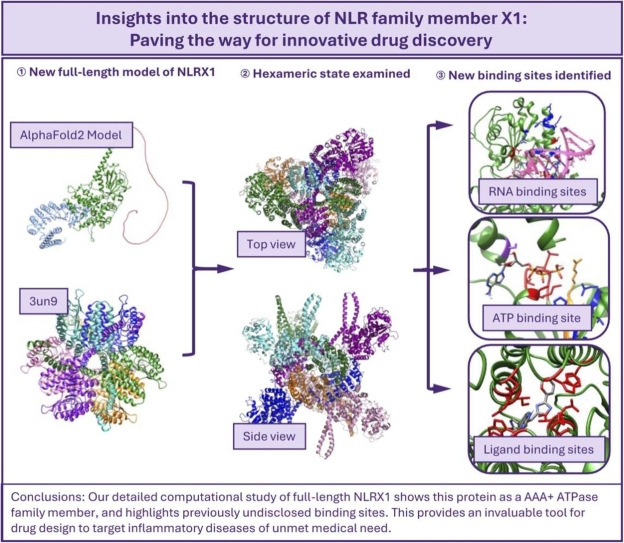
Abstract
Nucleotide-binding oligomerization domain, leucine rich repeat containing X1 (NLRX1) is a negative regulator of the nuclear factor kappa-light-chain-enhancer of activated B cells (NFκB) pathway, with a significant role in the context of inflammation. Altered expression of NLRX1 is prevalent in inflammatory diseases leading to interest in NLRX1 as a drug target. There is a lack of structural information available for NLRX1 as only the leucine-rich repeat domain of NLRX1 has been crystallised. This lack of structural data limits progress in understanding function and potential druggability of NLRX1. We have modelled full-length NLRX1 by combining experimental, homology modelled and AlphaFold2 structures. The full-length model of NLRX1 was used to explore protein dynamics, mutational tolerance and potential functions. We identified a new RNA binding site in the previously uncharacterized N-terminus, which served as a basis to model protein-RNA complexes. The structure of the adenosine triphosphate (ATP) binding domain revealed a potential catalytic functionality for the protein as a member of the ATPase Associated with Diverse Cellular Activity family of proteins. Finally, we investigated the interactions of NLRX1 with small molecule activators in development, revealing a binding site that has not previously been discussed in literature. The model generated here will help to catalyse efforts towards creating new drug molecules to target NLRX1 and may be used to inform further studies on functionality of NLRX1.
Keywords: Inflammation, NLRX1, Inflammasome, ATPase
Graphical Abstract
1. Introduction
Modulating activity of the NOD-like receptor (NLR) class of proteins is an area of intense interest in the drug discovery community due to unprecedented therapeutic potential, across a plethora of inflammatory diseases, many of which have significant unmet medical need. This has been highlighted through the plentiful publications, patents, new companies, lucrative acquisitions, and clinical trials surrounding small molecule modulators of the well-studied, inflammasome forming, NLR protein NLRP3 [1]. Other NLR proteins are less well understood but no less significant in their potential therapeutic role. Herein we focus on Nucleotide-binding oligomerization domain, leucine rich repeat containing X1 (NLRX1), a non-inflammasome forming NLR protein, unique amongst the NLR family as it localises to the mitochondria [2]. We have investigated, in detail, the structure of NLRX1 in the hope that these structural insights will facilitate further drug discovery efforts, to complement the increasing number of, extensively reviewed, biological studies which demonstrate the significant role of NLRX1 in disease [3], [4], [5], [6], [7].
Inflammation is a natural and essential component of innate immunity, as a protective response to infection and tissue damage [8]. Immune cells detect pathogen associated molecular patterns (PAMPs) and damage associated molecular patterns (DAMPs) through pattern recognition receptors (PRRs) [9]. There are several types of PRRs including toll-like receptors, retinoic acid-inducible gene I (RIG-1)-like receptors, NOD-like receptors (NLR), C-type Lectin like receptors and cytosolic DNA sensors, all of which have different downstream effects upon activation [10]. One of the pathways to activation of PRR, is via the NFκB pathway. NFκB is an inducible transcription factor that regulates various pro-inflammatory genes and is central to inflammatory processes [10]. NLRX1 is a negative regulator of NFκB pathway [11]. In the resting state NFκB is inhibited by IκB, but when activated IκB kinase (IKK) phosphorylates IκB causing release of NFκB and the inflammatory cascade ensues. Polyubiquitination of NLRX1 causes binding of its leucine rich repeat (LRR) domain to IKK kinase domain preventing phosphorylation of IkB, which remains complexed to NFκB, thereby inhibiting the inflammatory cascade [11]. NLRX1 is also involved in viral infection where interaction between NLRX1 and mitochondrial antiviral signalling protein (MAVS) prevents downstream activation of RIG-1 and ultimately NFκB [12]. In the clinic, altered expression of NLRX1 has been linked to increased inflammatory markers and poor prognosis in various inflammatory diseases including irritable bowel disease (IBD), cancer, arthritis, metabolic disease, diseases of the central nervous system and brain injury [3].
Despite the growing interest, there is limited understanding of NLRX1, and structural data is lacking. To date, only the C-terminal end of NLRX1 (cNLRX1) has been determined through X-ray crystallography (PDBcode: 3un9) [13]. The N-terminal end remains elusive; the lack of characterization of the N-terminus of NLRX1 (nNLRX1 - residues 77–628) is the reason that this protein acquired the X in its name [14]. Herein we have harnessed advances in structural modelling to characterise the full molecular structure of NLRX1, established likely oligomerisation states, predicted binding sites, and modelled various protein interactions. NLRX1 has a predicted ATP binding site which we have investigated in detail and conclude that NLRX1 is an ATPase Associated with Diverse Cellular Activity (AAA+) where ATP binding facilitates significant structural change to an active state. Consistent with this, we show full-length NLRX1 forms a stable centrosymmetric homo-hexameric structure, classically adopted by AAA+ ATPases. NLRX1 is known to bind RNA; we found that RNA-protein interaction occurs only with the NLRX1 hexamer, and that the trimer interface is critical for RNA recognition. Furthermore, we identified a positively charged RNA binding pocket formed by the helices of the previously unexplored N-terminus, surrounding the centre of the hexamer. Finally, we highlight two likely binding sites, one of which has not been suggested previously, for small molecule NLRX1 agonists NX-13, NX-64–3 and LABP-72–38 currently in clinical and pre-clinical development [15], [16], [17]. This paper lays the groundwork required to expedite design of further novel modulators of NLRX1 for therapeutic resolution of inflammatory disease.
2. Results
2.1. Homology model of NLRX1
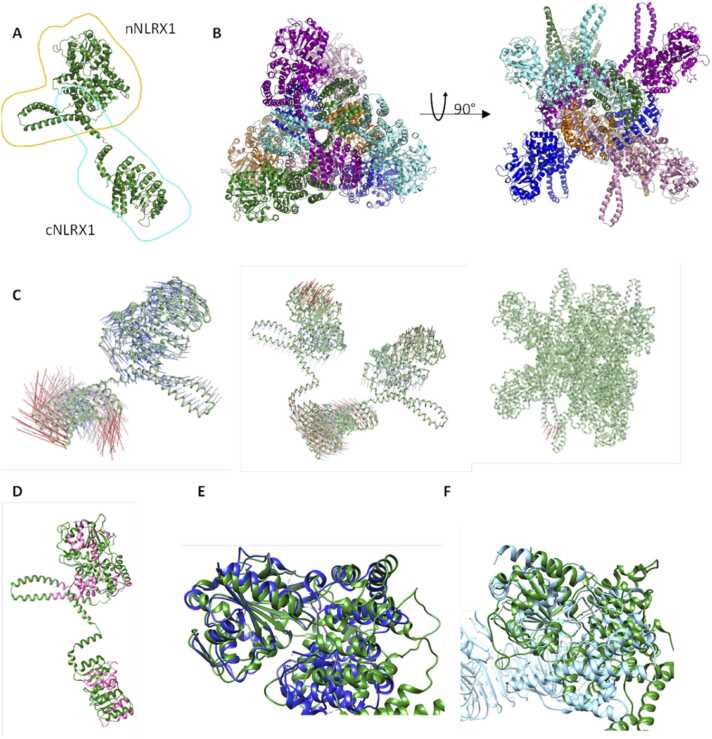
cNLRX1 (residues 629–975), comprising the LRR domain, exists primarily as a monomer and homo-hexamer by size exclusion chromatography and analytical ultracentrifugation [13]. In the hexameric state, NLRX1 exists as a dimer of trimers. The first 76 amino acid residues of NLRX1 are responsible for mitochondrial localization, making NLRX1 unique amongst the NLR family [2], [18], [19]. This flexible region is cleaved upon arrival at the mitochondria and lacks secondary structure [2] therefore the first 76 amino acid residues were excluded from our model of NLRX1. We successfully modelled the remaining 899 amino acid residues of NLRX1 as a monomer (Fig. 1A), dimer (Fig. 1B, chain A and F), and hexamer (Fig. 1B, chains A-F).
Fig. 1.
Structure of NLRX1. (A) Monomer of NLRX1, where cyan is cNLRX1, based on the crystal structure 3un9 and orange is nNLRX1, based on the AlphaFold model of NLRX1. (B) Top and side view of the hexamer model of NLRX1, where chains are coloured: A green, B aqua, C purple, D pink, E blue and F orange. (C) Dynamut1 output for NMA analysis of the monomer, dimer, and hexamer of NLRX1. (D) MTR output for NLRX1 were residues that were scored higher than 1 are coloured pink. (E) nNLRX1 (green) overlay with the NACHT domain of NLRP3 7vtq (blue). (F) nNLRX1 (green) overlay with the NACHT domain of NLRC4 4kxf (light blue).
Although NLRX1 has low overall sequence identity (less than 30 %) to other NLR proteins, there is a high degree of structural homology within specific regions, particularly for murine NLRP3 (PDBcode: 7vtq [20]), human NLRP3 (PDBcode: 7pzc [21]), murine NLRC4 (PDBcode: 4kxf [22] and 3jbl [23]) and baculovirus IAP repeat containing protein 1E (PDBcode: 5yud [24]). A high degree of structural homology is specifically present in the LRR domain, which was expected based on their familial tie (Table 1). The Cα RMSD values of the LRR domain between NLRX1 and aforementioned NLR proteins range from 1.9 to 4.6 Å where NLRP3 is the most similar.
Table 2.
ATP binding site analysis of the Cα RMSD (Å) of p-loop between nNLRX1 and other available structures.
| Protein name (PDBcode) | p-loop (166-173) | Sequence identity (%) |
|---|---|---|
| murine NLRP3 (7vtq) | 0.4 | 25.4 |
| human NLRP3 (7pzc) | 0.5 | 24.4 |
| mouse NLRC4 (4kxf) | 0.4 | 21.6 |
| Baculoviral IAP repeat-containing protein 1e (5yud) | 0.7 | 21.9 |
| Scytonema hofmannii TnsC (7plh) | 0.6 | 24.5 |
| cell division control protein 6 (2qby) | 0.5 | 19.5 |
| Mycobacterium tuberculosis proteasomal ATPase Mpa (5kzf) | 1.1 | 22.0 |
| human mitochondrial small subunit assembly intermediate (8csp) | 0.4 | 22.8 |
| 26S proteasome WT-Ubp6-UcVS complex (7qo4) | 0.9 | 23.5 |
| Mycobacterium smegmatis 0858 (5e7p) | 0.3 | 24.5 |
| wheat coiled-coil domain containing NLR (CNL) Sr35 (7xc2) | 0.3 | 21.8 |
| Drosophila origin recognition complex subunit 2(7jgr) | 0.6 | 23.4 |
Table 1.
Cα RMSD (Å) between NLRX1 (our model) and available NLR proteins.
| Protein name (PDBcode) | LRR domain (695-899) |
NACTH domain (160-483) |
Sequence identity (%) |
|---|---|---|---|
| NLR proteins | |||
| murine NLRP3 (7vtq) | 1.9 | 2.4 | 25.4 |
| human NLRP3 (7pzc) | 2.7 | 3.1 | 24.4 |
| mouse NLRC4 (4kxf) | 4.6 | 4.8 | 21.6 |
| mouse NLRC4 (3jbl) | 3.1 | 5.4 | 21.6 |
| Baculoviral IAP repeat-containing protein 1e (5yud) | 3.9 | 4.5 | 21.9 |
nNLRX1 and the NACHT domain of murine NLRP3, human NLRP3, Baculoviral IAP repeat-containing protein 1e (PDBcode: 5yud [24]), and mouse NLRC4 have high structural homology (Fig. 1E, F) [20], [22] Cα RMSD ranging from 2.4 to 5.4 Å.
Normal mode analysis was used to examine the relative stability of the monomer, dimer and hexamer models of NLRX1. The resulting porcupine plots indicated much greater movement in the monomer and dimer compared to the hexamer (Fig. 1C). This is not surprising, given that the hexamer inherently restricts itself from excessive fluctuation, restricting the freedom of movement of individual monomeric units. These findings, coupled with the oligomerization state experimentally observed for cNLRX1 indicate that NLRX1 likely functions as a hexamer [13].
2.2. Mutational tolerance analysis
Missense tolerance ratio (MTR) scores indicate how well a missense mutation is tolerated. Neutral mutations are given a score of 1, while a score greater than 1 indicates positive selection and less than 1 indicates that there is purifying selection against the mutation [25]. Analysis of the MTR scores indicated that NLRX1 does not have any disease mutations associated with it, with little evidence of purifying selection (Fig. 1D) [26]. Consurf results showed consistency with the MTR prediction where conserved regions have more cases with low MTR scores (less than 0.8). There were 50 residues in the variable region (Consurf colour scale 1 and 2), among which only one had MTR less than 0.8. There were 512 residues in the average region (Consurf colour scale from 3 to 7), among which 11 residues had MTR less than 0.8. There were 337 residues in the conserved region (Consurf colour scale from 8 to 9), where 19 residues had MTR less than 0.8. Notably, sections of the LRR domain involved in small molecule recognition were highly conserved as well as sites within the proposed ATP binding domain.
2.3. Interactions of NLRX1 with ATP
NLRX1 was proposed to contain an ATP binding site in the N-terminal region (GLY166-SER173) [19], however, due to the lack of higher order structural data, the ATP binding site of NLRX1 has not previously been investigated. On examining the structure of nNLRX1 the phosphate-binding loop (p-loop, or Walker A motif, residues 166 to 173), commonly observed in ATP binding proteins, was identified [18]. Furthermore, this was conserved across each of the homologous proteins described herein.
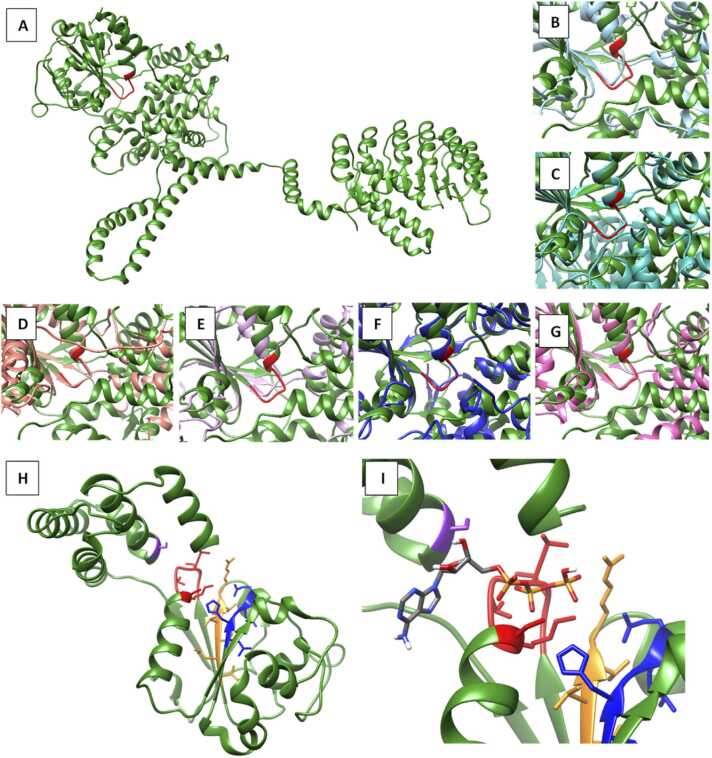
We sought to determine if the ATP binding site in NLRX1 was functional and if it resembled any other known ATP binding sites. The structurally homologous analysis from DALI indicated that there were twelve ATP interacting proteins with conserved p-loop, where NLRX1 had similar structure to murine NLRC4, murine NLRP3, baculovirus IAP repeat containing protein 1E and Scytonema hofmannii TnsC, cell division control protein 6 (PDBcode: 2qby [27]), human mitochondrial small subunit assembly intermediate (PDBcode: 8csp [28]), 26S proteasome WT-Ubp6-UcVS complex (PDBcode: 7qo4 [29]), Mycobacterium smegmatis 0858 (PDBcode: 5e7p [30]), wheat coiled-coil domain containing NLR (CNL) Sr35 (PDBcode: 7xc2 [31]) and Drosophila origin recognition complex bound (PDBcode: 7jgr [32]). Superimposing the solved structures of the ATP binding proteins with our model of NLRX1 showed a great deal of structural similarity with the Cα RMSD of the p-loop between nNLRX1 and other available structures ranging from 0.3 to 0.9 Å, suggesting that the site could act as a functional ATP binding site. Further, of those ATP binding proteins, nNLRX1 was found to have structural similarity to members of the AAA+ ATPase family (PDBcode: 5kzf, 7qo4, 5e7p, 7jgr and 7vtq) (Fig. 2B-G).
Fig. 2.
Investigation of the NLRX1 ATP binding site. (A) NLRX1 (A chain) is green, and the ATP binding site is highlighted in red. (B) Overlay of NLRX1 with 5kzf chain A in light blue. (C) Overlay of NLRX1 with 5e7p chain A in aqua. (D) Overlay of NLRX1 with 7jgr chain A in peach. (E) Overlay of NLRX1 with 7qo4 chain I in lilac. (F) Overlay of NLRX1 with 7vtq chain A in blue (G) Overlay of NLRX1 with 2qby chain A in pink. (H) ATP domain of NLRX1 (SER147-PHE406) where the Walker A motif is coloured red, the Walker B motif is coloured blue, Sensor 1 is coloured orange and SER372 is coloured purple. (I) AutoDock Vina output for NLRX1 interaction with ATP.
Having established the structural homology of nNLRX1 with ATP binding proteins, specifically the AAA+ ATPase family, we proposed that interaction is possible between ATP and NLRX1. Docking was performed to identify the location of ATP in the ATP binding site (Fig. 2I). The output indicated that the site could accommodate ATP; ATP was orientated in the binding pocket in a similar manner to ADP in the structure of murine NLRP3.
2.4. Interaction of NLRX1 with RNA
As a regulator of natural immunity, NLRX1′s interactions with RNA are thought to be pivotal to its function. There are no published models or available structures of NLRX1-RNA interactions and no proteins which are homologous to NLRX1 with a crystal structure containing RNA, we therefore progressed with docking RNA into NLRX1 [33], [34]. Literature suggests that NLRX1 has superior affinity for dsRNA (Kd 0.1 – 0.2 μM) compared to ssRNA [13]. The initial studies showed that dsRNA docking scores were significantly higher compared to that of the ssRNA, correlating well with the suggestion from the literature. All subsequent experiments were conducted with dsRNA.
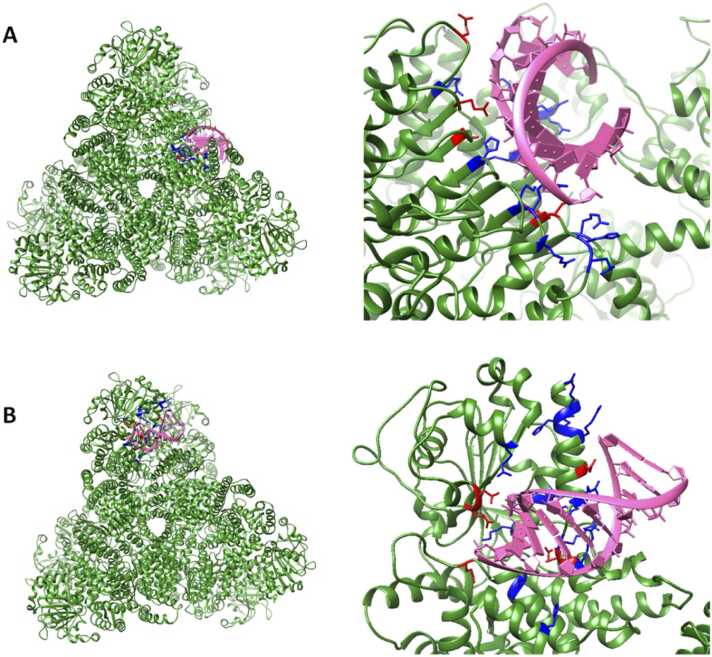
Hong et al. showed that residue ARG699 was key in protein-RNA interactions [13]; we therefore used this as a starting point for modelling the protein-RNA interactions. Our model was built to facilitate an interaction between dsRNA and ARG699, ensuring that the N-terminus could not block this region and fill the space that dsRNA would occupy. We conducted our studies with NLRX1 in the monomer, dimer, and hexamer states to determine if oligomerization had any implications on RNA interaction. The monomer did not exhibit a particular affinity for the dsRNA at position ARG699. In fact, we saw greater affinity for position ARG771 which has previously been shown to be shielded from RNA interactions [13]. The monomer does not shield itself from this interaction. When docking to the dimer (chains A and F), we faced similar issues, where the RNA sequence was found to preferentially bind at ARG771. It was not until we modelled the interaction with a dimer of chains D and F, which make up two parts of the trimer interface, that we saw the formation of favourable interactions between the experimentally validated position ARG699 and dsRNA. When modelling the interaction of dsRNA with the hexamer we observed dsRNA preferentially harbouring an interaction with ARG699, attaining docking scores of −302.50 (Fig. 3A). In addition to ARG699, we suspected that there may be additional binding sites in the previously unexplored N-terminus. When modelling RNA interaction with the hexamer, we identified a second possible RNA binding site that formed around the central helices located between ARG84-GLN99. This interaction was also observed in the monomer and dimer, though it was observed to be favoured in the hexamer, attaining a docking score of −253.84 (Fig. 3B). Several positively charged residues ARG81, HIS82, ARG83, ARG84, ARG92, ARG95, ARG98, ARG177, LYS178, ARG219, ARG403, and HIS634 facilitate the interaction via a positively charged binding pocket.
Fig. 3.
Modelling the interactions of dsRNA with NLRX1. Positively charged residues (LYS, ARG, and HIS) are coloured blue, while negatively charged residues (ASP, and GLU) are coloured red. (A) The interaction of dsRNA sequence with the previously identified binding site of NLRX1, where ARG699 is significant in protein-RNA interaction. (B) A novel RNA binding site located in the N terminal of NLRX1, where a positively charged pocket facilitates the protein-RNA interaction.
2.5. Predicting binding pockets in NLRX1 and docking of small molecule agonists
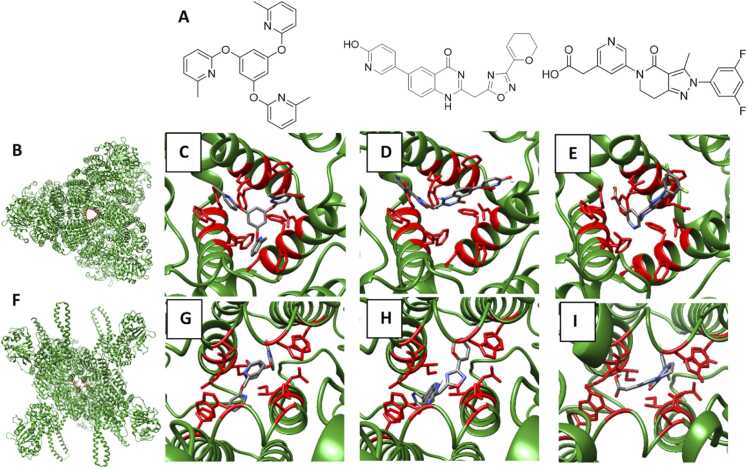
Given the therapeutic potential of NLRX1, we determined all, potentially druggable, small molecule binding sites. GHECOM output predicted six binding sites in nNLRX1 and three binding sites in cNLRX1. We investigated and prioritized these sites based on their affinity for patented small molecule NLRX1 agonists. There are only three published patents to date covering small molecule NLRX1 agonists and these are all from Landos Biopharma, recently acquired by AbbVie. The three patents claim three distinct molecular series where NX-13, NX-64–3, and LABP-72–38 are representative and are the compounds with the most supporting biological data in the patents. NX-13 is the most advanced compound in development having successfully completed two clinical trials (NCT04458805, NCT04862741). The first clinical trial investigated safety, tolerability and pharmacokinetics of NX-13 using oral dosing, while the second trial investigated treatment of active ulcerative colitis [15], [35], [36], [37]. A phase 2 clinical trial (NCT05785715) is now recruiting patients to assess NX-13 for efficacy in treatment of moderate to severe ulcerative colitis. NX-64–3 [17] and LABP-72–38 [16] are pre-clinical but have been proposed as potential therapeutics for inflammatory conditions ranging from asthma to neurodegenerative diseases such as Parkinson’s disease and Alzheimer’s disease [38], [39], [40].
The GHECOM output showed a binding site at the NLRX1 trimer interface. This is consistent with a previously published modelling study using cNLRX1 (PDBcode: 3un9) where this NLRX1 site associates with lipids docosahexaenoic acid (DHA), eleostearic acid (ESA) and punicic acid (PUA) [41]. Lu et al. determined the residues ASP677, PHE680, PHE681 and GLU684 were important for ligand recognition (Fig. 4B) [41]. The C3 symmetry of NX-13 suggests that it may interact with NLRX1 at the trimer interface, forming π-π interactions with the PHE residues. We docked NX-13 to the trimer interface of our NLRX1 model in its hexamer state; we report an affinity of −11.54 kcal/mol at this site (Fig. 4C). NX-64–3 lacks the C3 symmetry of NX-13, however docking score indicates a similar binding affinity of −11.05 kcal/mol, as its aromatic rings form favourable interactions with the PHE residues (Fig. 4D). Finally, LABP-72–38 had the lowest affinity for the trimer binding site, at −7.09 kcal/mol (Fig. 4E). Despite the presence of aromatic rings that could engage in π-π stacking with the PHE residues, as we observed with the other small molecules, the relative planar structure of the molecule prevents it from filling the space of the trimer binding site, resulting in a relatively low binding affinity at this site.
Fig. 4.
Interaction of NLRX1 with targeted ligands developed by Landos Biopharma. (A) Small molecule activators of NLRX1: NX-13, NX-64–3, and LABP-72–38. (B) Trimer binding site were the amino acids involved in protein-ligand interaction are highlighted in red. (C) Interaction of NX-13 with the trimer interface of NLRX1. (D) Interaction of NX-64–3 with the trimer interface of NLRX1. (E) Interaction of LABP-72–38 with the trimer interface of NLRX1. (F) The dimer interface of NLRX1 where the residues highlighted in red are those involved in protein-ligand interactions. (G) NX-13 interacting with the dimer interface of NLRX1. (H) NX-64–3 interacting with the dimer interface of NLRX1. (I) LABP-72–38 interacting with the dimer interface of NLRX1.
The output from GHECOM also suggests a binding site in cNLRX1 located at the dimer interface of chains A and F, composed of the two sets of α-helix bundles located at the C-terminus of NLRX1. These α-helix bundles do not have a known functionality but were proposed to play a sensory role [13]. In our docking study, residues ILE913, GLU916, ASN920, TRP924 and HIS932 are important for the protein-ligand interaction (Fig. 4F); interestingly, these residues fall within a sequence that is highly conserved (LYS870–ARG926), as determined by MTR. When we docked NX-13 at this dimer site, we found that affinity was similar to that of the trimer site but this time reporting −11.07 kcal/mol (Fig. 4G). NX-64–3 had a slightly higher affinity for the dimer site compared to the trimer site, at −11.85 kcal/mol (Fig. 4H). LABP-72–38 had much improved binding for the dimer site, compared to the trimer, as we report −11.06 kcal/mol as the binding affinity (Fig. 4I).
We also performed a blind docking with the known NLRX1 agonists, encompassing the entirety of the nNLRX1 to identify if there was any hierarchy to the sites that had been identified by GHECOM. Results showed that NX-13, NX-64–3 and LABP-72–38 docked exclusively within the ATP binding site. We then focused on each of the sites that had been proposed by GHECOM; low binding affinities were reported for each of the binding sites for all ligands. As such, it is our interpretation that there are no ligand binding domains located in the N-terminal end capable of interacting with the small molecules investigated in this work.
3. Discussion
For the first time, we present a model of the full-length structure of NLRX1, excluding only the first 76 amino acid residues, known to be associated with mitochondrial localization [2]. The homology model of NLRX1 is the product of the crystal structure of cNLRX1 and the AlphaFold output of nNLRX1 [13]. NMA analysis indicates that the protein has the highest level of stability in the hexamer state, which is supported by previous literature where 3un9 was proposed as a dimer of trimers [13]. MTR analysis showed NLRX1 did not have any disease mutations associated with the protein, though it did indicate regions of NLRX1 that are highly conserved and likely have significance, including the dimer binding site and the ATP binding domain.
NLR proteins are part of the AAA+ ATPase protein super family and typically oligomerise [42], [43]. In agreement with this, our results show that NLRX1 forms a, relatively stable, centrosymmetric hexamer. Through careful examination of NLRX1 including oligomerisation, ATP binding site, and comparison to closely homologous proteins, we conclude that NLRX1 is a AAA+ ATPase, consistent with other NLRs [43]. The ATP domain of AAA+ ATPase is usually 200–250 amino acids in length and commonly comprises two sub domains, where the C-terminal sub domain has numerous α-helixes, forming a lid over the N-terminal end. The N-terminus has a characteristic β-sheet arrangement of β5-β1-β4-β3-β2, with an α-helixes connecting to each β-sheet [44]. NLRX1 ATP domain (residues SER147-PHE406) retains these characteristic features of AAA+ ATPases in each domain. In the C-terminal sub domain, there are five α-helixes while the N-terminal sub domain has a narrow triangular shape, and the β-sheet pattern is exactly consistent with AAA+ ATPase (Fig. 2H).
The presence of both Walker A and B motifs are critical to the functionality of AAA+ ATPases. The Walker A motif, involved in γ-phosphate coordination, is commonly denoted by the sequence GXXXXGK(S/T), where X is any amino acid, and this presents in the ATP domain of NLRX1 (GTVGTGKS) [42], [44]. Further, the orientation of the docked ATP molecule (Fig. 2I) supports a γ-phosphate coordination to the Walker A site. The Walker B motif, however, has a sequence of hhhh(D/E), where h are hydrophobic residues, and is responsible for the coordination of Mg2+ involved in ATP hydrolysis. In NLRX1 we find LHGLE as the sequence of this region, which does not conform to the conventional definition of the Walker B motif because of the hydrophilic residue HIS242. However, histidine was previously found to be critical in coordination of Mg2+ in ATP hydrolysis and could, in this instance, be performing a similar function [45].
In addition to the Walker motifs, AAA+ ATPases have two sensor regions: Sensor 1 is characterized by the sequence hhh(T/S)(T/S)R and is present in NLRX1 (ILVTTR). Additionally, this sequence is highly conserved, as determined in our MTR analysis. Sensor 2, involved in ATP coordination and hydrolysis, is commonly denoted by a single lysine or arginine residue but in NLRX1, the residue that maps to that position is SER372. This does not exclude NLRX1 from the AAA+ ATPase family, as several NLRP proteins, for example: NLRP4, 6, 8, 11, 13 and 14, also lack this residue [44]. Instead, another region of NLRX1 may function as an alternative Sensor 2 motif such as ASP472, GLU409 or GLU96, all of which fall outside of the ATP binding site domain.
The identification of an ATP binding site and the potential for ATPase activity as a member of the AAA+ ATPase family opens an array of potential functions for NLRX1. Though we are not able to present any further answers here, it is possible and has been previously speculated that ATP could facilitate the oligomerization processes in a similar manner to oligomerisation of NLRP3 where ATP binding correlates with large conformational changes triggering an active state of the sensory protein [43], [46].
NLRX1 is known for its recognition of dsRNA, where interaction stimulates reactive oxygen species activation [13], [47]. Hong et al. previously proposed a positively charged patch in cNLRX1 for RNA recognition [13]. Our data supports the location of this RNA binding site and further suggests that NLRX1 binds RNA when in its hexameric form and that the trimer interface is critical for RNA recognition. The ability of NLRX1 to preferentially bind dsRNA supports the former conclusion that NLRX1 outcompetes protein kinase R for dsRNA interaction, resulting in early innate immune antiviral responses [48]. Moreover, we also identified a positively charged binding pocket in the previously unexplored nNLRX1 capable of interacting with RNA. Our data suggests preferential binding of dsRNA at this site which is not dependent on oligomerization state of the protein.
In order to facilitate drug discovery efforts, it is important to understand if there are any additional binding pockets, beyond the ATP and RNA binding sites. From the GHECOM output there were 9 small molecule binding sites predicted. One of these sites is at the centre of the trimer while a second new site was found at the dimer interface. We succeeded in validating these sites using the available ligands from Landos Biopharma. NX-13 is likely to bind to the trimer site but may also bind the dimer interface. NX-64–3 and LABP-72–38 are more likely to bind at the newly established dimer site. Landos Biopharma suggest in each of their patents that there is a second binding site in the C-terminus, however the location of this site was not reported.
With use of state-of-the-art modelling techniques, we have constructed a detailed representation of the structure of NLRX1 in various oligomerization states and conclude that a hexameric active form is most likely. Not surprisingly, NLRX1 was found to have high structural homology with several members of the larger NLR family, despite less than 30 % sequence identity. Mutational tolerance analysis showed no strong evidence of disease mutations associated with NLRX1 but provided insights into regions of the protein that have high levels of conserved sequences. Our model has allowed us to define NLRX1 as a AAA+ ATPase through confirming and examining the ATP binding site. We have provided the first model of interactions with dsRNA showing the most likely points of interaction. Finally, we have added structural validation for the three series of currently disclosed NLRX1 targeted small molecules. There is a great deal of work that is still required to understand the function and intricacies that surround NLRX1. We hope that this paper may serve as a catalyst to encourage others to pursue and apply their own knowledge to advance the field for therapeutic benefit.
4. Materials and methods
4.1. Modelling the structure of NLRX1
The model of NLRX1 was built in MODELLER v10.2 [49] using cNLRX1 (PDBcode: 3un9 [13]), mouse NLRP3 (PDBcode: 7vtq [20] - 25.4 % sequence identity) and the apo monomeric AlphaFold2 model [50] as templates.
Ten models were generated, extensively minimised and the model with lowest Discrete Optimized Protein Energy (DOPE) score [51] chosen. The experimental structure of the C-terminus fragment of NLRX1 suggested that it forms a homo-hexamer [13]; the full-length hexamer model was generated using cNLRX1 (PDBcode: 3un9) as a guide for the core of full-length NLRX1 protein (excluding residue 1 to 76).
4.2. Relative stability of NLRX1
To determine the relative stability of NLRX1, we performed normal mode analysis (NMA) using Dynamut [52]. Porcupine plots were generated to illustrate the movement in the protein.
4.3. Mutational tolerance analysis[13]
Missense Tolerance Ratio (MTR) score - a measure to quantify the amount of purifying selection on missense variation was calculated using MTR-Viewer [26].
4.4. Structural homology search
DALI [53] was used to determine structural homology to experimentally characterised proteins.
4.5. Conservation calculation
Consurf [54], [55] was performed to calculate the conservation of NLRX1 protein. All the parameters were used as default values.
4.6. Binding site prediction
GHECOM [38] was used to predict potential binding pockets in NLRX1.
4.7. Docking
4.7.1. NLRX1 and RNA
HDOCK [33], [56] was used to perform the docking between NLRX1 and RNA. The web server version was used with default parameters and an intrinsic scoring function [34]. Initially, we conducted the docking with both double-stranded (ds) and single-stranded (ss) RNA to see which one has better binding affinity to NLRX1. All subsequent experiments were conducted with dsRNA (GGCGCGCGCC) which was obtained from its complex with dsRNA-binding domain (PDBcode: 1DI2 [57].
4.7.2. NLRX1 and small molecules
AutoDock Vina [58] was used to determine the interaction between NLRX1 and small molecules, such as ATP and NLRX1 activators. In the AutoDock configuration file, the exhaustiveness parameter was set to 24. Ten poses were generated in the output and the pose with lowest binding affinity was chosen. The box size was 20 Å * 20 Å * 20 Å. When investigating the ATP binding stie the centroid of the box was defined by the residues at the interaction site of the mouse NLRP3 (PDBcode: 7vqt). When investigating the trimer binding site, the centroid of the box was definied by the residues ASP677, PHE680, PHE681, and GLU684 [41]. When investigating the identified dimer binding site, the centroid of the box was defined by the residues at the interaction sites identified by GHECOM.
4.8. Figure generation
All protein structure figures were generated in Pymol [The PyMOL Molecular Graphics System, Version 1.2r3pre, Schrödinger, LLC.].
Funding
SJ is supported by a University of Queensland Research Training PhD scholarship funded by The University of Queensland. TBN and DBA were supported by the Investigator Grant from the National Health and Medical Research Council (NHMRC) of Australia (GNT1174405 to DBA). Supported in part by the Victorian Government's Operational Infrastructure Support Program.
CRediT authorship contribution statement
Shannon Jewell: Writing – review & editing, Writing – original draft, Visualization, Validation, Investigation, Formal analysis, Data curation. Thanh Binh Nguyen: Writing – review & editing, Supervision, Methodology, Investigation, Formal analysis. David Ascher: Writing – review & editing, Supervision, Software, Project administration, Conceptualization. Avril Alexis Barbara Robertson: Writing – review & editing, Writing – original draft, Supervision, Resources, Methodology, Funding acquisition, Conceptualization.
Declaration of Competing Interest
AABR is inventor of intellectual property (WO2016131098, WO2017140778, WO2018215818), granted in multiple jurisdictions, which discloses NLRP3 inhibitors for treatment of inflammatory diseases. These patents are licensed to Inflazome Ltd., which sold to Roche in 2020, and the compounds disclosed are progressing through clinical trials. AABR has no IP associated with NLRX1 which is the subject of this publication. SJ, TBN, DBA have no relevant financial or non-financial interests to disclose.
Acknowledgements
None.
Contributor Information
David B. Ascher, Email: d.ascher@uq.edu.au.
Avril A.B. Robertson, Email: a.robertson3@uq.edu.au.
References
- 1.Mangan M.S.J., et al. Targeting the NLRP3 inflammasome in inflammatory diseases. Nat Rev Drug Discov. 2018;17(9):588. doi: 10.1038/nrd.2018.97. [DOI] [PubMed] [Google Scholar]
- 2.Arnoult D., et al. An N-terminal addressing sequence targets NLRX1 to the mitochondrial matrix. J Cell Sci. 2009;122(17):3161–3168. doi: 10.1242/jcs.051193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pickering R.J., Booty L.M. NLR in eXile: emerging roles of NLRX1 in immunity and human disease. Immunology. 2021;162(3):268. doi: 10.1111/imm.13291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leber A., et al. NLRX1 modulates immunometabolic mechanisms controlling the host-gut microbiota interactions during inflammatory bowel disease. Front Immunol. 2018;9:363. doi: 10.3389/fimmu.2018.00363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu B., et al. NOD-like receptor X1 functions as a tumor suppressor by inhibiting epithelial-mesenchymal transition and inducing aging in hepatocellular carcinoma cells. J Hematol Oncol. 2018;11(1):28. doi: 10.1186/s13045-018-0573-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Koblansky A.A., et al. The innate immune receptor NLRX1 functions as a tumor suppressor by reducing colon tumorigenesis and key tumor-promoting signals. Cell Rep. 2016;14(11):2562. doi: 10.1016/j.celrep.2016.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh K., et al. NLRX1 regulates TNF-alpha-induced mitochondria-lysosomal crosstalk to maintain the invasive and metastatic potential of breast cancer cells. Biochim Biophys Acta Mol Basis Dis. 2019;1865(6):1460. doi: 10.1016/j.bbadis.2019.02.018. [DOI] [PubMed] [Google Scholar]
- 8.Chen L., et al. Inflammatory responses and inflammation-associated diseases in organs. Oncotarget. 2018;9(6):7204. doi: 10.18632/oncotarget.23208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jewell S., Herath A.M., Gordon R. Inflammasome activation in Parkinson’s disease. J Park Dis. 2022;12(s1):S113. doi: 10.3233/JPD-223338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu T., et al. NF-kappaB signaling in inflammation. Sig Transduct Target Ther. 2017;2:17023. doi: 10.1038/sigtrans.2017.23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xia X., et al. NLRX1 negatively regulates TLR-induced NF-kappaB signaling by targeting TRAF6 and IKK. Immunity. 2011;34(6):843. doi: 10.1016/j.immuni.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Allen I.C., et al. NLRX1 protein attenuates inflammatory responses to infection by interfering with the RIG-I-MAVS and TRAF6-NF-kappaB signaling pathways. Immunity. 2011;34(6):854. doi: 10.1016/j.immuni.2011.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hong M., Yoon S.I., Wilson I.A. Structure and functional characterization of the RNA-binding element of the NLRX1 innate immune modulator. Immunity. 2012;36(3):337. doi: 10.1016/j.immuni.2011.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nagai-Singer M.A., Morrison H.A., Allen I.C. NLRX1 is a multifaceted and enigmatic regulator of immune system function. Front Immunol. 2019;10:2419. doi: 10.3389/fimmu.2019.02419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bassaganya-Riera J., Leber A., Hontecillas R. NLRX1 Ligands. Landos Biopharma, US10,487,057B1, 2019.
- 16.Bassaganya-Riera J., Hontecillas R., Tubau-Juni N., Leber A. Tetrahydropyrazolopyridine-analog ligands of NLRX1 and uses thereof. Landos Biopharma, WO2023018682A1, 2023.
- 17.Bassaganya-Riera J., Leber A., Tubau-Juni N., Hontecillas R. NLRX1 Ligands. Landos Biopharma, WO2022061224A1, 2022.
- 18.Moore C.B., et al. NLRX1 is a regulator of mitochondrial antiviral immunity. Nature. 2008;451(7178):573. doi: 10.1038/nature06501. [DOI] [PubMed] [Google Scholar]
- 19.Tattoli I., et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9(3):293. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ohto U., et al. Structural basis for the oligomerization-mediated regulation of NLRP3 inflammasome activation. Proc Natl Acad Sci USA. 2022;119(11) doi: 10.1073/pnas.2121353119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hochheiser I.V., et al. Structure of the NLRP3 decamer bound to the cytokine release inhibitor CRID3. Nature. 2022;604(7904):184. doi: 10.1038/s41586-022-04467-w. [DOI] [PubMed] [Google Scholar]
- 22.Hu Z., et al. Crystal structure of NLRC4 reveals its autoinhibition mechanism. Science. 2013;341(6142):172. doi: 10.1126/science.1236381. [DOI] [PubMed] [Google Scholar]
- 23.Zhang L., et al. Cryo-EM structure of the activated NAIP2-NLRC4 inflammasome reveals nucleated polymerization. Science. 2015;350(6259):404. doi: 10.1126/science.aac5789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yang X., et al. Structural basis for specific flagellin recognition by the NLR protein NAIP5. Cell Res. 2018;28(1):35. doi: 10.1038/cr.2017.148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G.C., Forster-Benson E.T.C., Sanders C.R. Genetic intolerance analysis as a tool for protein science. Biochim Biophys Acta Biomembr. 2020;1862(1) doi: 10.1016/j.bbamem.2019.183058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Silk M., Petrovski S., Ascher D.B. MTR-Viewer: Identifying regions within genes under purifying selection. Nucleic Acids Res. 2019;47(W1) doi: 10.1093/nar/gkz457. W121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dueber E.L., et al. Replication origin recognition and deformation by a heterodimeric archaeal Orc1 complex. Science. 2007;317(5842):1210. doi: 10.1126/science.1143690. [DOI] [PubMed] [Google Scholar]
- 28.Harper N.J., Burnside C., Klinge S. Principles of mitoribosomal small subunit assembly in eukaryotes. Nature. 2023;614(7946):175. doi: 10.1038/s41586-022-05621-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hung K.Y.S., et al. Allosteric control of Ubp6 and the proteasome via a bidirectional switch. Nat Commun. 2022;13:838. doi: 10.1038/s41467-022-28186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Unciuleac M.C., Smith P.C., Shuman S. Crystal structure and biochemical characterization of a Mycobacterium smegmatis AAA-Type nucleoside triphosphatase phosphohydrolase (Msm0858) J Bacteriol. 2016;198(10):1521. doi: 10.1128/JB.00905-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Forderer A., et al. A wheat resistosome defines common principles of immune receptor channels. Nature. 2022;610(7932):532. doi: 10.1038/s41586-022-05231-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt J.M., Bleichert F. Structural mechanism for replication origin binding and remodeling by a metazoan origin recognition complex and its co-loader Cdc6. Nat Commun. 2020;11(1):4263. doi: 10.1038/s41467-020-18067-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li H., et al. HDOCK update for modeling protein-RNA/DNA complex structures. Protein Sci. 2022;31(11) doi: 10.1002/pro.4441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huang S.Y., Zou X. A knowledge-based scoring function for protein-RNA interactions derived from a statistical mechanics-based iterative method. Nucleic Acids Res. 2014;42(7) doi: 10.1093/nar/gku077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Leber A., et al. Activation of NLRX1 by NX-13 alleviates inflammatory bowel disease through immunometabolic mechanisms in CD4(+) T cells. J Immunol. 2019;203(12):3407. doi: 10.4049/jimmunol.1900364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Leber A., et al. Exploratory studies with NX-13: Oral toxicity and pharmacokinetics in rodents of an orally active, gut-restricted first-in-class therapeutic for IBD that targets NLRX1. Drug Chem Toxicol. 2019;45(1):209. doi: 10.1080/01480545.2019.1679828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Verstockt B., et al. The safety, tolerability, pharmacokinetics, and clinical efficacy of the NLRX1 agonist NX-13 in active ulcerative colitis: results of a phase 1b study. J Crohns Colitis. 2024;18(5):762. doi: 10.1093/ecco-jcc/jjad192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kawabata T. Detection of cave pockets in large molecules: Spaces into which internal probes can enter, but external probes from outside cannot. Biophys Physicobiol. 2019;16:391. doi: 10.2142/biophysico.16.0_391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kawabata T. Detection of multiscale pockets on protein surfaces using mathematical morphology. Proteins. 2010;78(5):1195. doi: 10.1002/prot.22639. [DOI] [PubMed] [Google Scholar]
- 40.Kawabata T., Go N. Detection of pockets on protein surfaces using small and large probe spheres to find putative ligand binding sites. Proteins. 2007;68(2):516. doi: 10.1002/prot.21283. [DOI] [PubMed] [Google Scholar]
- 41.Lu P., et al. Modeling-enabled characterization of novel NLRX1 ligands. PLoS One. 2015;10(12) doi: 10.1371/journal.pone.0145420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanson P.I., Whiteheart S.W. AAA+ proteins: Have engine, will work. Nat Rev Mol Cell Biol. 2005;6(7):519. doi: 10.1038/nrm1684. [DOI] [PubMed] [Google Scholar]
- 43.Brinkschulte R., et al. ATP-binding and hydrolysis of human NLRP3. Commun Biol. 2022;5(1):1176. doi: 10.1038/s42003-022-04120-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.MacDonald J.A., et al. Biochemical and structural aspects of the ATP-binding domain in inflammasome-forming human NLRP proteins. IUBMB Life. 2013;65(10):851. doi: 10.1002/iub.1210. [DOI] [PubMed] [Google Scholar]
- 45.Rosler K.S., et al. Histidine 114 is critical for ATP hydrolysis by the universally conserved ATPase YchF. J Biol Chem. 2015;290(30):18650. doi: 10.1074/jbc.M114.598227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiao T.S., Ting J.P. NLRX1 has a tail to tell. Immunity. 2012;36(3):311. doi: 10.1016/j.immuni.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tattoli I., et al. NLRX1 is a mitochondrial NOD-like receptor that amplifies NF-kappaB and JNK pathways by inducing reactive oxygen species production. EMBO Rep. 2008;9(3):293. doi: 10.1038/sj.embor.7401161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Feng H., et al. NLRX1 promotes immediate IRF1-directed antiviral responses by limiting dsRNA-activated translational inhibition mediated by PKR. Nat Immunol. 2017;18(12):1299. doi: 10.1038/ni.3853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webb B., Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Bioinform. 2016;54:5.6.1. doi: 10.1002/cpbi.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jumper J., et al. Highly accurate protein structure prediction with AlphaFold. Nature. 2021;596(7873):583. doi: 10.1038/s41586-021-03819-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shen M.Y., Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci. 2006;15(11):2507. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rodrigues C.H., Pires D.E., Ascher D.B. DynaMut: Predicting the impact of mutations on protein conformation, flexibility and stability. Nucleic Acids Res. 2018;46(W1):W350. doi: 10.1093/nar/gky300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Holm L., et al. DALI shines a light on remote homologs: one hundred discoveries. Protein Sci. 2023;32(1):e4519. doi: 10.1002/pro.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ben Chorin A., et al. ConSurf-DB: An accessible repository for the evolutionary conservation patterns of the majority of PDB proteins. Protein Sci. 2020;29(1):258. doi: 10.1002/pro.3779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ashkenazy H., et al. ConSurf 2016: An improved methodology to estimate and visualize evolutionary conservation in macromolecules. Nucleic Acids Res. 2016;44(W1):W344. doi: 10.1093/nar/gkw408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yan Y., et al. The HDOCK server for integrated protein-protein docking. Nat Protoc. 2020;15(5):1829. doi: 10.1038/s41596-020-0312-x. [DOI] [PubMed] [Google Scholar]
- 57.Ryter J.M., Schultz S.C. Molecular basis of double-stranded RNA-protein interactions: structure of a dsRNA-binding domain complexed with dsRNA. EMBO J. 1998;17(24):7505. doi: 10.1093/emboj/17.24.7505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Trott O., Olson A.J. AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]