using BioNJ, rooted on SHV-1. Tips are labelled with the SHV allele name and coloured to indicate the mutation profile (black, WT; red, orange and pink, ESBL profiles; and blue and purple, BLI-resistant profiles). For alleles classed as non-WT, the class-modifying mutation is included in the label (e.g. 238S indicates substitution of serine at Ambler site 238 in the encoded protein; ‘omegaINS’ refers to a 6-aa insertion in the omega-loop between Ambler codons 167 and 168). Shading indicates clusters of alleles referred to in the text, which may share class-modifying mutations via vertical inheritance.
Fig. 3. A comparison of the genomic context of SHV allele clusters. Upstream (10 kbp) and downstream (10 kbp) sequences of each blaSHV were extracted and aligned. The 7585 bp chromosomal SHV collinear block is highlighted in yellow. BlaSHV is coloured in blue, while mobile genetic elements, such as insertion sequences and transposons, are illustrated in pink and green, respectively. Percent identity between the genes is shown by the gradient scale bar.
ESBL alleles were distributed throughout the cladogram (pink, red and orange in Fig. 2), consistent with at least 19 independent mutation events [n = 12 in Ambler codon 238, n = 1 in codon 148, n = 2 in codon 179, n = 2 others in the omega-loop and n = 2 (SHV-31.v1 and SHV-31.v2) in codon 240] [41]. Some ESBL alleles formed clusters that appear to share a resistance-conferring mutation (238S) via inheritance from a common ancestor (shading, Fig. 2). These include two pairs of alleles (blaSHV-3/blaSHV-4 and blaSHV-55/blaSHV-106) and three larger clusters centred around blaSHV-2/blaSHV-5 (n = 7), blaSHV-12 (n = 10) and blaSHV-7/blaSHV-30 (n = 8). Mobilization of blaSHV-2 and blaSHV-12 by IS26 are well documented [1,10]. BlaSHV-3 and blaSHV-4 are also known to be plasmid borne [43] and found in species outside Klebsiella [10], although we could not identify a complete plasmid sequence in which to explore the specific genetic context of the mobilized region. The members of the blaSHV-7 cluster, including blaSHV-7 [44], blaSHV-30 [45] and blaSHV-34 [46], have been reported as plasmid borne and found in Enterobacter. In the KlebNET-GSP collection, blaSHV-7 (n = 8) was identified in seven plasmids and one chromosome, whilst blaSHV-154 (n = 1) was identified in a chromosome (Table S5). Amongst the blaSHV-7 cluster, only blaSHV-30 was detected amongst complete K. pneumoniae genome sequences in the NCBI. This allele was identified in two similar plasmid sequences [99.99% identity over 55 821 bp of shared sequence (85% coverage), detected in ST2938 (accession NZ_CP032170.1) and ST45 (accession NZ_CP017936.1)], where it was flanked by IS903 and Tn5403 (Fig. 3). This provides a potentially novel, non-IS26-mediated, mobility mechanism for this ESBL cluster, which was found in a total of 13 K. pneumoniae genomes belonging to seven STs (and one K. variicola genome) in our genome collection. We could find no evidence to support that blaSHV-55/blaSHV-106 have been mobilized out of the K. pneumoniae chromosome. NCBI blast did not identify these alleles outside K. pneumoniae, nor in any complete K. pneumoniae genomes. We identified a single instance in our genome collection (blaSHV-106 in an ST14 genome); read analysis indicated a copy number of one, suggesting this was the only copy of blaSHV in the genome, and assembly graph analysis and plasmid prediction analysis supported its location in the chromosome (Table S5). Four of the five ESBL allele clusters therefore appear to be plasmid borne and likely reflect the diversification of ESBL alleles following mobilization from the K. pneumoniae chromosome.
BLI-resistant alleles (n = 8) were distributed throughout the cladogram (blue and purple in Fig. 2), consistent with at least six independent mutation events (n = 3 in Ambler codon 69, n = 2 in 234 and n = 1 in 235). These alleles have not been reported outside of K. pneumoniae, and sequence searches of the NCBI and CARD did not detect evidence of them in non-K. pneumoniae genomes. The original reports of blaSHV-56 and blaSHV-49 confirmed these variants as chromosomally located [19], and we also found blaSHV-52 and blaSHV-56 in draft genome sequences where assembly graph inspection and plasmid prediction software confirmed that they were located on chromosomal contigs (Table S5). The other alleles blaSHV-72, blaSHV-73 and blaSHV-92 were not found in our genome collection or in NCBI genomes via blastn search. The original report of blaSHV-92 states that it was detected in a transconjugant, suggesting that it was plasmid borne [47]; however, we found no evidence of any other BLI-resistant alleles being mobile. These data suggest that the currently reported BLI-resistant blaSHV alleles have arisen in WT chromosomal blaSHV backgrounds. With the exception of blaSHV-92, these BLI-resistant alleles have not yet been mobilized to plasmids, which is consistent with the low prevalence of the phenotype reported in K. pneumoniae isolates.
Genotype–phenotype relationships
We compared blaSHV alleles with AST phenotypes for 3GCs and BLIs in a set of n = 3999 K. pneumoniae genomes that carried at least one blaSHV allele and no other β-lactamase (Tables S2–S4). Within these genomes, we identified 70 of the known 181 blaSHV alleles [38% of those in the Kleborate (v2.2.0)].
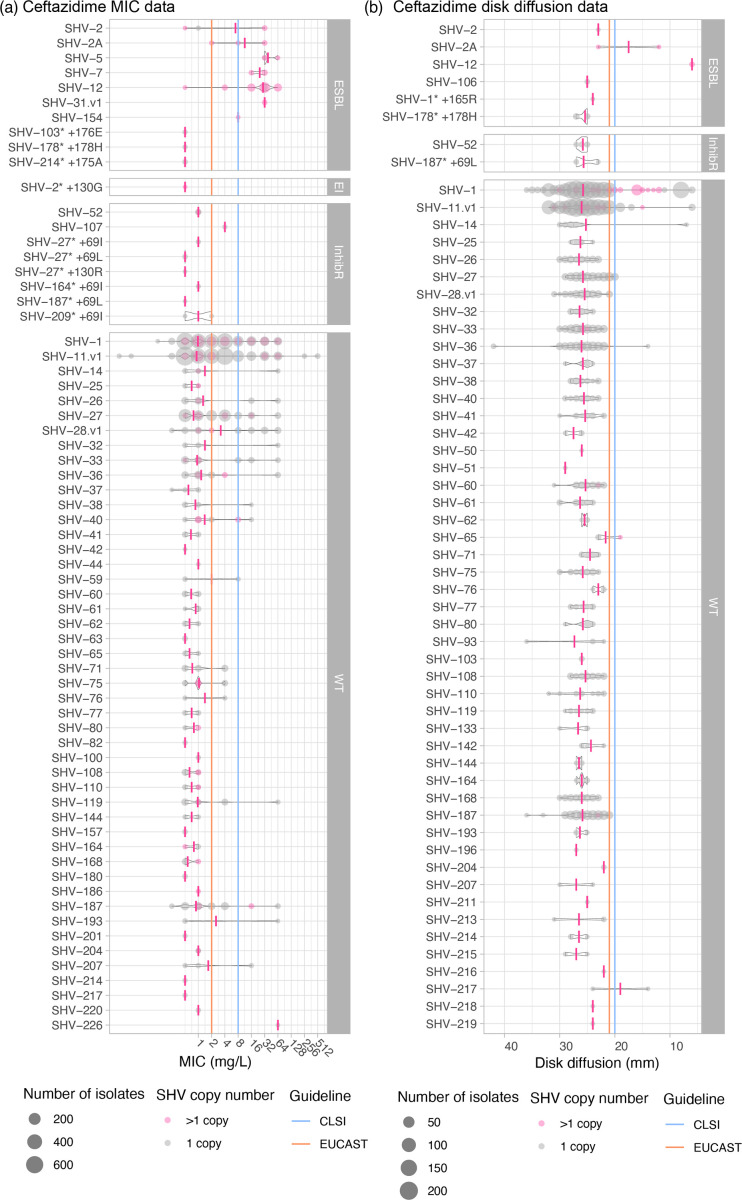
Eight known ESBL protein variants (classified as such in the literature and here) were identified in isolates that were tested for susceptibility to ceftazidime and all but one (sole representative of SHV-106) showed evidence for resistance (see Table 1, Fig. 4). All of these protein variants have at least a 238S substitution, with the exception of SHV-31.v1, which had both 35Q and 240K substitutions. All isolates representing the remaining protein variants and for which data were available also showed evidence of resistance to ceftriaxone. However, resistance to cefotaxime was more variable. Eleven other isolates carried blaSHV alleles with a non-synonymous mutation in the omega-loop but were not previously reported as ESBL alleles (blaSHV-51 and four novel alleles, see Table 1); all tested susceptible to ceftazidime (Table 1, Fig. 4) and all other 3GCs for which they were tested (Table 1).
Table 1. 3GC susceptibility phenotypes for ESBL-assigned alleles.
| Allele | Ceftazidime (R/n, %R) | Cefotaxime (R/n, %R) | Ceftriaxone (R/n, %R | Mutation/s |
| Exact matches to known alleles assigned here as ESBL | ||||
| SHV-2 | 3/6 (50%) | 1/2 (50%) | 3/3 (100%) | 238S |
| SHV-2A | 5/6 (83.3%) | 2/3 (66.7%) | 2/2 (100%) | 35Q, 238S |
| SHV-5 | 4/4 (100%) | 1/1 (100%) | 3/3 (100%) | 238S, 240K |
| SHV-7 | 8/8 (100%) | – | 8/8 (100%) | 238S, 240K |
| SHV-12 | 81/83 (97.6%) | 2/2 (100%) | 82/82 (100%) | 35Q, 238S, 240K |
| SHV-31.v1 | 2/2 (100%) | – | 2/2 (100%) | 35Q, 240K |
| SHV-106 | 0/1 (0%) | 0/1 (0%) | – | 238S |
| SHV-154 | 1/1 (100%) | – | 1/1 (100%) | 238S, 240K |
| Other alleles with omega-loop mutations | ||||
| SHV-1* +165R | 0/1 (0%) | 0/1 (0%) | – | 165R |
| SHV-103* +176E | 0/1 (0%) | – | 0/1 (0%) | 176E |
| SHV-178* +178 h | 0/6 (0%) | 0/6 (0%) | 0/1 (0%) | 35Q, 178 h |
| SHV-214* +175A | 0/2 (0%) | – | 0/2 (0%) | 175A |
| SHV-51 (175A) | 0/1 (0%) | 0/1 (0%) | – | 175A |
| Exact matches to alleles reported in the literature as ESBL but assigned here as WT | ||||
| SHV-27 | 14/304 (4.6%) | 4/106 (3.8%) | 12/211 (5.7%) | 156D |
| SHV-38 | 1/21 (4.8%) | 0/14 (0%) | 1/7 (14.3%) | 146V |
| SHV-40 | 3/31 (9.7%) | 0/15 (0%) | 3/30 (10%) | 35Q |
| SHV-41 | 0/17 (0%) | 0/9 (0%) | 1/8 (12.5%) | – |
| SHV-42 | 0/3 (0%) | 0/3 (0%) | – | 25S |
| SHV-65 | 1/6 (16.7%) | 1/3 (33.3%) | 0/4 (0%) | – |
| SHV-164 | 0/7 (0%) | 0/5 (0%) | 0/2 (0%) | – |
| SHV-187 | 3/135 (2.2%) | 0/87 (0%) | 4/51 (7.8%) | – |
Novel alleles identified in this study have been highlighted with an ‘*’.
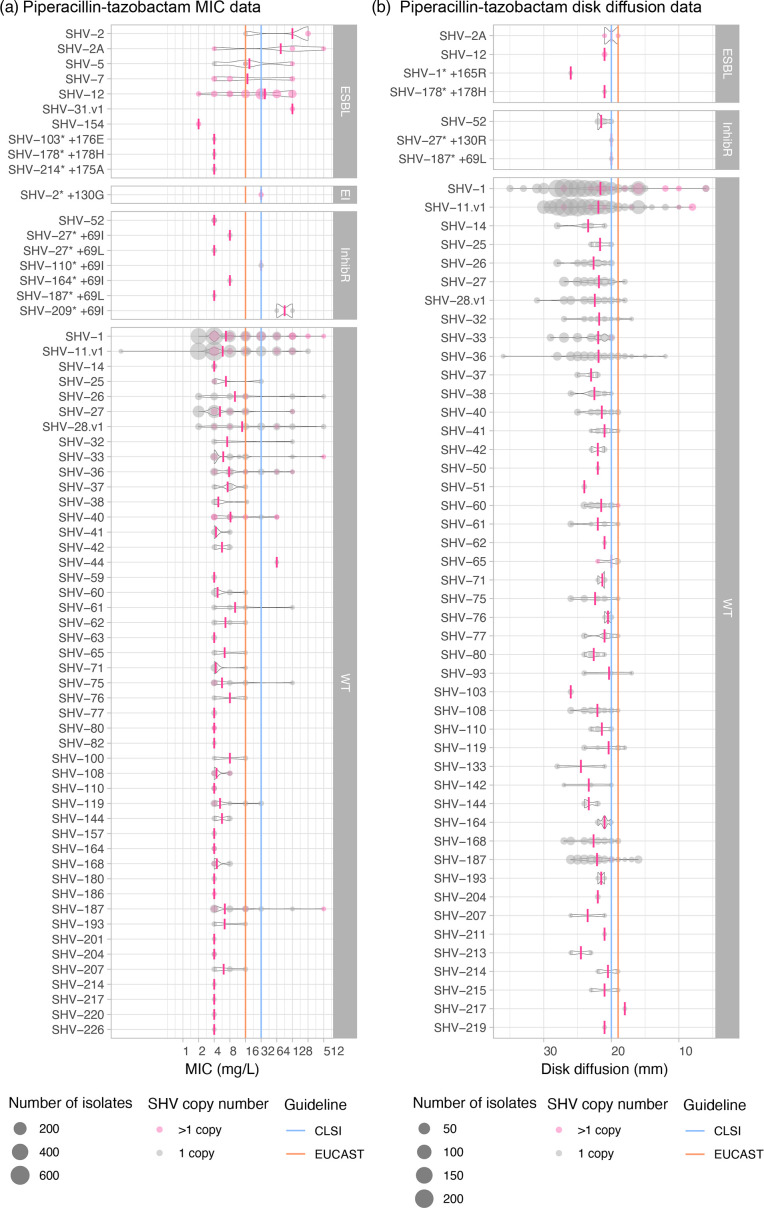
Fig. 4. AST value distributions for ceftazidime. The size of each circle represents the number of isolates with an SHV allele and no other acquired β-lactamase. (a) MIC and (b) disc diffusion measurements show the distribution of phenotypes for each SHV allele. Average MIC and disc diffusion measurements per SHV allele are indicated by a pink vertical line. Grey circles indicate 1 SHV copy, whilst pink circles indicate >1 SHV copy. SHV alleles are grouped based on ESBL, ESBL- and BLI resistant (EI), BLI resistant (inhibR) and WT phenotype classifications. EUCAST (v13.0) or CLSI (M100 33rd edition) S/I breakpoints are indicated using orange and blue lines, respectively. SHV-187* +69L is SHV-132 in Kleborate v2.4.1. For MIC values, larger values indicate increased resistance; for disc diffusion results, larger zone sizes indicate increased susceptibility.
We identified n = 533 isolates with alleles that were initially reported as ESBL and assigned as such in the NCBI’s Reference Gene Catalog and/or BLDB but do not carry any causative mutations and were therefore classified in our database as WT (encodes SHV-27, SHV-38, SHV-40, SHV-41, SHV-42, SHV-65, SHV-164 and SHV-187). Our phenotype data support the assignment to WT for all these alleles (see Table 1). A summary of the comparison with BLDB and the NCBI’s Reference Gene Catalog’s class assignments is given in Table S1, available in the online version of this article.
Two BLI-resistant variants were identified in isolates that were tested for susceptibility to piperacillin-tazobactam and/or amoxicillin-clavulanic acid: SHV-52 (which harbours 69I) and SHV-107 (which harbours 235A) (see Table 2). The two isolates carrying SHV-107 came from the same study and were resistant to amoxicillin-clavulanic acid as expected (MIC 32 mg l−1 via the automated Vitek platform; piperacillin-tazobactam results were not available). All isolates carrying SHV-52 and tested for piperacillin-tazobactam were susceptible (n = 12, from five different studies using either disc diffusion or MIC via Vitek); n = 9 of these isolates were also tested for amoxicillin-clavulanic acid, and all were susceptible. The closely related allele SHV-92, which shares the 69I mutation and clusters with blaSHV-52 in the cladogram (differing from it at a single nt, see Fig. 1), was not present in our dataset, so we could not assess its phenotype directly; the original report of this allele also did not assess phenotype [47]. Nine isolates carried novel variants harbouring a substitution at Ambler site 69; six of these tested susceptible to BLIs and three tested resistant to piperacillin-tazobactam and amoxicillin-clavulanic acid (Table 2, Fig. 5). The resistant isolates were as follows: n = 1 carrying a novel variant closest to SHV-110 with additional mutation 69I (accession: SRR15097887) and n = 2 (from different studies [48,49], accessions: SRR15098057 and ERR486441) harbouring a novel variant closest to SHV-209 with additional mutation 69I (Tables 2 and S4). Two isolates were identified with novel alleles carrying mutations at codon 130, one (carrying SHV-27 plus 130R) was tested for susceptibility to BLIs but showed susceptibility to both piperacillin-tazobactam and amoxicillin-clavulanic acid via disc diffusion (Table 2, Fig. 5). The other isolate (carrying SHV-2 plus 130G) was resistant to both piperacillin-tazobactam and amoxicillin-clavulanic acid via agar dilution (Table 2, Fig. 5).
Table 2. BLI susceptibility phenotypes for inhibitor resistance-assigned alleles.
| Allele | Piperacillin-tazobactam (R/n, %R) | Amoxicillin-clavulanic acid (R/n, %R) | Mutation/s |
| Exact matches to known alleles assigned as inhibitor resistant | |||
| SHV-52 | 0/12 (100%) | 0/9 (100%) | 35Q, 69I |
| SHV-107 | – | 2/2 (100%) | 235A |
| Other alleles with mutations at Ambler site 69, 130, 234 or 235 | |||
| SHV-2 +130G | 1/1 (100%) | 1/1 (100%) | 130G, 238S |
| SHV-27* +69I | 0/1 (0%) | 0/1 (0%) | 69I |
| SHV-27* +69L | 0/2 (0%) | 0/2 (0%) | 69L |
| SHV-27* +130R | 0/1 (0%) | 0/1 (0%) | 130R |
| SHV-110* +69I | 1/1 (100%) | 1/1 (100%) | 35Q, 69I, 156D |
| SHV-164* +69I | 0/1 (0%) | 0/1 (0%) | 69I |
| SHV-187* +69L† | 0/2 (0%) | 0/1 (0%) | 69L |
| SHV-209* +69I | 2/2 (100%) | 1/1 (100%) | 35Q, 69I |
| Exact matches to alleles reported in the literature as inhibitor resistant but assigned here as WT | |||
| SHV-26 | 13/63 (20.6%) | 10/59 (16.9%) | – |
Novel alleles identified in this study have been highlighted with an ‘*’.
†SHV-187* +69L is SHV-132 in Kleborate v2.4.1.
Fig. 5. AST value distributions for piperacillin-tazobactam. The size of each circle represents the number of isolates with an SHV allele and no other acquired β-lactamase. (a) MIC and (b) disc diffusion measurements show the distribution of phenotypes for each SHV allele. The average MIC and disc diffusion measurements per SHV allele are indicated by a pink vertical line. Grey circles indicate 1 SHV copy, whilst pink circles indicate >1 SHV copy. SHV alleles are grouped based on ESBL, ESBL and BLI resistant (EI), BLI resistant (inhibR),and WT phenotype classifications. EUCAST (v13.0) or CLSI (M100 33rd edition) S/I breakpoints are indicated using orange and blue lines, respectively. SHV-187* +69L is SHV-132 in Kleborate v2.4.1. For MIC values, larger values indicate increased resistance; for disc diffusion results, larger zone sizes indicate increased susceptibility.
We also identified n = 63 isolates carrying SHV-26, which we assigned as WT due to lacking functional mutations but is classified as BLI resistant (2br) in BLDB. The original report of SHV-26 [50] described it as harbouring a mutation at Ambler site 187 and reduced susceptibility to amoxicillin-clavulanic acid (to ‘intermediate/susceptible, increased exposure’ levels). However, this mutation (A187T) was tested by Neubauer et al. [26] who found no effect on BLI susceptibility and concluded that the phenotype was likely incorrectly assigned. Our data show some evidence of a BLI-resistant phenotype [(n = 13/63 (20.6%) to piperacillin-tazobactam and n = 10/59 (17%) resistant to amoxicillin-clavulanic acid], but with majority support for WT (Table 2).
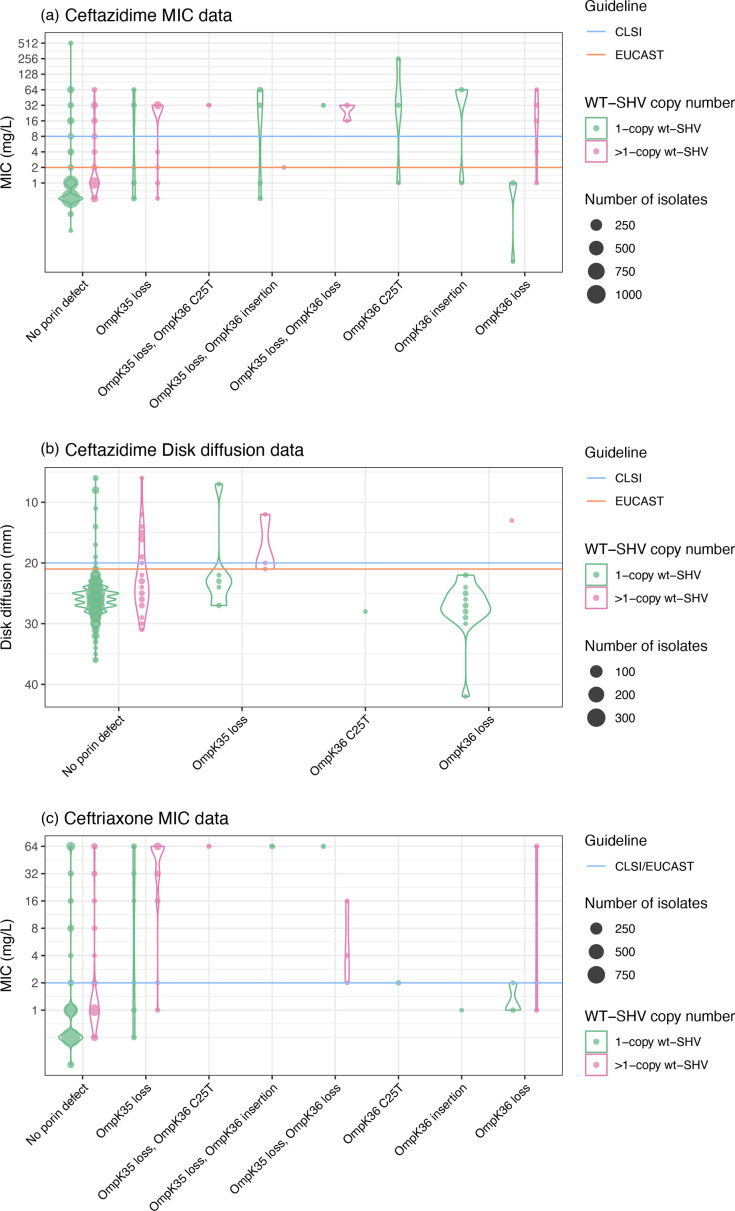
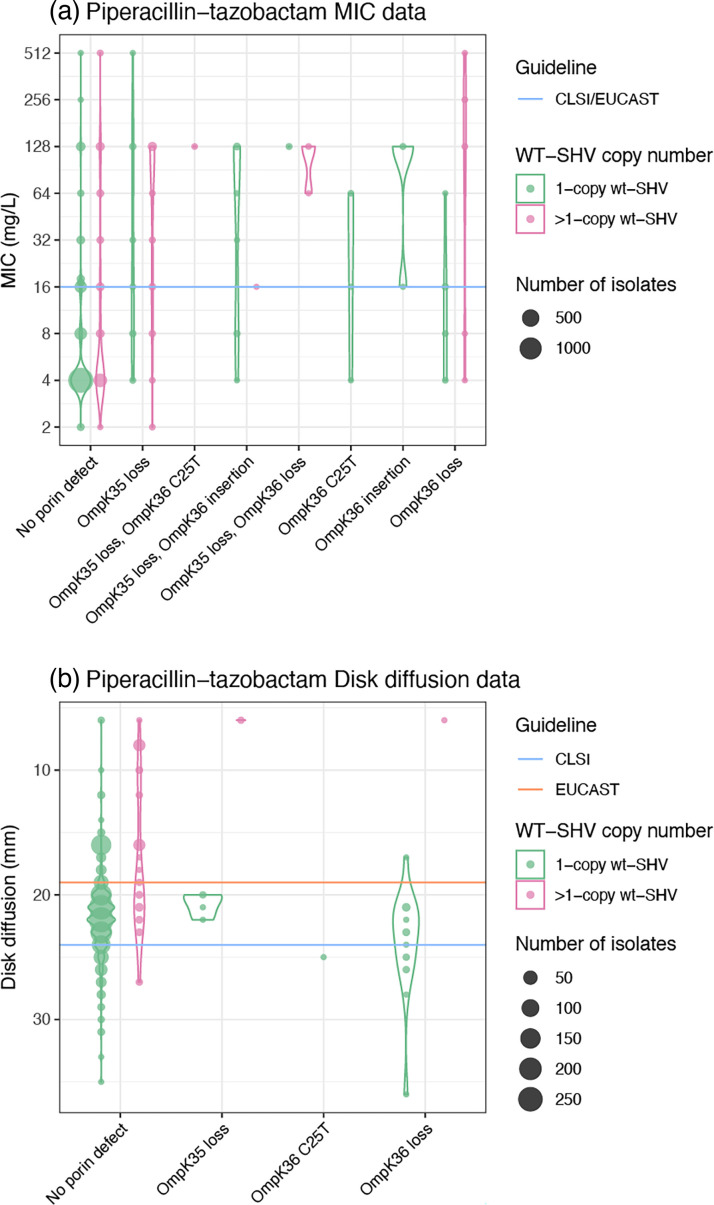
Sixty alleles classified as WT were detected in the genome collection (total n = 3858 isolates), and the WT phenotype was supported in all cases. Thirty-six of these alleles (60%) were found only in 3GC-susceptible isolates. One allele (blaSHV-59) was found in one resistant isolate and one susceptible (both ST76 with no other resistance determinants detected). The remaining n = 23 WT-classified alleles were primarily found in susceptible strains (66.7–98.0% susceptible, per allele). These include alleles blaSHV-1 and blaSHV-11, the most common and well-known WT alleles. We hypothesized that the increased copy number of blaSHV and/or porin mutations could explain 3GC and BLI resistance in isolates with WT-assigned blaSHV alleles and no other acquired β-lactamases. Amongst isolates with a WT-assigned blaSHV allele, blaSHV copy number was indeed significantly associated with ceftazidime MIC (correlation = 0.21, P < 1×10−15 using linear regression on log2 MIC), disc diffusion zone diameter (correlation = −0.76, P < 1×10−15 using linear regression) and clinical resistance (mean 2.7 vs 1.1 copies, P = 2×10−6 using the Wilcoxon rank sum test). Similarly, blaSHV copy number in isolates with WT blaSHVs was significantly associated with piperacillin-tazobactam MIC (correlation = 0.25, P < 1×10−15 using linear regression on log2 MIC), disc diffusion zone diameter (correlation = −0.10, P < 1×10−15 using linear regression) and clinical resistance (mean 2.3 vs 1.1 copies, P = 2×10−6 using the Wilcoxon rank sum test). The presence of two or more copies of blaSHVs was significantly associated with ceftazidime resistance (odds ratio (OR) 3.6, P = 6×10−13 amongst isolates with a WT-assigned blaSHV allele), accounting for 25.6% of the resistance observed amongst these isolates. A further 12.8% of ceftazidime resistance could potentially be explained by porin defects in isolates with a single blaSHV copy (see Fig. 6). We also investigated the presence of insertion sequences upstream of WT blaSHV (with no other acquired β-lactamases) in genomes of phenotypically 3GC-resistant isolates that could potentially explain the phenotype [3], but there were none identified. For piperacillin-tazobactam, the presence of two or more copies of blaSHV was significantly associated with resistance (OR 4.78, P < 1×10−15 amongst isolates with a WT-assigned blaSHV allele), accounting for 34.1% of unexplained resistance, with a further 9.5% potentially explained by porin defects (see Fig. 7).
Fig. 6. Presence of porin defects and copy number effects amongst isolates with WT-assigned alleles with genomes tested against 3GCs. Violin plots show the distribution of susceptibility testing measures, coloured by copy number, for WT SHV alleles (n = 1659 isolates tested against ceftazidime and n = 1937 isolates tested against ceftriaxone). EUCAST (v13.0) or CLSI (M100 33rd edition) I/R breakpoints are indicated using orange and blue lines, respectively.
Fig. 7. Presence of porin defects and copy number effects amongst isolates with WT-assigned alleles with genomes tested against piperacillin-tazobactam. Violin plots show the distribution of susceptibility testing measures, coloured by copy number, for n = 2268 isolates with WT SHV alleles. EUCAST (v13.0) or CLSI (M100 33rd edition) I/R breakpoints are indicated using orange and blue lines, respectively.
Discussion
BlaSHV alleles have been studied since their discovery in 1972 and were first explored phylogenetically in 1990 to study the context of blaSHV-2 and its relationships with other β-lactamase genes [51]. As new blaSHV variants are discovered, phylogenetic trees were inferred to explore their ancestry and relationships with each other [2,10, 52]. Most recently, Liakopoulos et al. inferred a maximum likelihood tree with 149 SHV-type β-lactamases, but it was unclear which blaSHV was the likely ancestral variant [10]. It has been assumed that SHV-1 is the ancestral variant since it was the first blaSHV discovered, and our cladogram, pairwise distance data and minimum-spanning tree also support blaSHV-1 as the ancestral variant (Figs 2 and S1). There is also support from Chaves et al. [53] and Haeggman et al. [54] who show that blaSHV-1 is predominantly species specific to K. pneumoniae and has a long evolutionary history as a stable chromosomal gene, suggesting that even the ancestor of blaSHV-1 is also from the K. pneumoniae chromosome.
Our phylogenetic and comparative genomic analyses support that ESBL- and BLI-resistant variants of blaSHV have evolved multiple times independently through parallel substitution mutations (Fig. 2) and that many of these variants have been mobilized out of the K. pneumoniae chromosome via independent events (Fig. 3), enabling them to spread between lineages, species and genera. We found evidence of mobilization for most ESBL variants but only one BLI-resistant conferring variant (SHV-92). Consistent with this, most 3GC-resistant K. pneumoniae carrying ESBL variants and no other β-lactamases were found to have multiple copies of SHV (presumably a chromosomal copy with WT activity plus a plasmid-borne copy with ESBL activity).
We have reviewed the classification of blaSHV alleles into functional classes to better support the interpretation of genomic data. Our work builds on the experimental study of Neubauer et al., which provided evidence of the role of specific mutations in enzyme activity. By systematically assigning alleles to functional classes based on the presence of specific mutations associated with enzyme activity (Fig. 1), rather than the presence in an ESBL- or BLI-resistant isolate (which may confuse mobile and chromosomal variants), we propose re-classification of 20 blaSHV alleles from ESBL to WT (n = 12 changes vs the NCBI’s Reference Gene Catalog and n = 14 changes vs BLDB, see Table S1).
We used matched genotype–phenotype data, for 3999 K. pneumoniae carrying blaSHV and no other acquired β-lactamases, to assess the predictability of phenotype based on blaSHV alleles (Figs4 5, Tables1 2). For this, we used our Kleborate tool to identify and type blaSHV alleles and specific SHV mutations associated with a change in enzyme activity. This analysis provided additional support for the role of 238S and 179G [13,14] in ESBL activity and consequent 3GC resistance but suggests that most changes in the omega-loop do not result in a change in activity. These data also support our classification of variants SHV-27, SHV-38, SHV-40, SHV-41, SHV-42, SHV-65, SHV-164 and SHV-187 – which lack mutations at site 238 or any other mutations associated experimentally with resistance – as WT.
Mutations 69I and 69V have been thought to explain BLI resistance of variants SHV-49, SHV-52, SHV-92 and SHV-203, respectively, and were found by Neubauer et al. to confer resistance to piperacillin-tazobactam. Interestingly, our data do not support a simple association between Ambler site 69 mutations and BLI resistance in K. pneumoniae, whether in the SHV-52 variant (n = 0/12 resistant to piperacillin-tazobactam or amoxicillin-clavulanic acid) or arising de novo in other SHV backgrounds (SHV-27, SHV-110, SHV-164, SHV-187 and SHV-209) (n = 3/9 resistant). We identified two genomes with a mutation at Ambler site 235 (both SHV-107, which carry mutation 235A) which were both resistant to amoxicillin-clavulanic acid (piperacillin-tazobactam was not tested), providing support for the role of this mutation, which was confirmed by Neubauer et al. [26].
The approach and results outlined here have been implemented in Kleborate v2.4.1, along with all new alleles identified in this study, and all those available in public databases as of 7 November 2023. In the Kleborate v2.4.1 database, known blaSHV alleles classified as ESBL are those with aa substitutions at Ambler site 238 (n = 36 alleles), 179 (SHV-8), 169 (SHV-57), 148 (SHV-70) and 240K+35Q (SHV-31) or insertion in the omega-loop (SHV-16). SHV variants are classified as BLI resistant if they possess mutations at Ambler site 69, 130, 234 or 235. Where exact nt or protein matches are found to a known allele, these are reported in the relevant column (Bla_ESBL, Bla_inhibR, Bla_ESBL_inhibR and Bla_chr) based on the classification in the Kleborate database. As the mutations noted above are considered causative of a change of enzyme activity (class modifying), Kleborate checks for these mutations in all SHV sequences and reports them in a separate column, SHV_mutations. If a class-modifying mutation is detected in an otherwise WT-classified allele background, the novel allele will be reported in the relevant functional column, i.e. Bla_ESBL, Bla_ESBL_inhibR or Bla_inhibR rather than Bla_chr, and labelled with the mutation. Kleborate will also report any mutation in the omega-loop (sites 164–179) in the SHV_mutations column, as it is theoretically possible that any modification disrupting the omega-loop structure could impact function [16,55, 56]. However, the detection of these mutations will not change the class assignment in Kleborate since most changes are likely to be non-functional and all novel omega-loop mutants we identified in our study tested susceptible to 3GCs (Table 2). Our phenotype data also do not support a simple association between mutations at Ambler site 69 and clinical resistance to piperacillin-tazobactam or amoxicillin-clavulanic acid (Table 2). However, our numbers are small (n = 9 isolates, of which three tested resistant), and the functional evidence for BLI resistance associated with mutations at this site is convincing [18,26, 57]; therefore, we consider it appropriate to distinguish alleles with Ambler site 69 mutations from WT alleles in the Kleborate database and reporting.
The KlebNET-GSP matched genotype–phenotype dataset yielded coverage of 40% of known blaSHV alleles in otherwise β-lactamase-free backgrounds, which is essential to interpret the role of blaSHV specifically. Despite other alleles being in unfavourable genomic contexts, our approach enabled a systematic assessment of how blaSHV alleles are assigned to functional classes in the public AMR gene databases and provides evidence that some existing assignments are incorrect (Table S1). In turn, this helped us to implement a more transparent and consistent approach to detecting and reporting known and novel blaSHV alleles in K. pneumoniae genomes, via Kleborate v2.4.1. In addition, the diversity of this dataset (isolates from 24 different countries across 2001–2021 and collected from humans, animals and environments) avoids the very often local epidemiological effects that could bias results. Additional insights into the role of genetic background, expression and co-expression of SHV variants and/or other β-lactamases on resistance mechanisms will help to further clarify the impact of individual variants and lead to better interpretation of genotypes and prediction of phenotypes.
This study exemplifies the importance of sharing AST data together with genome data and the potential role for global collaboration such as KlebNET-GSP to utilize these data to enhance the understanding of resistance mechanisms. This is particularly relevant in cases like blaSHV, where complex evolutionary processes have contributed to the emergence and mobilization of resistant variants within and between the originating species. As the KlebNET-GSP isolate collection grows, we intend to regularly update this analysis to support the growing evidence for blaSHV phenotypes, to explore genotype–phenotype variation in the homologous enzymes of other members of the K. pneumoniae species complex (blaOKP in K. quasipneumoniae and blaLEN in K. variicola) and to undertake similar analyses to inform the understanding of the mechanisms of resistance to other drug classes relevant to the treatment of K. pneumoniae infection.
supplementary material
Acknowledgements
KlebNET-GSP AMR Genotype-Phenotype Group shares this work on behalf of the Kp-T7 and Kp-MDR studies, the Norwegian Study Group on Klebsiella pneumoniae, Kp-NORM study, JPI-AMR consortium SpARK, NIHR Global Health Research Unit Genomic Surveillance of Antimicrobial Resistance, the ‘Controlling Superbugs’ flagship study and the Victorian Carbapenemase-Producing Enterobacterales (CPE) programme, Vietnam ICU WGS study, GHRU-GSA-Ibadan and REDUCEAMU project. KlebNET-GSP AMR Genotype-Phenotype Group membership includes all named authors and the following individuals: Gherard Batisti Biffignandi, Ben Cooper, Jennifer Cornick, Annapaula Correia, Derrick Crook, Vo Thi Trang Dai, Nicholas Feasey, Maria Laura Ferrando, Ebenezer Foster-Nyarko, Giovanna Graziano, Yukino Gütlin, Marisa Haenni, Eva Heinz, Nguyen Thi Hoa, Suchita Shrestha Joshi, John Lees, Le Thi Lien, Giovanni Lorenzin, Catrin Moore, Patrick Musicha, Nguyen Van Kinh, Charlene Rodrigues, Francesca Saluzzo, Andrea Spitaleri, Paul Turner, Jon Van Aartsen and Nguyen Hoang Vu. We acknowledge Michael Feldgarden for his valuable comments on the preprint.
Abbreviations
- AMR
antimicrobial resistance
- AST
antimicrobial susceptibility testing
- BLI
β-lactamase inhibitor
- CLSI
Clinical and Laboratory Standards Institute
- ESBL
extended-spectrum β-lactamase
- EUCAST
European Committee on Antimicrobial Susceptibility Testing
- 3GCs
third-generation cephalosporins
- MIC
minimum inhibitory concentration
- SHV
sulfhydryl variable
Footnotes
Funding: This work was supported, in whole or in part, by the Bill and Melinda Gates Foundation (OPP025280). Under the grant conditions of the foundation, a Creative Commons Attribution 4.0 Generic License has already been assigned to the author accepted manuscript version that might arise from this submission. This work was also supported financially by the MedVetKlebs project from the European Joint Programme One Health, which has received funding from the European Union’s Horizon 2020 Research and Innovation Programme under Grant Agreement No. 773830.
Author contributions: Conceptualization: K.E.H. Methodology: M.M.C.L., K.K.T., K.E.H. Software: M.M.C.L., K.E.H. Validation: M.M.C.L., K.K.T., K.E.H. Formal analysis: K.K.T. Investigation: K.K.T. Resources: K.E.H., M.B., S.B., K.B., S.B., A.C., D.M.C., J.C., M.C., A.C., A.C., N.D., P.D., A.E., R.F., E.J.F., A.F., C.L.G., Y.G., B.H., M.A.K.H., L.N.M.H., L.T.H., B.H., O.I., A.W.J.J., H.K., F.K., T.L., I.H.L., S.W.L., G.L., M.L., A.J.M., A.G.M., G.N., A.O.O., I.N.O., H.P., J.P., M.H.P., F.P., N.R., A.R., K.L.R.K., L.R., C.R., Ø.S., K.S., D.S., H.S., V.S., N.L.S., S.S., A.S., N.S., M.S., A.S., P.N.T., N.T., H.A.T., E.T., V.D.T., N.V.T., J.V., T.W., B.W., H.W., G.D.W., K.L.W. Data curation: M.M.C.L., K.K.T. Project administration: K.E.H. Supervision: K.E.H. Writing – original draft: K.K.T. Writing – review and editing: K.K.T., M.M.C.L., R.R.W., K.L.W., M.B., S.B., K.B., S.B., A.C., D.M.C., J.C., M.C., A.C., A.C., N.D., P.D., A.E., R.F., E.J.F., A.F., C.L.G., Y.G., B.H., M.A.K.H., L.N.M.H., L.T.H., B.H., O.I., A.W.J.J., H.K., F.K., T.L., I.H.L., S.W.L., G.L., M.L., A.J.M., A.G.M., G.N., A.O.O., I.N.O., H.P., J.P., M.H.P., F.P., N.R., A.R., K.L.R.K., L.R., C.R., Ø.S., K.S., D.S., H.S., V.S., N.L.S., S.S., A.S., N.S., M.S., A.S., P.N.T., N.T., H.A.T., E.T., V.D.T., N.V.T., J.V., T.W., B.W., H.W., G.D.W., K.E.H. Visualization: K.K.T. Funding acquisition: K.E.H.
Contributor Information
Kara K. Tsang, Email: Kara.Tsang@lshtm.ac.uk.
Margaret M. C. Lam, Email: margaret.lam@monash.edu.
Ryan R. Wick, Email: rrwick@gmail.com.
Kelly L. Wyres, Email: kelly.wyres@monash.edu.
Michael Bachman, Email: mikebach@med.umich.edu.
Stephen Baker, Email: sgb47@cam.ac.uk.
Katherine Barry, Email: KEB3RM@hscmail.mcc.virginia.edu.
Sylvain Brisse, Email: sbrisse@pasteur.fr.
Susana Campino, Email: Susana.Campino@lshtm.ac.uk.
Alexandra Chiaverini, Email: a.chiaverini@izs.it.
Daniela Maria Cirillo, Email: cirillo.daniela@hsr.it.
Taane Clark, Email: Taane.Clark@LSHTM.ac.uk.
Jukka Corander, Email: jukka.corander@medisin.uio.no.
Marta Corbella, Email: m.corbella@smatteo.pv.it.
Alessandra Cornacchia, Email: a.cornacchia@izs.it.
Aline Cuénod, Email: aline.cuenod@mcgill.ca.
Nicola D'Alterio, Email: n.dalterio@izs.it.
Federico Di Marco, Email: dimarco.federico@hsr.it.
Pilar Donado-Godoy, Email: pidonado@agrosavia.co.
Adrian Egli, Email: aegli@imm.uzh.ch.
Refath Farzana, Email: refath.farzana@zoo.ox.ac.uk.
Edward J. Feil, Email: e.feil@bath.ac.uk.
Aasmund Fostervold, Email: aasmund.fostervold@sus.no.
Claire L. Gorrie, Email: gorrie.c@unimelb.edu.au.
Brekhna Hassan, Email: brekhnahassan@hotmail.com.
Marit Andrea Klokkhammer Hetland, Email: marit.andrea.klokkhammer.hetland@sus.no.
Le Nguyen Minh Hoa, Email: hoalenguyenminh@gmail.com.
Le Thi Hoi, Email: lehoi2003@gmail.com.
Benjamin Howden, Email: bhowden@unimelb.edu.au.
Odion O. Ikhimiukor, Email: odionikh@gmail.com.
Adam W. J. Jenney, Email: A.Jenney@alfred.org.au.
Håkon Kaspersen, Email: hakon.kaspersen@vetinst.no.
Fahad Khokhar, Email: fak31@cam.ac.uk.
Thongpan Leangapichart, Email: winmicro40@hotmail.com.
Małgorzata Ligowska-Marzęta, Email: MALM@ssi.dk.
Iren Høyland Löhr, Email: iren.hoyland.lohr@sus.no.
Scott W. Long, Email: SWLong@houstonmethodist.org.
Amy J. Mathers, Email: Ajm5b@virginia.edu.
Andrew G. McArthur, Email: mcarthua@mcmaster.ca.
Geetha Nagaraj, Email: geetha.ndri@gmail.com.
Anderson O. Oaikhena, Email: andersonose@yahoo.co.uk.
Iruka N. Okeke, Email: iruka.n.okeke@gmail.com.
João Perdigão, Email: jperdigao@campus.ul.pt.
Hardik Parikh, Email: parikhhi12@gmail.com.
My H. Pham, Email: mp29@sanger.ac.uk.
Francesco Pomilio, Email: f.pomilio@izs.it.
Niclas Raffelsberger, Email: niclas.raffelsberger@unn.no.
Andriniaina Rakotondrasoa, Email: aina@pasteur.mg.
K. L. Ravi Kumar, Email: klravikumar@gmail.com.
Leah W. Roberts, Email: leah@ebi.ac.uk.
Carla Rodrigues, Email: carla.parada-rodrigues@pasteur.fr.
Ørjan Samuelsen, Email: orjan.samuelsen@unn.no.
Kirsty Sands, Email: kirsty.sands@zoo.ox.ac.uk.
Davide Sassera, Email: davide.sassera@unipv.it.
Helena Seth-Smith, Email: hsethsmith@imm.uzh.ch.
Varun Shamanna, Email: varunshamanna4@gmail.com.
Norelle L. Sherry, Email: norelle.sherry@unimelb.edu.au.
Sonia Sia, Email: sonia.sia@ritm.gov.ph.
Anton Spadar, Email: anton.spadar@lshtm.ac.uk.
Nicole Stoesser, Email: nicole.stoesser@ndm.ox.ac.uk.
Marianne Sunde, Email: marianne.sunde@vetinst.no.
Arnfinn Sundsfjord, Email: arnfinn.sundsfjord@uit.no.
Pham Ngoc Thach, Email: phamngocthachnhtd@gmail.com.
Nicholas R. Thomson, Email: nrt@sanger.ac.uk.
Harry A. Thorpe, Email: harry.thorpe@medisin.uio.no.
M. Estée Torok, Email: et317@cam.ac.uk.
Van Dinh Trang, Email: vandinhtrang.nhtd@gmail.com.
Nguyen Vu Trung, Email: nguyen.vu.trung@gmail.com.
Jay Vornhagen, Email: jayvornh@iu.edu.
Timothy Walsh, Email: timothy.walsh@zoo.ox.ac.uk.
Ben Warne, Email: bw252@cam.ac.uk.
Hayley Wilson, Email: hjw58@cam.ac.uk.
Gerard D. Wright, Email: wrightge@mcmaster.ca.
Kathryn E. Holt, Email: kat.holt@lshtm.ac.uk.
References
- 1.Ford PJ, Avison MB. Evolutionary mapping of the SHV beta-lactamase and evidence for two separate IS26-dependent blaSHV mobilization events from the Klebsiella pneumoniae chromosome. J Antimicrob Chemother. 2004;54:69–75. doi: 10.1093/jac/dkh251. [DOI] [PubMed] [Google Scholar]
- 2.Heritage J, M’Zali FH, Gascoyne-Binzi D, Hawkey PM. Evolution and spread of SHV extended-spectrum beta-lactamases in gram-negative bacteria. J Antimicrob Chemother. 1999;44:309–318. doi: 10.1093/jac/44.3.309. [DOI] [PubMed] [Google Scholar]
- 3.Hammond DS, Schooneveldt JM, Nimmo GR, Huygens F, Giffard PM bla. bla(SHV) genes in Klebsiella pneumoniae: different allele distributions are associated with different promoters within individual isolates. Antimicrob Agents Chemother. 2005;49:256–263. doi: 10.1128/AAC.49.1.256-263.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pitton J-S. Mechanisms of bacterial resistance to antibiotics. Ergebnisse der Physiol Rev Physiol. 1972;65:15–93. doi: 10.1007/3-540-05814-1_2. [DOI] [PubMed] [Google Scholar]
- 5.Matthew M, Hedges RW, Smith JT. Types of beta-lactamase determined by plasmids in gram-negative bacteria. J Bacteriol. 1979;138:657–662. doi: 10.1128/jb.138.3.657-662.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nugent ME, Hedges RW. The nature of the genetic determinant for the SHV-1 beta-lactamase. Mol Gen Genet . 1979;175:239–243. doi: 10.1007/BF00397222. [DOI] [PubMed] [Google Scholar]
- 7.Kliebe C, Nies BA, Meyer JF, Tolxdorff-Neutzling RM, Wiedemann B. Evolution of plasmid-coded resistance to broad-spectrum cephalosporins. Antimicrob Agents Chemother. 1985;28:302–307. doi: 10.1128/AAC.28.2.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knothe H, Shah P, Krcmery V, Antal M, Mitsuhashi S. Transferable resistance to cefotaxime, cefoxitin, cefamandole and cefuroxime in clinical isolates of Klebsiella pneumoniae and Serratia marcescens. Infection. 1983;11:315–317. doi: 10.1007/BF01641355. [DOI] [PubMed] [Google Scholar]
- 9.Barthélémy M, Péduzzi J, Ben Yaghlane H, Labia R. Single amino acid substitution between SHV-1 beta-lactamase and cefotaxime-hydrolyzing SHV-2 enzyme. FEBS Lett. 1988;231:217–220. doi: 10.1016/0014-5793(88)80734-8. [DOI] [PubMed] [Google Scholar]
- 10.Liakopoulos A, Mevius D, Ceccarelli D. A review of SHV extended-spectrum β-lactamases: neglected yet ubiquitous. Front Microbiol. 2016;7:1374. doi: 10.3389/fmicb.2016.01374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alcock BP, Huynh W, Chalil R, Smith KW, Raphenya AR, et al. CARD 2023: expanded curation, support for machine learning, and resistome prediction at the comprehensive antibiotic resistance database. Nucleic Acids Res. 2023;51:D690–D699. doi: 10.1093/nar/gkac920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Podbielski A, Schönling JA, Melzer B, Warnatz K. Molecular cloning and nucleotide sequence of a new plasmid-coded Klebsiella pneumoniae beta-lactamase gene (SHV-2a) responsible for high-level cefotaxime resistance. Zentralbl Bakteriol. 1991;275:369–373. doi: 10.1016/s0934-8840(11)80302-6. [DOI] [PubMed] [Google Scholar]
- 13.Rasheed JK, Jay C, Metchock B, Berkowitz F, Weigel L, et al. Evolution of extended-spectrum beta-lactam resistance (SHV-8) in a strain of Escherichia coli during multiple episodes of bacteremia. Antimicrob Agents Chemother. 1997;41:647–653. doi: 10.1128/AAC.41.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kurokawa H, Yagi T, Shibata N, Shibayama K, Kamachi K, et al. A new SHV-derived extended-spectrum beta-lactamase (SHV-24) that hydrolyzes ceftazidime through A single-amino-acid substitution (D179G) in the -loop. Antimicrob Agents Chemother. 2000;44:1725–1727. doi: 10.1128/AAC.44.6.1725-1727.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ma L, Alba J, Chang F-Y, Ishiguro M, Yamaguchi K, et al. Novel SHV-derived extended-spectrum beta-lactamase, SHV-57, that confers resistance to ceftazidime but not cefazolin. Antimicrob Agents Chemother. 2005;49:600–605. doi: 10.1128/AAC.49.2.600-605.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Arpin C, Labia R, Andre C, Frigo C, El Harrif Z, et al. SHV-16, a beta-lactamase with a pentapeptide duplication in the omega loop. Antimicrob Agents Chemother. 2001;45:2480–2485. doi: 10.1128/AAC.45.9.2480-2485.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prinarakis EE, Miriagou V, Tzelepi E, Gazouli M, Tzouvelekis LS. Emergence of an inhibitor-resistant beta-lactamase (SHV-10) derived from an SHV-5 variant. Antimicrob Agents Chemother. 1997;41:838–840. doi: 10.1128/AAC.41.4.838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dubois V, Poirel L, Arpin C, Coulange L, Bebear C, et al. SHV-49, a novel inhibitor-resistant beta-lactamase in a clinical isolate of Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:4466–4469. doi: 10.1128/AAC.48.11.4466-4469.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dubois V, Poirel L, Demarthe F, Arpin C, Coulange L, et al. Molecular and biochemical characterization of SHV-56, a novel inhibitor-resistant beta-lactamase from Klebsiella pneumoniae. Antimicrob Agents Chemother. 2008;52:3792–3794. doi: 10.1128/AAC.00387-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mendonça N, Manageiro V, Robin F, Salgado MJ, Ferreira E, et al. The Lys234Arg substitution in the enzyme SHV-72 is a determinant for resistance to clavulanic acid inhibition. Antimicrob Agents Chemother. 2008;52:1806–1811. doi: 10.1128/AAC.01381-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Manageiro V, Ferreira E, Cougnoux A, Albuquerque L, Caniça M, et al. Characterization of the inhibitor-resistant SHV β-lactamase SHV-107 in a clinical Klebsiella pneumoniae strain coproducing GES-7 enzyme. Antimicrob Agents Chemother. 2012;56:1042–1046. doi: 10.1128/AAC.01444-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Feldgarden M, Brover V, Gonzalez-Escalona N, Frye JG, Haendiges J, et al. AMRFinderPlus and the reference gene catalog facilitate examination of the genomic links among antimicrobial resistance, stress response, and virulence. Sci Rep. 2021;11:12728. doi: 10.1038/s41598-021-91456-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Naas T, Oueslati S, Bonnin RA, Dabos ML, Zavala A, et al. Beta-lactamase database (BLDB) - structure and function. J Enzyme Inhib Med Chem. 2017;32:917–919. doi: 10.1080/14756366.2017.1344235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bradford PA, Bonomo RA, Bush K, Carattoli A, Feldgarden M, et al. Consensus on β-lactamase nomenclature. Antimicrob Agents Chemother. 2022;66:e0033322. doi: 10.1128/aac.00333-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin T-L, Tang S-I, Fang C-T, Hsueh P-R, Chang S-C, et al. Extended-spectrum beta-lactamase genes of Klebsiella pneumoniae strains in Taiwan: recharacterization of shv-27, shv-41, and tem-116. Microb Drug Resist. 2006;12:12–15. doi: 10.1089/mdr.2006.12.12. [DOI] [PubMed] [Google Scholar]
- 26.Neubauer S, Madzgalla S, Marquet M, Klabunde A, Büttner B, et al. A Genotype-phenotype correlation study of SHV β-lactamases offers new insight into SHV resistance profiles. Antimicrob Agents Chemother. 2020;64:e02293-19. doi: 10.1128/AAC.02293-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lam MMC, Wick RR, Watts SC, Cerdeira LT, Wyres KL, et al. A genomic surveillance framework and genotyping tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12:4188. doi: 10.1038/s41467-021-24448-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fajardo-Lubián A, Ben Zakour NL, Agyekum A, Qi Q, Iredell JR. Host adaptation and convergent evolution increases antibiotic resistance without loss of virulence in a major human pathogen. PLOS Pathog. 2019;15:e1007218. doi: 10.1371/journal.ppat.1007218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hasman H, Saputra D, Sicheritz-Ponten T, Lund O, Svendsen CA, et al. Rapid whole-genome sequencing for detection and characterization of microorganisms directly from clinical samples. J Clin Microbiol. 2014;52:139–146. doi: 10.1128/JCM.02452-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katoh K, Misawa K, Kuma K, Miyata T. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 2002;30:3059–3066. doi: 10.1093/nar/gkf436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhou Z, Alikhan N-F, Sergeant MJ, Luhmann N, Vaz C, et al. GrapeTree: visualization of core genomic relationships among 100,000 bacterial pathogens. Genome Res. 2018;28:1395–1404. doi: 10.1101/gr.232397.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu G. Using ggtree to Visualize data on tree‐like structures. CP Bioinform. 2020;69 doi: 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
- 33.Zhu Q, Gao S, Xiao B, He Z, Hu S. Plasmer: an accurate and sensitive bacterial plasmid prediction tool based on machine learning of shared k-mers and genomic features. Microbiol Spectr. 2023;11:e0464522. doi: 10.1128/spectrum.04645-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Darling ACE, Mau B, Blattner FR, Perna NT. Mauve: multiple alignment of conserved genomic sequence with rearrangements. Genome Res. 2004;14:1394–1403. doi: 10.1101/gr.2289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seemann T. Prokka: rapid prokaryotic genome annotation. Bioinformatics. 2014;30:2068–2069. doi: 10.1093/bioinformatics/btu153. [DOI] [PubMed] [Google Scholar]
- 36.Gilchrist CLM, Chooi Y-H. Clinker & clustermap.js: automatic generation of gene cluster comparison figures. Bioinformatics. 2021;37:2473–2475. doi: 10.1093/bioinformatics/btab007. [DOI] [PubMed] [Google Scholar]
- 37.Thorn AV, Aarestrup FM, Munk P. Flankophile: a bioinformatic pipeline for prokaryotic genomic synteny analysis. Microbiol Spectr. 2024;12:e0241323. doi: 10.1128/spectrum.02413-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Siguier P, Perochon J, Lestrade L, Mahillon J, Chandler M. ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34:D32–6. doi: 10.1093/nar/gkj014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inouye M, Dashnow H, Raven L-A, Schultz MB, Pope BJ, et al. SRST2: rapid genomic surveillance for public health and hospital microbiology labs. Genome Med. 2014;6:90. doi: 10.1186/s13073-014-0090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mazzariol A, Roelofsen E, Koncan R, Voss A, Cornaglia G. Detection of a new SHV-type extended-spectrum beta-lactamase, SHV-31, in a Klebsiella pneumoniae strain causing a large nosocomial outbreak in The Netherlands. Antimicrob Agents Chemother. 2007;51:1082–1084. doi: 10.1128/AAC.00909-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ling B-D, Liu G, Xie Y-E, Zhou Q-X, Zhao T-K, et al. Characterisation of a novel extended-spectrum beta-lactamase, SHV-70, from a clinical isolate of Enterobacter cloacae in China. Int J Antimicrob Agents. 2006;27:355–356. doi: 10.1016/j.ijantimicag.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 43.Nicolas MH, Jarlier V, Honore N, Philippon A, Cole ST. Molecular characterization of the gene encoding SHV-3 beta-lactamase responsible for transferable cefotaxime resistance in clinical isolates of Klebsiella pneumoniae. Antimicrob Agents Chemother. 1989;33:2096–2100. doi: 10.1128/AAC.33.12.2096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Levison ME, Mailapur YV, Pradhan SK, Jacoby GA, Adams P, et al. Regional occurrence of plasmid-mediated SHV-7, an extended-spectrum beta-lactamase, in Enterobacter cloacae in Philadelphia Teaching Hospitals. Clin Infect Dis. 2002;35:1551–1554. doi: 10.1086/344178. [DOI] [PubMed] [Google Scholar]
- 45.Szabó D, Melan MA, Hujer AM, Bonomo RA, Hujer KM, et al. Molecular analysis of the simultaneous production of two SHV-type extended-spectrum beta-lactamases in a clinical isolate of Enterobacter cloacae by using single-nucleotide polymorphism genotyping. Antimicrob Agents Chemother. 2005;49:4716–4720. doi: 10.1128/AAC.49.11.4716-4720.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Heritage J, Chambers PA, Tyndall C, Buescher ES. SHV-34: an extended-spectrum beta-lactamase encoded by an epidemic plasmid. J Antimicrob Chemother. 2003;52:1015–1017. doi: 10.1093/jac/dkh017. [DOI] [PubMed] [Google Scholar]
- 47.Lavilla S, González-López JJ, Sabaté M, García-Fernández A, Larrosa MN, et al. Prevalence of qnr genes among extended-spectrum beta-lactamase-producing enterobacterial isolates in Barcelona, Spain. J Antimicrob Chemother. 2008;61:291–295. doi: 10.1093/jac/dkm448. [DOI] [PubMed] [Google Scholar]
- 48.Moradigaravand D, Martin V, Peacock SJ, Parkhill J. Evolution and epidemiology of multidrug-resistant Klebsiella pneumoniae in the United Kingdom and Ireland. mBio. 2017;8:e01976-16. doi: 10.1128/mBio.01976-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sherry NL, Lane CR, Kwong JC, Schultz M, Sait M, et al. Genomics for molecular epidemiology and detecting transmission of carbapenemase-producing Enterobacterales in Victoria, Australia, 2012 to 2016. J Clin Microbiol. 2019;57 doi: 10.1128/JCM.00573-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang FY, Siu LK, Fung CP, Huang MH, Ho M. Diversity of SHV and TEM beta-lactamases in Klebsiella pneumoniae: gene evolution in Northern Taiwan and two novel beta-lactamases, SHV-25 and SHV-26. Antimicrob Agents Chemother. 2001;45:2407–2413. doi: 10.1128/AAC.45.9.2407-2413.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Huletsky A, Couture F, Levesque RC. Nucleotide sequence and phylogeny of SHV-2 beta-lactamase. Antimicrob Agents Chemother. 1990;34:1725–1732. doi: 10.1128/AAC.34.9.1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Newire EA, Ahmed SF, House B, Valiente E, Pimentel G. Detection of new SHV-12, SHV-5 and SHV-2a variants of extended spectrum beta-lactamase in Klebsiella pneumoniae in Egypt. Ann Clin Microbiol Antimicrob. 2013;12:16. doi: 10.1186/1476-0711-12-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chaves J, Ladona MG, Segura C, Coira A, Reig R, et al. SHV-1 beta-lactamase is mainly a chromosomally encoded species-specific enzyme in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2001;45:2856–2861. doi: 10.1128/AAC.45.10.2856-2861.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Haeggman S, Löfdahl S, Paauw A, Verhoef J, Brisse S. Diversity and evolution of the class A chromosomal beta-lactamase gene in Klebsiella pneumoniae. Antimicrob Agents Chemother. 2004;48:2400–2408. doi: 10.1128/AAC.48.7.2400-2408.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kuzin AP, Nukaga M, Nukaga Y, Hujer AM, Bonomo RA, et al. Structure of the SHV-1 beta-lactamase. Biochemistry. 1999;38:5720–5727. doi: 10.1021/bi990136d. [DOI] [PubMed] [Google Scholar]
- 56.Sampson JM, Ke W, Bethel CR, Pagadala SRR, Nottingham MD, et al. Ligand-dependent disorder of the Omega loop observed in extended-spectrum SHV-type beta-lactamase. Antimicrob Agents Chemother. 2011;55:2303–2309. doi: 10.1128/AAC.01360-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Giakkoupi P, Miriagou V, Gazouli M, Tzelepi E, Legakis NJ, et al. Properties of mutant SHV-5 β-lactamases constructed by substitution of isoleucine or valine for methionine at position 69. Antimicrob Agents Chemother . 1998;42:1281–1283. doi: 10.1128/AAC.42.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.