Abstract
The human immunodeficiency virus type 1 (HIV-1) and human T-cell leukemia virus type 1 (HTLV-1) capsid proteins (CA) display similar structures formed by two independently folded N-terminal (NTD) and C-terminal (CTD) domains. To characterize the functions harbored by the HTLV-1 CA domains in particle formation, 12 sites scattered throughout the protein were mutated. The effects of the mutations on Gag membrane binding, proteolytic processing, and virus-like particle secretion were analyzed. It appears that the NTD is the major partner of indirect or direct Gag-Gag interactions. In particular, most of the NTD mutations impaired virion morphogenesis, and no mutation located in the NTD could be fully rescued by coexpression of wild-type Gag. In contrast, the CTD seems not to be involved in Gag-Gag interactions. Nevertheless, an unknown function required for particle formation is located in the CTD. Thus, despite an overall structural similarity between the HIV-1 and HTLV-1 CA proteins, their NTDs and CTDs exhibit different functions.
Human T-cell leukemia virus type 1 (HTLV-1) is the etiologic agent of adult T-cell leukemia and a neuromyelopathy referred to as tropical spastic paraparesis/HTLV-1-associated myelopathy (14, 31, 34, 46). The HTLV-1 is a retrovirus that belongs to the HTLV-bovine leukemia virus group.
As with all retroviruses, the HTLV-1 gag gene encodes the major structural proteins of the virion core and the pro gene encodes the viral protease. The HTLV-1 gag-pro genes are expressed as two polyproteins, Pr53Gag and Pr76Gag-Pro precursors, respectively. Synthesis of the Pr76Gag-Pro precursor requires one ribosomal frameshift that occurs between the gag and pro reading frames (17, 28). The viral protease generated from the Pr76Gag-Pro cleaves the Gag precursors into mature proteins: the matrix protein (MA) p19, the capsid protein (CA) p24, and the nucleocapsid protein p15 (18, 28).
The HTLV-1 virion assembly pattern is defined by targeting, accumulation, and association of the different Gag precursors at the inner face of the plasma membrane. Then, budding out of the cell leads to the release of enveloped particles. During or just after budding, viral protease activity leads to the maturation of the Gag precursors. This morphogenesis process resembles that of both type C retroviruses and lentiviruses (for review, see references 13 and 38).
Studies done on HTLV-2, bovine leukemia virus, human immunodeficiency virus type 1 (HIV-1), Rous sarcoma virus (RSV), and murine leukemia virus (MuLV) have shown that the expression of the gag gene alone is sufficient for membrane targeting and binding of Gag proteins, and assembly, budding, and release of immature virus-like particles (VLPs). Simultaneous expression of the gag-pro genes leads to secretion of mature VLPs (15, 20, 23, 38, 39, 44). The process that leads to obtention of the VLPs mimics the natural particle formation process and provides a tool that has been used to identify Gag determinants that are important for HIV-1, MuLV, and RSV virion formation (for review, see references 9 and 43). For most but not all retroviruses, myristylation of the N-terminal glycine of the Gag proteins is one of the determinants required for effective membrane binding of Gag precursors (for review, see reference 38). For MuLV, myristylation and membrane binding are prerequisites for Gag-Gag interaction and assembly (36). In contrast, for HIV-1, nonmyristylated Gag-Pol proteins can interact with wild-type Gag proteins, allowing assembly and secretion of particles containing both wild-type Gag and nonmyristylated Gag-Pol proteins (32, 37). Thus, HIV-1 Gag proteins, but not MuLV Gag proteins, seem to be able to interact, either directly or indirectly, in the cytosol. Nevertheless, for both viruses, myristylation of some or all of the Gag proteins is required in order to target and allow Gag assembly at the membrane for particle release. For most retroviruses, a rule of thumb is that membrane binding of Gag precursors would be required for efficient Gag-Gag interactions, probably by increasing local Gag concentrations.
Regarding HTLV-1, the Pr53Gag determinants involved in virion morphogenesis are poorly understood because of (i) the lack, until recent years, of an infectious proviral molecular clone, (ii) the very low infectious potential of HTLV-1 virions (5, 48), and (iii) the fact that only two studies attempting to identify the functions of HTLV-1 Gag proteins have been published. The first study revealed that expression of the gag gene alone is sufficient for secretion of the HTLV-1 Gag protein precursor in yeast and that this precursor is myristylated (19). The second study reported the role of different amino acids of the HTLV-1 MA protein in a cell-to-cell infectivity system and, particularly, that the N-terminal glycine is required for membrane binding and infectivity (25).
The CA protein that contains the major homology region (MHR) of 20 highly conserved amino acids (33, 43) is the most conserved mature Gag protein in retroviruses. For HIV-1, the C-terminal third of the CA that includes the MHR is required for assembly and release of virus particles, but the N-terminal half, which is possibly involved in virus morphology, is not (2, 7, 30, 35, 40, 42, 47). Three-dimensional (3D) structural models of the N- and C-terminal parts and of the entire HIV-1 CA protein have been published (1, 10, 11, 16, 27). The lentivirus crystal structure of the equine infectious anemia virus (EIAV) CA has also been determined, and recently the solution structure of the HTLV-1 CA protein has been reported (22, 24). All of these CA structures appear to be very similar and fold into two independent domains, the N-terminal domain (NTD) and the C-terminal domain (CTD).
The present study elaborates the role of the HTLV-1 CA protein in particle formation. A comparison of the amino acid sequences of the HTLV-1 CA with that of HIV-1 reveals that they have a number of amino acids in common, some of which have been previously determined to be involved in HIV-1 particle formation (4, 21, 26, 35, 41). In order to determine if analogous HTLV-1 sites function similarly in HTLV-1 particle formation, some of these sites were mutated. The published experiments attempting to chararacterize the functions of the HIV-1 CA domains have introduced either small or large deletions, insertions, or substitutions into the Gag protein. In these experiments, some mutations have led to informative phenotypes, but numerous mutations have led to no obvious phenotypes, indicating that the Gag protein is highly mutation tolerant. For instance, comparison of conservative and nonconservative substitutions of the same residues revealed that only nonconservative mutations give rise to informative phenotypes (26). Moreover, neither large deletions (7, 12) nor insertions (35, 40) exert a drastic effect on particle release. It is evident that at least nonconservative substitutions must be introduced into the HTLV-1 CA in order to obtain informative phenotypes. Thus, nonconservative substitutions and insertions were introduced into 12 sites scattered throughout the sequence of the HTLV-1 CA. The mutated HTLV-1 gag or gag-pro genes were then expressed in 293T cells. Thus, the amino acids of the HTLV-1 CA protein involved in VLP formation were identified. In addition, two functional domains were characterized, corresponding to the N-terminal and the C-terminal structural domains. Their respective importance for particle formation was analyzed.
MATERIALS AND METHODS
Amino acid sequence and HTLV-1 CA structure.
The gag gene amino acid sequence was translated from the pMT-2 plasmid (originated from MT-2 cells [3]) sequenced in our laboratory. The 3D structure of the HTLV-1 CA protein (24) was obtained from the Protein Data Bank (no. 1QRJ) and the Molecular Modelling Data Base (no. 10943). The HTLV-1 CA 3D structure was viewed using the Cn3D 2.5 program.
Construction of mutated HTLV-1 gag and gag-pro genes.
The gag and gag-pro genes were inserted into the multiple cloning site of the pBSM13+, leading to plasmids pBSgag wild type (wt) and pBSgag-pro wt, respectively. Mutagenesis was performed on the pBSgag wt according to the manufacturer (Transformer; Clontech). Nucleotide numbering starts at the A of the gag ATG. Oligonucleotides used are described in Table 1. CA insertion mutations (CAi) were obtained by mutagenesis that simultaneously introduced mutated codons flanked by duplicated endonuclease restriction sites. Successive digestion of the duplicated site by the endonuclease and ligation of each pBSgag CAi mutant led to eight CA substitution mutants, CAs 1 to 5 and 10 to 12. The Myr0 substitution and four CAs substitutions, CAs 6, 7, and 9 and C63A, were obtained by single, regular mutagenesis. To obtain the gag-pro constructs with Myr0 or CA mutations, the BstXI-EcoRI (nucleotide 1163 to the 3′ multiple cloning site) pro fragment was transferred from pBSgag-pro wt to each mutated pBSgag plasmid.
TABLE 1.
Mutation name, sequence, and endonuclease restriction sites of oligonucleotides used for site-directed mutagenesis
 |
i, insertion; s, substitution.
Numbering starts at the A of the gag ATG initiator codon. Mutated sequences are in bold type. Introduced endonuclease restriction sites are underlined, and the restriction endonucleases used are indicated.
The entire gag and gag-pro mutated genes were transferred from pBSgag and pBSgag-pro to the expression vector pKCR3 (29), under the control of the simian virus 40 promoter. The resulting constructs were named pKgag and pKgag-pro followed by the mutant name.
Cell culture and transfections.
The 293T cells were cotransfected by the calcium phosphate precipitation technique with 2.5 μg of DNA constructs and 0.25 μg of the IIITatORF vector expressing HTLV-1 Tax and Rex proteins (6). For each transfection experiment, a β-Gal-expressing vector was used to control the transfection efficiency; this efficiency varied from 10 to 40%. For the complementation assays, cells were cotransfected with 1.25 μg of pKgag wt and 1.25 μg of mutated pKgag-pro vector.
Metabolic labeling of transfected cells and viral protein analysis.
Two days posttransfection, the transfected cells were labeled with 100 μCi of [35S]methionine (Promix; Amersham Pharmacia Biotech AB) per ml for 5 h. The cells were then lysed in radioimmunoprecipitation assay (RIPA) buffer (0.5% deoxycholate, 0.5% NP-40, 1 mM Na2HPO4, 1 mM EDTA, 10 mM Tris [pH 7.8]) containing 0.2 mM phenylmethylsulfonyl fluoride for 30 min at 4°C and centrifuged for 15 min at 9,000 × g at 4°C. The supernatant was adjusted to 0.1% sodium dodecyl sulfate (SDS) and was clarified again.
In order to analyze the released viral proteins, the medium was clarified (10 min at 9,000 × g) and then centrifuged for 2 h at 36,000 × g at 4°C through a 20% sucrose cushion in TNE buffer (10 mM Tris, 100 mM NaCl, 1 mM EDTA [pH 7.9]). The pelleted proteins (particulate material) were disrupted in RIPA buffer containing 0.1% SDS, and the supernatant (soluble proteins) was adjusted to 0.5% NP-40 and 0.1% SDS. Both samples were immunoprecipitated. Exceptionally, the particulate material was directly subjected to SDS-polyacrylamide gel electrophoresis (SDS-PAGE).
Subcellular fractionation.
Transfected 293T cells were lysed in hypotonic buffer (10 mM Tris [pH 7.4], 1 mM MgCl2, 0.2 mM phenylmethylsulfonyl fluoride) at 4°C for 15 min and broken using 30 strokes of a Dounce homogenizer type B pestle. Cell lysates were adjusted to 1 mM EDTA–0.15 M NaCl, and a low-speed centrifugation (1,000 × g) for 10 min led to a pellet containing intact cells, cell fragments, and nuclei and to a supernatant containing cytoplasm and cell membranes. Centrifugation at 100,000 × g for 30 min at 4°C led to supernatants with soluble proteins and to pellets with proteins associated with membranes.
Immunoprecipitation of viral proteins.
The radioactive cell lysate and proteins released were immunoprecipitated at 4°C for 16 h with antibodies coupled to protein A-Sepharose (Amersham Pharmacia Biotech AB). A pool of three mouse monoclonal antibodies was used (Valbiotech anti-HTLV-1 CA and MA and Advanced BioScience Laboratories anti-HTLV-1 CA). These antibodies also recognized the Pr53Gag and Pr76Gag-Pro precursors. Immunoprecipitated proteins were separated by SDS-PAGE. The dried gel was analyzed by fluorography or by using an Instant Imager (Packard).
Secretion of immature or mature VLPs.
The total amount of Pr53Gag was evaluated by adding the radioactivity of both the Pr53Gag and CA proteins. The percentage of secretion of the mutated proteins was calculated relative to both the quantity of mutated Pr53Gag precursors within the cell lysate and the quantity of wt Pr53Gag or CA secreted proteins according to the following equation: [(secreted Pr53Gag or CA mutants /total mutated Pr53Gag in cells and VLPs)/(wt secreted Pr53Gag or CA/total wt Pr53Gag in cells and VLPs)] × 100.
RESULTS
Alignment of HTLV-1 and HIV-1 CA proteins and construction of HTLV-1 CA mutants.
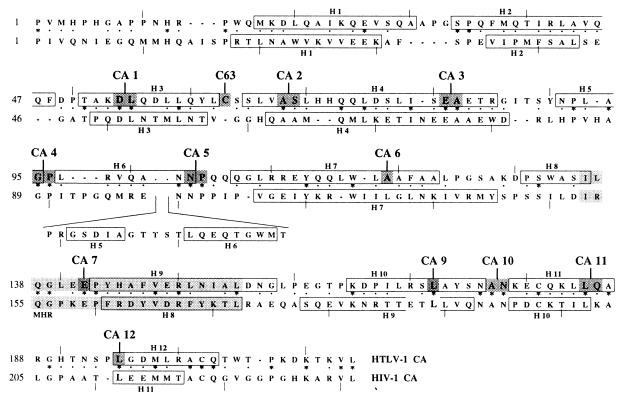
The amino acid sequence of the HTLV-1 CA protein was compared with that of the HIV-1 CA protein (Fig. 1). The two proteins had 24% identity and 43% similarity. The similarity level was homogeneously distributed throughout the CA sequence. In contrast, 19 and 31% identities were found, respectively, for the N-terminal half (residues 1 to 130) and the C-terminal half (residues 131 to 215). Based on this alignment and on the HIV-1 CA 3D structure, 12 sites were selected, CA 1 to 7 and 9 to 12 (Fig. 1), and mutated. In addition, a unique cysteine in position 63 (C63) of the HTLV-1 CA, which had no counterpart in HIV-1 CA, was substituted for an alanine (C63A).
FIG. 1.
Alignment of HIV-1 and HTLV-1 CA proteins. ∗ and · indicate perfectly and well-conserved amino acids, respectively. Light gray background indicates MHR. Open boxes indicate helices characterized in the 3D structures. A dark gray background indicates C63 and CA 1 to 7 and 9 to 12 mutated sites.
For eight of the mutated sites (CA 1 to 5 and 10 to 12), two types of mutations were introduced by using a single-shot site-directed mutagenesis, leading to insertion mutants, named CAi, and to substitution mutants, named CAs (Table 2). Four single-amino acid-substitution mutants, CAs 6, 7, and 9 and C63A, were constructed (Table 2). All of these CA mutants were subcloned in gag and gag-pro genetic backgrounds and were inserted into the eukaryotic expression vector pKCR3.
TABLE 2.
Amino acid sequences of HTLV-1 CA mutants
| Mutated site | Wild-type sequencea | Mutated sequences
|
|
|---|---|---|---|
| CAi | CAs | ||
| CA 1 | 51TAKDLQDL | 51TAKRSAGRSQDL | 51TAKRSQDL |
| C63 | 60QYLCSSL | 60QYLASSL | |
| CA 2 | 65SLVASLHH | 65SLVRSAGRSLHH | 65SLVRSLHH |
| CA 3 | 78LISEAETR | 78LISRSAGRSETR | 78LISRSETR |
| CA 4 | 92PLAGPLRV | 92PLADIAGDILRV | 92PLADILRV |
| CA 5 | 100QANNPQQQ | 100QANRSAGRSQQQ | 100QANRSQQQ |
| CA 6 | 116LWLAAFA | 116LWLPAFA | |
| CA 7 | 139GLEEPYH | 139GLEKPYH | |
| CA 9 | 169LRSLAYS | 169LRSSAYS | |
| CA 10 | 174YSNANKEC | 174YSNRSAGRSKEC | 174YSNRSKEC |
| CA 11 | 182QKLLQARG | 182QKLRSAGRSARG | 182QKLRSARG |
| CA 12 | 192NSPLGDM | 192NSPRGPRGDM | 192NSPRGDM |
The wt amino acids of the HTLV-1 CA mutated sites and their corresponding insertion (CAi) and/or substitution (CAs) mutations are underlined. Amino acid numbering starts at the first amino acid of the HTLV-1 CA protein.
Effect of HTLV-1 CA mutations on VLP formation and egress.
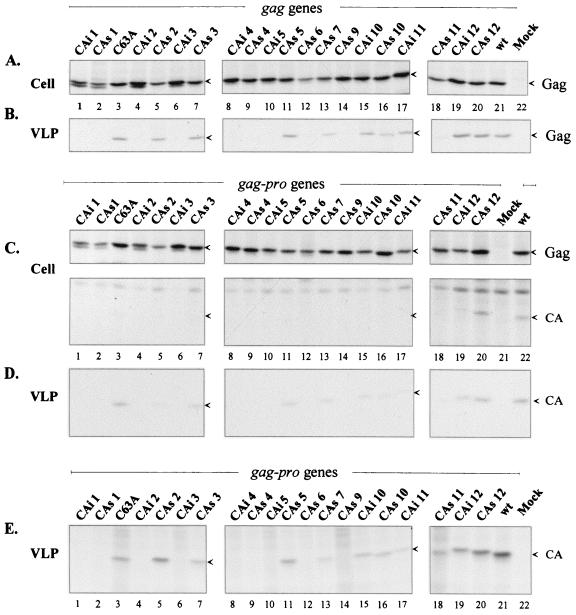
To examine the ability of the various mutated gag genes or gag-pro genes to lead to the release of immature or mature VLPs, the corresponding DNAs were transfected in 293T cells and the expressed proteins were labeled with [35S]methionine. Cell lysates were immunoprecipitated and analyzed by SDS-PAGE. The culture medium was centrifuged through a 20% sucrose cushion, and the VLPs present in the pellet and the soluble proteins remaining in the supernatant were analyzed. No Gag protein was visualized in the soluble fractions (data not shown).
In the cell lysates, the expression of the wt HTLV-1 gag gene alone and of the wt HTLV-1 gag-pro genes led to synthesis of Pr53Gag (Fig. 2A, lane 21, and C, lane 22, top). Pr76Gag-Pro, which was translated through one frameshift event, was difficult to visualize but was readily produced, as some mature CA proteins were specifically immunoprecipitated, indicating the presence of an active viral protease (Fig. 2C, lane 22, bottom). Expression of the mutated gag and gag-pro genes also led to the obtention of Pr53Gag and CA proteins in cell lysates, but in variable amounts (Fig. 2A and C, lanes 1 to 20). These heterogeneous expression patterns of the mutated Pr53Gag were taken into account in the following quantitative evaluation of the VLP egress obtained for each Gag mutant.
FIG. 2.
Expression of the HTLV-1 CA mutants in gag or gag-pro genetic context. 293T cells were transfected by the wild-type or the mutated pKgag (A and B) or pKgag-pro vectors (C, D and E). Two days posttransfection, cells were metabolically labeled with [35S]methionine. (A and C) Cell-associated proteins were analyzed by immunoprecipitation and fluorography. VLP-associated proteins were either analyzed by immunoprecipitation (B and D) or directly subjected to SDS-PAGE analysis (E). The Pr53Gag and the CA proteins are indicated by arrows.
The expression of the wt HTLV-1 gag gene alone led to the release of immature pelletable VLPs containing Pr53Gag (Fig. 2B, lane 21). When the wt gag-pro genes were expressed, only the mature CA protein with the expected 24-kDa molecular mass was revealed in the VLPs (Fig. 2D, lane 22) because neither MA nor nucleocapsid proteins contain methionine; neither Pr76Gag-Pro, Pr53Gag, nor intermediate cleavage products were detected in the pelleted VLPs (data not shown). This indicated that the expression of the wt HTLV-1 gag-pro genes in 293T cells led to the secretion of fully mature VLPs containing active protease.
The VLP secretion patterns of the mutants are presented in Fig. 2B (lanes 1 to 20) for gag and Fig. 2D (lanes 1 to 20) for gag-pro gene expressions. In a gag genetic context, 11 mutated Pr53Gag proteins were released as VLPs (Fig. 2B, lanes 3, 5, 7, 11, 13, and 15 to 20). For the remaining mutants, no Pr53Gag was detected (Fig. 2B, lanes 1, 2, 4, 6, 8 to 10, 12, and 14).
In a gag-pro context, when the CA mutations did not impair VLP secretion, these VLPs contained only mature proteins (Fig. 2D, lanes 1 to 20); neither immature Pr76Gag-Pro, Pr53Gag, nor intermediate cleavage products were present (data not shown). This observation revealed that the processing of mutated Pr76Gag-Pro and Pr53Gag had been achieved. The same 11 CA mutants that led to VLP secretion in the gag context gave rise, in the gag-pro context, to VLPs containing CA proteins (Fig. 2D, lanes 3, 5, 7, 11, 13, and 15 to 20), whereas, as in the gag context, the nine remaining mutations did not allow detectable viral protein secretion (Fig. 2D, lanes 1, 2, 4, 6, 8 to 10, 12, and 14).
The lack of CA detection of some mutants may have been due to a failure of recognition of the mutated CA proteins by the antibodies. This was unlikely, because the monoclonal antibodies recognized all the different mutated Gag proteins in cell lysates, indicating that probably none of the mutations led to misfolding of the CA proteins. To rule out the possibility of such an artifact, the release of mutated CA proteins was examined by subjecting VLPs directly to SDS-PAGE analysis, i.e., without an immunoprecipitation step. The results obtained without immunoprecipitation (Fig. 2E, lanes 3, 5, 7, 11, 13, and 15 to 20) were the same as those obtained using an immunoprecipitation step (Fig. 2D, compare to E). These results confirmed that the absence of mutated CA detection within some VLPs was effectively due to impairment of particle formation and not to immunoprecipitation artifacts related to an eventual CA misfolding.
From the above experiments, the amount of particulate Pr53Gag and CA released into the medium was determined. Because of the heterogeneous expression of the mutated Gag proteins, the values obtained were corrected with respect to the total (intracellular plus extracellular) amount of the corresponding Gag precursor. Then, the efficiency of VLP secretion by the CA mutants was expressed as a percentage of wt secretion level.
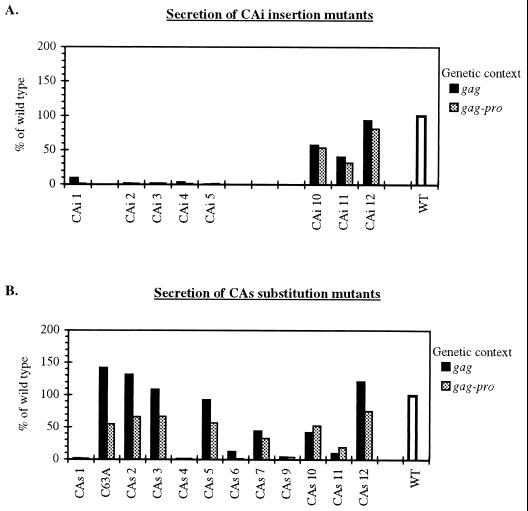
Concerning the CAi mutants, the levels of VLP release were similar for a given mutant expressed from either the gag or gag-pro genetic context (Fig. 3A). All insertions in the N-terminal half of the CA region (CAi mutants 1 to 5) drastically impaired secretion of both immature and mature VLPs. In strict contrast, those in the C-terminal portion displayed a less drastic defect, and VLP release varied from 31 to 93% for the CAi mutants 10 to 12.
FIG. 3.
VLP secretion levels of the HTLV-1 CA mutants expressed in gag and gag-pro genetic contexts. (A) CAi mutants; (B) CAs mutants. Dark bars indicate results in gag context (mean values of two independent experiments); gray bars indicate results in gag-pro context (mean values of three independent experiments). For each genetic context, all CAi and CAs mutants were analyzed in parallel.
For the CAs mutants, no such dramatic difference between the N and C domains was observed (Fig. 3B). For the CAs substitution mutants expressed in the gag genetic context, five mutated Pr53Gag proteins retained their competence for release of immature VLPs (C63A and CAs 2, 3, 5, and 12). Their secretion values were similar (92% for the CAs 5) or much higher (up to 142% for C63A) than the value observed for the wt construct. CAs mutants 7 and 10 led to a reduced immature VLP release of 44 and 42%, respectively. For the CAs 1, 4, 6, 9, and 11 mutants, secretion of immature VLPs was dramatically reduced and never exceeded 12%.
Concerning the CAs substitution mutants expressed in a gag-pro context, the levels of VLP secretion were reduced (Fig. 3B) compared to either the wt protein or the same mutants expressed in a gag context. The maximum value was 75% of the wt Gag value. For the CAs 1, 4, 6, 9, and 11 substitutions, secretion of mature VLPs was strongly impaired and never exceeded 19%. This seemed to indicate that when Pr76Gag-Pro polyprotein was present, particle formation was less tolerant of a defect in Gag.
The CAs substitutions were expected to have only a minor effect on VLP release, in comparison to the corresponding CAi insertions. This was the case for some (CAs 2, 3, and 5) but not all (CAs 1 and 4) substitutions located at the N-terminal half of the CA. Whatever the genetic context used, the CAs 2, 3, and 5 substitution mutants led to the release of VLPs, whereas the corresponding insertions, CAi 2, 3, and 5, strongly inhibited VLP production (Fig. 3A and B).
The particle formation defect phenotypes observed for some of the CA mutants could be due to major misfolding of the mutated precursors, impairing their membrane binding potential. This membrane binding function is crucial for capsid assembly, and thus a membrane binding defect of one of the above mutants would impair capsid assembly and subsequently VLP formation. To rule out this possibility, it is necessary to know (i) whether membrane binding is required for HTLV-1 indirect or direct Gag-Gag interactions (hereafter termed “Gag interactions”), and (ii) the membrane-binding potential of each Gag mutant.
Secretion defect of nonmyristylated Gag protein could not be rescued efficiently by coexpression of wild-type Gag precursor.
In an attempt to determine whether HTLV-1 Gag precursors can interact in the cytosol or if these interactions require membrane binding, the following experiments were performed using nonmyristylated and wt Gag proteins.
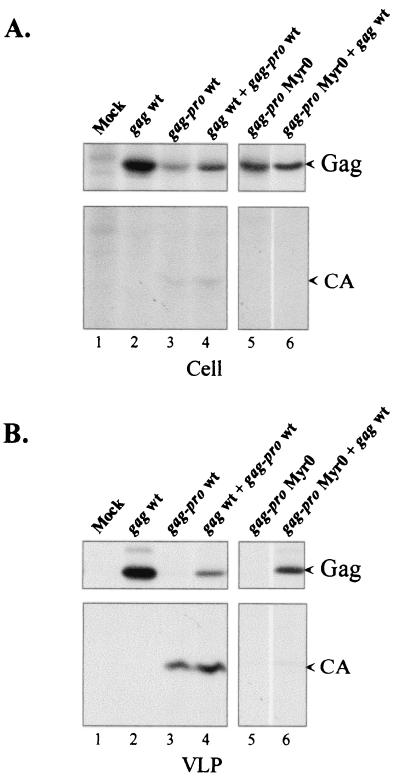
In the first step, the effective myristylation of wt and N-terminal glycine-mutated HTLV-1 Gag proteins expressed in 293T cells was verified. The N-terminal glycine of gag or gag-pro genes was mutated to alanine, and the resulting mutated genes were termed gagMyr0 and gag-proMyr0, respectively. A metabolic labeling with [3H]myristic acid indicated that wt Gag proteins expressed from wt gag or wt gag-pro genes were myristylated and that the mutated Gag Myr0 proteins expressed from gagMyr0 or gag-proMyr0 genes were not (data not shown). Additional experiments demonstrated that myristylation of the HTLV-1 Gag precursor was required for membrane association and VLP release (Fig. 4B, lane 5, and Fig. 5; also data not shown).
FIG. 4.
Complementation assay with the nonmyristylated HTLV-1 Gag precursor. The pKgag wt and the pKgag-proMyr0 vectors were cotransfected into 293T cells at a ratio of 1:1. As a control, cells were singly transfected with the pKgag wt or pKgag-pro wt vectors or with the pKgag-proMyr0 vector. Two days posttransfection, cells were metabolically labeled with [35S]methionine. Cell-associated (A) and VLP-associated (B) Gag proteins were analyzed by immunoprecipitation and fluorography. The Pr53Gag and CA proteins are indicated by arrows.
FIG. 5.
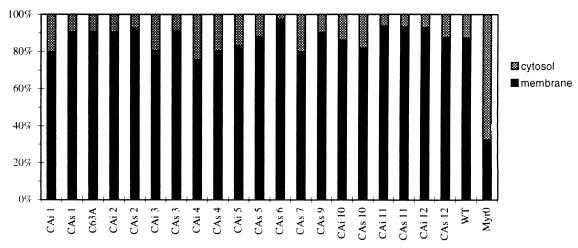
Subcellular localization of the Pr53Gag precursor of the HTLV-1 CA mutants. 293T cells were transfected with the wild-type or the Myr0 or CA mutated pKgag vectors. Two days posttransfection, cells were metabolically labeled with [35S]methionine and subjected to subcellular fractionation. Cells were broken by Dounce homogenization, and unbroken cells and nuclei were removed by low-speed centrifugation. Membrane (P100) and cytosolic (S100) fractions were separated by centrifugation (100,000 × g, 30 min). The two fractions were analyzed by immunoprecipitation and SDS-PAGE. For all CA mutants, the amount of Pr53Gag contained in each membrane (P100) and cytosolic (S100) fraction was quantified and reported relative to the total amount of Pr53Gag contained in the two fractions. Dark bars indicate membrane fraction, and shaded bars indicate cytosolic fraction. All CAi and CAs mutants were analyzed in parallel.
In the second step, a complementation experiment was performed in order to determine whether HTLV-1 Gag interactions require membrane binding. In this attempt, cells were cotransfected with gag-proMyr0 and wt gag DNAs. The cotransfection assays were based on the hypothesis that if the Gag Myr0 and Gag-Pro Myr0 precursors, which are unable to bind to membrane and to be secreted, were nevertheless able to interact with the wt Gag precursor molecules, they would then be dragged into budding particles by wt Gag. This would lead to secretion of chimeric VLPs, containing the wt Pr53Gag plus the mutated Pr53Gag and the mutated Pr76Gag-Pro. The presence of the protease would process the precursors, leading to the detection of VLPs containing mature CA. Conversely, if the mutated Pr76Gag-Pro did not interact with wt Pr53Gag, the mutated Pr76Gag-Pro would not be dragged into the particles by the wt Pr53Gag. Thus, the wt Gag would remain unprocessed. Furthermore, mature CA proteins would be detected in VLPs only if Gag interactions could occur in the absence of membrane binding. Control experiments confirmed that all the genes, either mutated or wt, were readily expressed in the cells (Fig. 4A). Transfection of wt gag and wt gag-pro genes led to VLPs containing Gag and mature CA proteins, respectively (Fig. 4B, lanes 2 and 3); coexpression of wt gag and wt gag-pro genes led to VLPs containing small amounts of nonprocessed Gag and large amounts of mature CA protein (Fig. 4B, lane 4). In this latter case, the amount of the protease produced was most probably restricted and did not permit the completion of Gag precursor processing. In contrast, gag-proMyr0 gene expression did not lead to VLP release (Fig. 4B, lane 5), confirming that the myristylation of HTLV-1 Gag is required for the particle formation. The coexpression of gag-proMyr0 and wt gag led to VLPs containing only immature Gag precursors and no or very low levels of mature CA proteins (Fig. 4B, lane 6). These results indicate that Gag-Pro Myr0-wt Gag interactions did not occur efficiently in the cytosol and that membrane binding of the Gag proteins is required for VLP formation.
All CA mutants possess full membrane-binding function.
The above experiments demonstrated that the Gag precursor must be able to bind to the membrane in order to allow Gag precursor interactions that lead to Gag assembly and VLP secretion. As a consequence, the membrane-binding potential of each Gag mutant was characterized. In this experiment, cells were transfected with either wt or mutated gag DNA. After labeling, cell extracts were prepared in the absence of detergent and were submitted to fractionation experiments. The amounts of each Gag protein contained in either the cytosol or in the membrane fractions were analyzed by SDS-PAGE. These amounts were quantified, and the results obtained were expressed as the percentage of Gag protein contained in one fraction per total Gag proteins contained in the two fractions (Fig. 5). As expected, the major part (87%) of the wt Gag proteins was membrane associated and the major part (68%) of nonmyristylated Gag Myr0 proteins was in the cytosol (Fig. 5). Concerning the Gag proteins mutated on the CA, they were mainly (76 to 98%) membrane associated (Fig. 5). These results indicated that all of the CA mutants were sufficiently well folded to bind to membrane. As a consequence, all of these mutant proteins could be targeted at the cell membrane, a step required for the HTLV-1 capsid assembling process. It is likely that the observed VLP secretion impairments described above were not due to misfolding that would impair the accumulation of Gag precursors at the cell membrane, a step required for HTLV-1 capsid assembly.
Most of the VLP secretion-defective mutants could not be rescued by coexpression of wild-type Gag precursor.
Knowing that all mutated and wt Gag proteins had no membrane binding defect, we next determined whether the mutations of the HTLV-1 CA proteins prevented VLP secretion by impairing Gag interactions. In this attempt, complementation experiments were performed. Based on the same hypothesis as above, the mutated gag-pro and wt gag genes were coexpressed in 293T cells. In brief, mature CA proteins were detected in VLPs only when the mutations did not concern a CA site involved in indirect or direct Gag interactions.
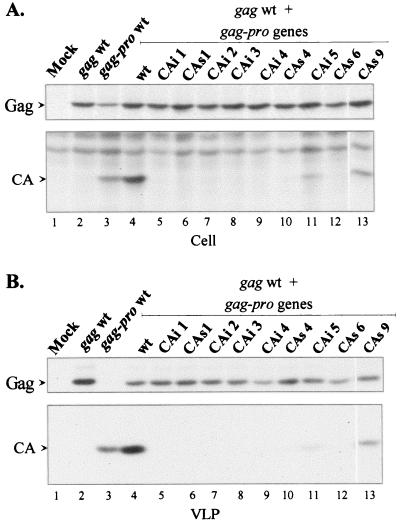
Each DNA sample of the pKgag-pro vectors harboring the VLP secretion-defective mutant genes (CAi 1 to 5 and CAs 1, 4, 6, and 9) and the DNA of the pKgag wt vector were cotransfected at a ratio of 1:1. Both cell lysates and released VLPs were immunoprecipitated and analyzed by SDS-PAGE. As a control, the cotransfection of two independent vectors harboring the wt gag and gag-pro genes led to the synthesis of the Pr53Gag and Pr76Gag-Pro precursors. This latter precursor was revealed by the presence of mature CA protein (Fig. 6A, lane 4). Both Pr53Gag precursor and CA protein were detected in the VLPs secreted from the cotransfected cells but, as shown above (Fig. 4B, lane 4), the amount of CA was larger than that of unprocessed Gag.
FIG. 6.
Complementation assay by coexpression of VLP secretion-defective mutants and the wt Gag precursor. The pKgag wt and the CA mutated pKgag-pro vectors were cotransfected into 293T cells at a ratio of 1:1. As a control, cells were transfected with pKgag wt or pKgag-pro wt vectors or cotransfected with the pKgag wt and pKgag-pro wt vectors. Two days posttransfection, cells were metabolically labeled with [35S]methionine. Cell-associated (A) and VLP-associated (B) proteins were analyzed by immunoprecipitation and fluorography (panel A, top and bottom, shows results from the 1- and 3-day exposures, respectively). The Pr53Gag and the CA proteins are indicated by arrows.
The results showed that only one of the nine VLP secretion-defective mutants, CAs 9, was well rescued by the wt Pr53Gag precursor (Fig. 6B, lanes 13 and Fig. 7). Although the CAi insertions were expected to cause serious alteration of the protein structure, one insertion mutant, CAi 5, was rescued, although with a much lower efficiency than that of the CAs 9 mutant (Fig. 6B, lane 11). The CAs 4 mutant was very poorly rescued, as only traces of mature CA protein were detected (Fig. 6B, lane 10).
FIG. 7.
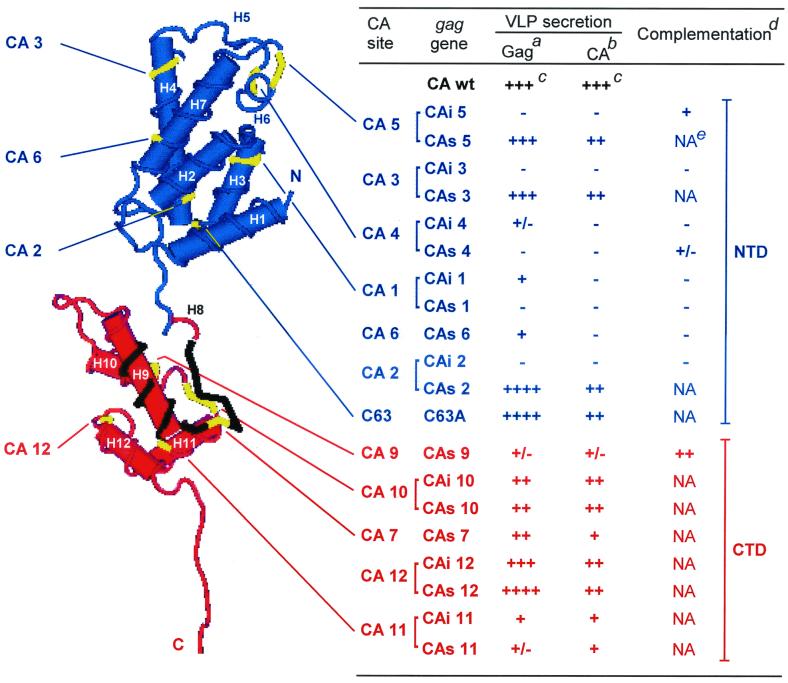
Mutated sites on the HTLV-1 CA 3D structure and the corresponding phenotypes of the CAi and CAs Gag mutants. The NTD and CTD are shown in blue and red, respectively. The MHR is in black. The helices are indicated as H1 to H12. The table summarizes the mutant phenotypes. The C63 and CA 1 to 6 mutated sites (yellow) are located in the NTD. The CA 7 and 9 to 12 mutated sites (yellow) are located in the CTD. Footnotes for table are as follows. aImmature VLPs released from gag transfected cells. bMature VLPs released from gag-pro transfected cells. cAmount of secreted VLPs relative to the wt protein: ++++, higher than 110%; +++, 86 to 110%; ++, 41 to 85%; +, 10 to 40%; +/−, below 10%; −, VLP secretion-defective mutant. dComplementation assay of the mutated gag-pro genes by the wt gag gene. ++, +, and +/− indicate well, poorly, and very poorly rescued VLP secretion-defective mutants, respectively, and − indicates unrescued mutants. eNA, not applicable.
In contrast, the CAi 1 to 4 and CAs 1 and 6 mutants were not rescued by coexpression of the wt Pr53Gag precursor, since no mature CA proteins were detected in the culture medium (Fig. 6B, lanes 5 to 9 and 12, and Fig. 7). This finding indicated that these mutations interfered dramatically with Gag interactions. Remarkably, the only CA mutant truly rescued (CAs 9) by coexpression of wt Gag corresponded to a mutation located in the C-terminal half of the HTLV-1 CA, i.e., the CTD in the HTLV-1 CA 3D structure (Fig. 7).
DISCUSSION
The work presented here demonstrates that two domains of the HTLV-1 CA protein can be defined from both sequence and functional analyses. The two functional domains correspond to the two structural domains, NTD and CTD, that have been identified for both the HIV-1 and HTLV-1 CA proteins. Here, we have demonstrated that the functions harbored by the HTLV-1 NTD and CTD differ markedly from those reported for the NTD and CTD of HIV-1.
The similarity between the overall structures of the HTLV-1 and HIV-1 CA proteins that was suggested by our alignment was recently confirmed by reports of their respective 3D structures (1, 24). Like the HIV-1 CA, the HTLV-1 CA protein folds into two independent subdomains linked by a linker peptide (Fig. 7). The NTD and CTD of the HTLV-1 CA protein stretch from residue 15 to the serine 127 and from the proline 131 to the threonine 206, respectively. The A128 K129 and D130 residues form the linker peptide.
Concerning the functional analysis, the VLP secretion of each of the CAi and CAs Gag mutants, expressed either from the gag gene alone or the gag-pro genes, was compared. The CAi insertions were expected to impair the Gag structure more drastically than their corresponding CAs substitutions. This study showed that this was true for only three out of eight mutated sites: the CAi 2, 3, and 5 mutations drastically impaired VLP secretion, whereas the corresponding CAs 2, 3, and 5 mutations had only moderate effects. For the remaining CA mutants, the CAi mutations caused a similar or, unexpectedly, a less negative effect on VLP secretion than the corresponding CAs mutations.
In this study, mutational analysis identified discrete sites of the CA domain of the Gag precursor that have critical functions in VLP formation, most of which are probably indirect or direct Gag-Gag interactions involved in capsid assembling. First of all, the N-terminally located insertions, CAi 1 to 5, inhibited the secretion of both immature and mature VLPs, whereas the C-terminally located insertions, CAi 10 to 12, had only moderate effects. These results indicate that the integrity of the NTD is essential for particle formation but not that of the CTD. In addition, the CAi mutants displayed similar phenotypes in either both gag- and gag-pro-transfected cells. In contrast, for most of the CAs mutants, VLP release was more severely impaired when the mutated Gag precursors were expressed from the gag-pro genes than when they were expressed from the gag gene alone. This finding suggests that the presence of Gag-Pro precursors in the HTLV-1 Gag assembly introduces new constraints in the assembly process.
Among the HTLV-1 CA mutated sites, the glutamic acid in position 142 (E142) at the CA 7 site is one of the three invariant residues of the MHR that are conserved among numerous retroviruses (26). Its exchange for a lysine (CAs 7) resulted in a significant reduction, but not total inhibition, of immature and mature HTLV-1 VLP secretion (44 and 33%, respectively). This indicates that the invariant E142 of the HTLV-1 MHR is not absolutely required for particle release but is required only for reaching a maximum of VLP release. On one hand, such an observation contrasts with the absolute requirement for virus production of the equivalent invariant glutamic acids E159 and E162 of HIV-1 and RSV, respectively (4, 26). On the other hand, the substitution E162G in RSV has been reported not to significantly alter RSV VLP egress from non-avian cells but only to slow down Gag precursor maturation (4). The 3D structures of the HIV-1 and HTLV-1 CA proteins reveal that this glutamic acid is involved in the hydrogen-bonding network which ensures the stability of the CTD structure (1, 11, 24). Moreover, the HIV-1 E159 takes part in in vitro membrane binding of the Gag precursor, a step required for virus budding (8). Our results demonstrate that the CAs 7 mutated Gag protein binds to cell membranes; thus, no similar role is attributed to the HTLV-1 E142.
The CA 1 site corresponding to the D54L55 dipeptide of the HTLV-1 CA is well conserved in numerous retroviruses. For HIV-1, the negative charge of the carboxyl side chain of the corresponding aspartic acid 51 (D51) forms a salt bridge with the positive charge of the nascent α NH2 of the N-terminal proline of the CA, which is released after MA-CA cleavage. In addition, this HIV-1 aspartic acid is critical for in vitro assembly and efficient in vivo virus production (41). In accordance with such a possible role, the CAi 1 and CAs 1 mutants failed to release detectable mature VLPs. Interestingly, they also failed to release detectable immature particles. The formation of the intramolecular salt bridge cannot account for the HTLV-1 D54L55-site requirement for release of the myristylated Gag precursor, a protein devoid of a free NH2 terminus. The drastic effect of the CAi 1 and CAs 1 mutations on nonprocessed Gag polyprotein release may also more possibly be attributed to the L55 substitution. Indeed, the L55 pertains to the 12 hydrophobic residues that promote the packing of NTD via interactions between H7 and H1-H3 α-helices (24). However, for HIV-1, a large insertion (DL52N→DFSSSRN) which removes the analogous leucine does not hamper secretion of the virion, despite the fact that it abolishes infectivity (35). Thus, the HTLV-1 L55 most likely plays no major role in particle formation, and the mutation of D54 may be entirely responsible for the absence of particle formation of CAs 1 and CAi 1 mutants.
Another HTLV-1 CA site required for particle formation is the G95P96 (CA 4), which is included in a conserved AGPL/I peptide of a small loop (24). At the same location, the HIV-1 CA 3D structure exhibits a very large loop (1). The CAi 4 and 5 insertions, which are located in the HTLV-1 CA small loop, block VLP release. These insertions increase the size of this loop and could lead to a steric hindrance that hampers Gag interactions. Indeed, one of the corresponding substitution (CAs 5) mutants is still able to allow particle formation (92 and 56% in the gag and gag-pro genetic contexts, respectively). In contrast, the double substitution, G95P96→D95I96 (CAs 4), prevents particle formation. Thus, in addition to the loop size limit, the glycine G95 and/or the proline P96 are probably necessary to give a particular structure to the CA protein, which would be critical for HTLV-1 Gag assembly and release. This seems to fit well with the EIAV CA assembly model proposed by Jin et al. (22). In that model, this loop pertains to a pore resulting from a CA NTD arrangement that is susceptible to the fivefold as well as the sixfold associations, leading to the folding axes required for icosahedron-based capsid structure. Several studies have demonstrated that the corresponding HIV-1 loop and particularly the proline 90 of the AGPL/I-conserved motif are involved in the association of the Gag precursor with cyclophilin A (10, 45). Nevertheless, this feature seems to be an intrinsic HIV-1 CA property, since incorporation of cyclophilin A into particles of other retroviral species has never been demonstrated. For example, Khorasanizadeh et al. have shown that recombinant HTLV-1 CA protein failed to bind cyclophilin A in vitro (24). Hence, if this situation is also true in vivo, the requirement of the G95P96 dipeptide is not related to cyclophilin A binding.
The HTLV-1 CA site CA 6 corresponds to the alanine 119 (A119). The mutation of this residue inhibits VLP formation; the same phenotype was observed for an HIV-1 L136P mutant expressed in insect cells (21).
Finally, the last identified residue required for HTLV-1 VLP egress is the leucine 172 (L172) of the CA 9 site. This leucine is well conserved between the HTLV-1 and HIV-1 CA proteins, in which it is included in an α helix. In HIV-1 CA dimers, several hydrophobic residues of this α helix are involved in CA dimerization. The same is likely to happen for EIAV CA (1, 11, 22). Interestingly, Khorasanizadeh et al. reported that, due to a small rotation of the HTLV-1 H10 α helix, the hydrophobic residues (such as the L172 of the CA 9 site) of the H10 α helix are buried and cannot establish intermolecular hydrophobic interactions to promote CTD dimerization (24). Thus, the requirement for L172 is probably not due to its possible role in direct Gag-Gag interactions during the assembling process.
The complementation assays revealed that only the C-terminal CAs 9 mutant is efficiently rescued for VLP release. In contrast, the N-terminal CA mutants CAs 4, CAi 5, CAi 1 to 4, and CAs 1 and 6 are rescued very poorly or not at all. Thus, no mutants located in the NTD could be fully rescued. In contrast, for HIV-1 the Gag mutant analogous to the CAs 6 mutant (L136P) is readily rescued by the wt Gag protein (21). These experiments revealed that, on one hand, the NTD is the major partner of direct or indirect Gag-Gag interactions, and, on the other hand, the CTD seems not to be involved in these Gag interactions, as the CAs 9 mutant of the CTD is very well rescued. Note that the CAs 9 mutation does not impair the Gag-Gag interactions; as seen in the complementation experiment, such interactions must happen between two CAs 9 mutated Gag-Pro precursors in order for protease dimerization and activation to occur. In view of the severe impairment phenotype of the CAs 9 mutant, this domain must instead be involved in an unknown step required for particle formation.
In summary, our results indicate that, despite an overall structural similarity between the HIV-1 and the HTLV-1 CA proteins (for review, see reference 9), their NTDs and CTDs exhibit different functions. For the HTLV-1, the NTD plays a major role in virion formation and unexpectedly, the CTD seems to play a marginal role, at least in Gag interactions. Such a conclusion is in accordance and is strengthened by the HTLV-1 CA 3D structure (24), which differs markedly from the structures of HIV-1 and EIAV CA proteins. In particular, the HTLV-1 CA is monomeric in solution and this could be due to important differences between the CTDs, a domain that promotes dimerization of lentivirus CAs. Further studies are needed to determine whether the differences observed between the NTDs and CTDs of the oncoretrovirus HTLV-1 and those of lentiviruses HIV-1 and EIAV are related to their overall capsid shapes, which are spherical and conical, respectively.
ACKNOWLEDGMENTS
We acknowledge Bernadette Trentin for critical review and insightful comments on the manuscript. We thank Kathryn Mayo for final review of the English of the manuscript.
This work was supported by grants from the Comité de la Gironde de la Ligue Nationale Contre le Cancer, the Etablissement Public Régional d'Aquitaine, and the Association pour la Recherche contre le Cancer. Fabienne Rayne is a recipient of fellowships from the Fondation pour la Recherche Médicale and the Association pour la Recherche contre le Cancer.
REFERENCES
- 1.Berthet-Colominas C, Monaco S, Novelli A, Sibai G, Mallet F, Cusack S. Head-to-tail dimers and interdomain flexibility revealed by the crystal structure of HIV-1 capsid protein (p24) complexed with a monoclonal antibody Fab. EMBO J. 1999;18:1124–1136. doi: 10.1093/emboj/18.5.1124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Borsetti A, Ohagen A, Gottlinger H G. The C-terminal half of the human immunodeficiency virus type 1 Gag precursor is sufficient for efficient particle assembly. J Virol. 1998;72:9313–9317. doi: 10.1128/jvi.72.11.9313-9317.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clarke M F, Gelmann E P, Reitz M S., Jr Homology of human T-cell leukaemia virus envelope gene with class I HLA gene. Nature. 1983;305:60–62. doi: 10.1038/305060a0. [DOI] [PubMed] [Google Scholar]
- 4.Craven R C, Leure-duPree A E, Weldon R A, Jr, Wills J W. Genetic analysis of the major homology region of the Rous sarcoma virus Gag protein. J Virol. 1995;69:4213–4227. doi: 10.1128/jvi.69.7.4213-4227.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Derse D, Mikovits J, Polianova M, Felber B K, Ruscetti F. Virions released from cells transfected with a molecular clone of human T-cell leukemia virus type I give rise to primary and secondary infections of T cells. J Virol. 1995;69:1907–1912. doi: 10.1128/jvi.69.3.1907-1912.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Dokhelar M C, Pickford H, Sodroski J, Haseltine W A. HTLV-1 p27rex regulates Gag and Env protein expression. J Acquir Immune Defic Syndr. 1989;2:431–440. [PubMed] [Google Scholar]
- 7.Dorfman T, Bukovsky A, Ohagen A, Hoglund S, Gottlinger H G. Functional domains of the capsid protein of human immunodeficiency virus type 1. J Virol. 1994;68:8180–8187. doi: 10.1128/jvi.68.12.8180-8187.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ebbets-Reed D, Scarlata S, Carter C A. The major homology region of the HIV-1 Gag precursor influences membrane affinity. Biochemistry. 1996;35:14268–14275. doi: 10.1021/bi9606399. [DOI] [PubMed] [Google Scholar]
- 9.Freed E O. HIV-1 Gag proteins: diverse functions in the virus life cycle. Virology. 1998;251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 10.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 11.Gamble T R, Yoo S, Vajdos F F, von Schwedler U K, Worthylake D K, Wang H, McCutcheon J P, Sundquist W I, Hill C P. Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein. Science. 1997;278:849–853. doi: 10.1126/science.278.5339.849. [DOI] [PubMed] [Google Scholar]
- 12.Garnier L, Ratner L, Rovinski B, Cao S X, Wills J W. Particle size determinants in the human immunodeficiency virus type 1 Gag protein. J Virol. 1998;72:4667–4677. doi: 10.1128/jvi.72.6.4667-4677.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gelderblom H R. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS. 1991;5:617–637. [PubMed] [Google Scholar]
- 14.Gessain A, Barin F, Vernant J C, Gout O, Maurs L, Calender A, de The G. Antibodies to human T-lymphotropic virus type-I in patients with tropical spastic paraparesis. Lancet. 1985;ii:407–410. doi: 10.1016/s0140-6736(85)92734-5. [DOI] [PubMed] [Google Scholar]
- 15.Gheysen D, Jacobs E, de Foresta F, Thiriart C, Francotte M, Thines D, De Wilde M. Assembly and release of HIV-1 precursor Pr55Gag virus-like particles from recombinant baculovirus-infected insect cells. Cell. 1989;59:103–112. doi: 10.1016/0092-8674(89)90873-8. [DOI] [PubMed] [Google Scholar]
- 16.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 17.Hatanaka M, Nam S H. Identification of HTLV-1 Gag protease and its sequential processing of the gag gene product. J Cell Biochem. 1989;40:15–30. doi: 10.1002/jcb.240400103. [DOI] [PubMed] [Google Scholar]
- 18.Hattori S, Kiyokawa T, Imagawa K, Shimizu F, Hashimura E, Seiki M, Yoshida M. Identification of gag and env gene products of human T-cell leukemia virus (HTLV) Virology. 1984;136:338–347. doi: 10.1016/0042-6822(84)90170-3. [DOI] [PubMed] [Google Scholar]
- 19.Hayakawa T, Miyazaki T, Misumi Y, Kobayashi M, Fujisawa Y. Myristoylation-dependent membrane targeting and release of the HTLV-1 Gag precursor, Pr53Gag, in yeast. Gene. 1992;119:273–277. doi: 10.1016/0378-1119(92)90282-t. [DOI] [PubMed] [Google Scholar]
- 20.Hertig C, Pye A D, Hyatt A D, Boyle D B. Retrovirus-like particles produced by vaccinia viruses expressing gag-pro-pol region genes of bovine leukaemia virus. J Gen Virol. 1994;75(Pt 9):2213–2221. doi: 10.1099/0022-1317-75-9-2213. [DOI] [PubMed] [Google Scholar]
- 21.Hong S S, Boulanger P. Assembly-defective point mutants of the human immunodeficiency virus type 1 Gag precursor phenotypically expressed in recombinant baculovirus-infected cells. J Virol. 1993;67:2787–2798. doi: 10.1128/jvi.67.5.2787-2798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jin Z, Jin L, Peterson D L, Lawson C L. Model for lentivirus capsid core assembly based on crystal dimers of EIAV p26. J Mol Biol. 1999;286:83–93. doi: 10.1006/jmbi.1998.2443. [DOI] [PubMed] [Google Scholar]
- 23.Karacostas V, Nagashima K, Gonda M A, Moss B. Human immunodeficiency virus-like particles produced by a vaccinia virus expression vector. Proc Natl Acad Sci USA. 1989;86:8964–8967. doi: 10.1073/pnas.86.22.8964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Khorasanizadeh S, Campos-Olivas R, Summers M F. Solution structure of the capsid protein from the human T-cell leukemia virus type-I. J Mol Biol. 1999;291:491–505. doi: 10.1006/jmbi.1999.2986. [DOI] [PubMed] [Google Scholar]
- 25.Le Blanc I, Rosenberg A R, Dokhelar M C. Multiple functions for the basic amino acids of the human T-cell leukemia virus type 1 matrix protein in viral transmission. J Virol. 1999;73:1860–1867. doi: 10.1128/jvi.73.3.1860-1867.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mammano F, Ohagen A, Hoglund S, Gottlinger H G. Role of the major homology region of human immunodeficiency virus type 1 in virion morphogenesis. J Virol. 1994;68:4927–4936. doi: 10.1128/jvi.68.8.4927-4936.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Momany C, Kovari L C, Prongay A J, Keller W, Gitti R K, Lee B M, Gorbalenya A E, Tong L, McClure J, Ehrlich L S, Summers M F, Carter C, Rossmann M G. Crystal structure of dimeric HIV-1 capsid protein. Nat Struct Biol. 1996;3:763–770. doi: 10.1038/nsb0996-763. [DOI] [PubMed] [Google Scholar]
- 28.Nam S H, Kidokoro M, Shida H, Hatanaka M. Processing of Gag precursor polyprotein of human T-cell leukemia virus type I by virus-encoded protease. J Virol. 1988;62:3718–3728. doi: 10.1128/jvi.62.10.3718-3728.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Hare K, Benoist C, Breathnach R. Transformation of mouse fibroblasts to methotrexate resistance by a recombinant plasmid expressing a prokaryotic dihydrofolate reductase. Proc Natl Acad Sci USA. 1981;78:1527–1531. doi: 10.1073/pnas.78.3.1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ono A, Demirov D, Freed E O. Relationship between human immunodeficiency virus type 1 Gag multimerization and membrane binding. J Virol. 2000;74:5142–5150. doi: 10.1128/jvi.74.11.5142-5150.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Osame M, Usuku K, Izumo S, Ijichi N, Amitani H, Igata A, Matsumoto M, Tara M. HTLV-1 associated myelopathy, a new clinical entity. Lancet. 1986;i:1031–1032. doi: 10.1016/s0140-6736(86)91298-5. [DOI] [PubMed] [Google Scholar]
- 32.Park J, Morrow C D. The nonmyristylated Pr160gag-pol polyprotein of human immunodeficiency virus type 1 interacts with Pr55gag and is incorporated into viruslike particles. J Virol. 1992;66:6304–6313. doi: 10.1128/jvi.66.11.6304-6313.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patarca R, Haseltine W A. A major retroviral core protein related to EPA and TIMP. Nature. 1985;318:390. doi: 10.1038/318390a0. [DOI] [PubMed] [Google Scholar]
- 34.Poiesz B J, Ruscetti F W, Gazdar A F, Bunn P A, Minna J D, Gallo R C. Detection and isolation of type C retrovirus particles from fresh and cultured lymphocytes of a patient with cutaneous T-cell lymphoma. Proc Natl Acad Sci USA. 1980;77:7415–7419. doi: 10.1073/pnas.77.12.7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Reicin A S, Paik S, Berkowitz R D, Luban J, Lowy I, Goff S P. Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J Virol. 1995;69:642–650. doi: 10.1128/jvi.69.2.642-650.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schultz A M, Rein A. Unmyristylated Moloney murine leukemia virus Pr65Gag is excluded from virus assembly and maturation events. J Virol. 1989;63:2370–2373. doi: 10.1128/jvi.63.5.2370-2373.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith A J, Srinivasakumar N, Hammarskjöld M-L, Rekosh D. Requirements for incorporation of Pr160gag-pol from human immunodeficiency virus type 1 into virus-like particles. J Virol. 1993;67:2266–2275. doi: 10.1128/jvi.67.4.2266-2275.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Swanstrom R, Wills J W. Synthesis, assembly, and processing of viral proteins. In: Coffin J M, Hughes S H, Varmus H E, editors. Retroviruses. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1997. pp. 263–334. [PubMed] [Google Scholar]
- 39.Takahashi R H, Nagashima K, Kurata T, Takahashi H. Analysis of human lymphotropic T-cell virus type II-like particle production by recombinant baculovirus-infected insect cells. Virology. 1999;256:371–380. doi: 10.1006/viro.1999.9655. [DOI] [PubMed] [Google Scholar]
- 40.von Poblotzki A, Wagner R, Niedrig M, Wanner G, Wolf H, Modrow S. Identification of a region in the Pr55Gag-polyprotein essential for HIV-1 particle formation. Virology. 1993;193:981–985. doi: 10.1006/viro.1993.1210. [DOI] [PubMed] [Google Scholar]
- 41.von Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang C T, Barklis E. Assembly, processing, and infectivity of human immunodeficiency virus type 1 Gag mutants. J Virol. 1993;67:4264–4273. doi: 10.1128/jvi.67.7.4264-4273.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wills J W, Craven R C. Form, function, and use of retroviral Gag proteins. AIDS. 1991;5:639–654. doi: 10.1097/00002030-199106000-00002. [DOI] [PubMed] [Google Scholar]
- 44.Wills J W, Craven R C, Achacoso J A. Creation and expression of myristylated forms of Rous sarcoma virus Gag protein in mammalian cells. J Virol. 1989;63:4331–4343. doi: 10.1128/jvi.63.10.4331-4343.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yoo S, Myszka D G, Yeh C, McMurray M, Hill C P, Sundquist W I. Molecular recognition in the HIV-1 capsid/cyclophilin A complex. J Mol Biol. 1997;269:780–795. doi: 10.1006/jmbi.1997.1051. [DOI] [PubMed] [Google Scholar]
- 46.Yoshida M, Miyoshi I, Hinuma Y. Isolation and characterization of retrovirus from cell lines of human adult T-cell leukemia and its implication in the disease. Proc Natl Acad Sci USA. 1982;79:2031–2035. doi: 10.1073/pnas.79.6.2031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhang W H, Hockley D J, Nermut M V, Morikawa Y, Jones I M. Gag-Gag interactions in the C-terminal domain of human immunodeficiency virus type 1 p24 capsid antigen are essential for Gag particle assembly. J Gen Virol. 1996;77:743–751. doi: 10.1099/0022-1317-77-4-743. [DOI] [PubMed] [Google Scholar]
- 48.Zhao T M, Robinson M A, Bowers F S, Kindt T J. Characterization of an infectious molecular clone of human T-cell leukemia virus type I. J Virol. 1995;69:2024–2030. doi: 10.1128/jvi.69.4.2024-2030.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]