Abstract
Infection of mice by murine gammaherpesvirus 68 (MHV-68) is an excellent small-animal model of gammaherpesvirus pathogenesis in a natural host. We have carried out comparative studies of another herpesvirus, murine herpesvirus 76 (MHV-76), which was isolated at the same time as MHV-68 but from a different murid host, the yellow-necked mouse (Apodemus flavicollis). Molecular analyses revealed that the MHV-76 genome is essentially identical to that of MHV-68, except for deletion of 9,538 bp at the left end of the unique region. MHV-76 is therefore a deletion mutant that lacks four genes unique to MHV-68 (M1, M2, M3, and M4) as well as the eight viral tRNA-like genes. Replication of MHV-76 in cell culture was identical to that of MHV-68. However, following infection of mice, MHV-76 was cleared more rapidly from the lungs. In line with this, there was an increased inflammatory response in lungs with MHV-76. Splenomegaly was also significantly reduced following MHV-76 infection, and much less latent MHV-76 was detected in the spleen. Nevertheless, MHV-76 maintained long-term latency in the lungs and spleen. We utilized a cosmid containing the left end of the MHV-68 genome to reinsert the deleted sequence into MHV-76 by recombination in infected cells, and we isolated a rescuant virus designated MHV-76(cA8+)4 which was ostensibly genetically identical to MHV-68. The growth properties of the rescuant in infected mice were identical to those of MHV-68. These results demonstrate that genetic elements at the left end of the unique region of the MHV-68 genome play vital roles in host evasion and are critical to the development of splenic pathology.
Infection of laboratory mice by murine gammaherpesvirus 68 (MHV-68) (formally designated murid herpesvirus 4) is an excellent model system for the study of gammaherpesvirus pathogenesis and for the development of therapeutic strategies against these viruses (31). Following intranasal inoculation of mice with MHV-68, productive infection initially occurs in the lung (34). The acute, productive infection is cleared from the lung by day 10 postinfection (p.i.) by CD8+ T cells (12), but the virus then persists in a latent form in epithelial cells at this site (32). MHV-68 spreads to the spleen during the subsequent viremia, where it becomes latent in B lymphocytes, macrophages, and dendritic cells (13, 35, 39, 44). Establishment of latency in the spleen is associated with a marked splenomegaly and a mononucleosis that resembles that caused by primary infection of humans by Epstein-Barr virus (36). Splenomegaly is driven by CD4+ T cells (12, 37) and is dependent on the presence of MHV-68-infected B cells in the spleen (39, 43). The resolution of splenomegaly is achieved by CD8+ T cells (12, 43), which are also important in the long-term control of persistent infection (5, 32, 43).
The MHV-68 genome consists of a unique region of 118,237 bp flanked on each side by multiple copies of a 1,213-bp terminal repeat (11, 42). The left end of the unique region has attracted considerable interest owing to the presence of four protein-coding genes (M1, M2, M3, and M4) and eight viral tRNA-like (vtRNA) genes, all of which lack counterparts in other gammaherpesviruses (4, 42). Of these, only the function of M3 has been elucidated. This gene encodes a secreted chemokine binding protein that is expressed during acute infection and persistence in mice (25, 38, 40, 41). M1 has been shown to be nonessential for lytic replication in vitro and in vivo for the maintenance and establishment of latency (6, 28). M2 is expressed during latency in vitro and in vivo (14, 41) and contains a CD8+ T-cell epitope, which is a target for the host immune response (14, 38). The vtRNA genes are abundantly expressed during lytic and latent infection (4), but their functional significance is not known.
Field studies in Slovakia aimed at identifying a small-animal vector for flaviviruses resulted in the isolation of five herpesviruses from two species of murid rodents (3). MHV-60, -68, and -72 were isolated from the bank vole (Clethrionomys glareolus), and MHV-76 and -78 were isolated from the yellow-necked wood mouse (Apodemus flavicollis). Further molecular studies of MHV-68 and sequencing of the genome showed that it belongs to the genus Rhadinovirus (often referred to as gamma-2 herpesviruses), which includes Kaposi's sarcoma-associated herpesvirus and herpesvirus saimiri (HVS) (10, 24, 42).
Of the other herpesvirus strains isolated alongside MHV-68 (3), only MHV-72 has been studied. This virus, which was isolated from the same host as MHV-68, appears to have similar biological properties in that it infects B lymphocytes (22, 26) and long-term infection is associated with tumorigenesis (21). In contrast, MHV-76 was isolated from a different murid species (3). Therefore, we hypothesized that it may have different biological characteristics and thus yield additional insights into gammaherpesvirus pathogenesis. In this paper, we describe the characterization of MHV-76. We demonstrate that the MHV-76 genome is essentially identical to that of MHV-68 except that it lacks M1, M2, M3, M4, and all of the vtRNA genes. We show that MHV-76 shares many biological features with MHV-68, including identical growth in vitro, productive infection in the lung, and the establishment and maintenance of latency in B lymphocytes. However, it is much less pathogenic in vivo than MHV-68, and reduced pathogenicity is associated with an increased inflammatory response. A rescuant virus was constructed by insertion of the left end of the MHV-68 genome into that of MHV-76. The MHV-76-derived rescuant regained the in vivo properties of MHV-68. We conclude that this region of the MHV-68 genome plays an important role in pathogenesis.
MATERIALS AND METHODS
Cell line and virus.
Viruses were grown and titrated using BHK-21 cells as described previously (34). MHV-68 was originally isolated during field studies from the bank vole, C. glareolus (3), and was subsequently plaque purified on BHK-21 cells to obtain clone g2.4 as described by Efstathiou et al. (11). MHV-76 was isolated from the yellow-necked mouse, A. flavicollis, during the same field study and was plaque purified thrice on BHK-21 cells to obtain a pure stock
Purification of viral DNA.
MHV-76 or MHV-68 DNA was prepared from purified virions as described previously (11). The DNAs migrated as a single band of high molecular weight by agarose gel electrophoresis and resulted in the production of infectious virus when transfected into BHK-21 cells (not shown). For some experiments, high-molecular-weight DNA was also prepared from infected cells.
Analyses of MHV-76 DNA.
Restriction endonuclease digestion and Southern blot hybridization were carried out using standard procedures. Cosmid clones of MHV-68 and MHV-76 DNAs were prepared as described previously (7) using infected-cell DNA and were identified by standard colony hybridization protocols. Pertinent cosmids were an MHV-68 cosmid (cA8) containing nucleotides 115165 to 26842 and an MHV-76 cosmid (cM1) containing the region equivalent to nucleotides 115587 to 26842. Both cosmids originated from circular or concatemeric forms and thus contain both ends of the genome. Nucleotide coordinates are given according to the genome sequence published by Virgin et al. (42).
The DNA sequence of cosmid M1 was determined by standard procedures of sonication, end repair, and cloning into bacteriophage M13mp19. Inserts were sequenced using an ABI Prism 377 DNA sequencer, and the data were assembled into the finished sequence using the Sequence Assembly Program (30).
Determination of viral DNA in tissue samples.
Viral DNA was extracted from blood and tissues using QIAmp DNA minikits (Qiagen) according to the manufacturer's instructions. DNA quantification was performed using a DNA fluorimeter (DyNA Quant 200; Pharmacia). PCR amplification was performed on 1 μg of high-molecular-weight DNA using primers specific for the gp150 or M3 gene as described below. PCR products (15 μl) were electrophoresed through 1.5% agarose gels and analyzed by Southern blot hybridization to confirm the specificity of the products. DNA was blotted onto Hybond N+ membranes (Amersham) and hybridized to 32P-labeled probes specific for gp150 or M3.
PCR analysis of viral genomes.
PCR amplification was performed using 1 μg of viral DNA as the template for 40 cycles under conditions described previously (38). In addition, negative control reactions for all primer sets were performed using water instead of template DNA (not shown). Primer sequences were as follows: for M1, 5′-GTT ACC TAG GAC ATA CAG TGG-3′ and 5′-CAG AAC CTT ACC AGT CAT GTG-3′ (product size, 279 bp); for M2, 5′-GCG GGA TCC ATG GCC CCA ACA CCC CCA C-3′ and 5′-GCG GAA TTC GTT ATG TTC TGC GTT AGC ACC-3′ (product size, 654 bp); for M3, 5′-TGG CAC TCA AAC TTG GTT GTG G-3′ and 5′-TAA CAG GCA GAT TGC CAT TCC C-3′ (product size, 381 bp); for M4, 5′-GCG CGG ATC CGA CAC CTG GAG AAG ATG ATG ATA TTC C-3′ and 5′-CGC GGA ATT CGG TTC TAG AAA GTC ATA AAT CTC AAT ACC-3′ (product size, 1,452 bp); for gp150, 5′-GCG CAA GCT TCG CCG CCA CCA TGT GTG GCG TTA AAT-3′ and 5′-CGC GCT CGA GTT ATT CAT GTA AAC ACA CAC AG-3′ (product size, 1,533 bp); and for ORF 74, 5′-GCC ACG ATG CTT GTC CTG CG-3′ and 5′-TTA GGA GCT TAG TCT ACA AAC TG-3′ (product size, 1,010 bp).
A modification was used to analyze viral plaques or viral stocks. The virus sample (10 μl; approximately 10 PFU) was added to PCR mix without Taq DNA polymerase. Proteinase K (Roche) was then added to 0.2 μg/μl. After incubation at 65°C for 15 min and then at 95°C for 10 min, 1 U of Taq DNA polymerase (Gibco/BRL) was added and 40 cycles of amplification were performed.
Construction of rescuant viruses.
MHV-76 genomic DNA and cosmid A8 were cotransfected into BHK-21 cells by electroporation. After 6 days, the monolayer was harvested and the supernatant was used to infect BALB/c mice. After 14 days, the spleens were harvested and analyzed by infective-center assay. Individual plaques containing rescued virus were identified by PCR amplification of the M2 and ORF 74 open reading frames (ORFs). Viruses were plaque purified by limiting-dilution assay on BHK-21 cells until they were homogeneous by PCR screening.
Southern analysis.
High-molecular-weight DNA was isolated from infected BHK-21 cells and cut with restriction enzymes according to the instructions of the manufacturer (Gibco/BRL). Southern analysis was then performed by established procedures (27), and probes were labeled with 32P using a random-primed DNA labeling kit (Roche).
In vitro infections.
Single-step growth curves were obtained by infecting subconfluent BHK-21 cells at a multiplicity of infection (MOI) of 5. After adsorption for 1 h, the wells were washed with medium to remove unbound virus and fresh medium was added. At various times p.i., the wells were harvested and infectious virus was quantified by plaque assay as described below. All experiments were carried out in duplicate.
Infection of mice and analysis of tissues.
BALB/c mice were purchased from Bantin and Kingman and infected when 4 to 6 weeks old. Mice were anesthetized with halothane and inoculated intranasally with 2 × 105 PFU of virus in 40 μl of sterile phosphate-buffered saline. At various times p.i., mice were euthanatized by cervical dislocation and tissues were harvested for analysis. Plaque assays were performed using BHK-21 cells to detect infectious virus as described previously (34). The limit of detection of the plaque assay was 10 PFU per organ. An infective-center assay was used to detect latent virus as described previously (34). The data were analyzed statistically using the two-sample t test. P values are indicated in Results.
Histopathology.
After euthanasia by CO2 asphyxiation, the lungs were perfused in situ via the trachea using 10% neutral buffered Formol saline. Portions of spleen were also fixed in 10% buffered Formol saline, and the tissues were processed routinely into paraffin wax-embedded sections. These were stained with hematoxylin and eosin and examined by light microscopy.
Nucleotide sequence accession number.
The sequence of the MHV-76 insert in cosmid M1 was deposited with the GenBank data library under accession number AF324455.
RESULTS
The MHV-76 genome is essentially identical to that of MHV-68, except for a 9,538-bp deletion at the left end of the unique region.
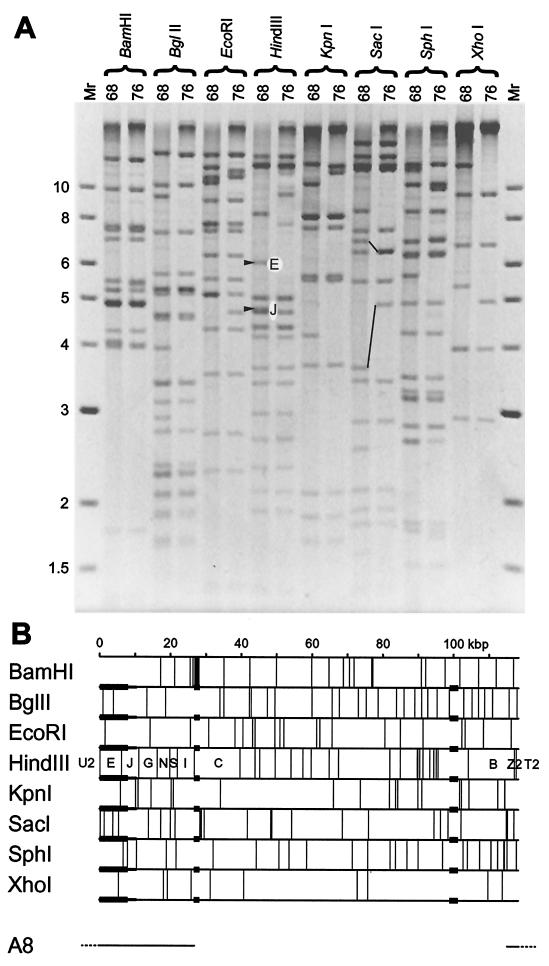
To assess the genetic differences between MHV-68 and MHV-76, we compared the restriction fragment banding patterns of the two strains. Figure 1A shows the products of MHV-68 and MHV-76 DNAs digested with eight restriction endonucleases cleaving at a total of 185 unique sites. Each enzyme produced several fragments of identical mobility from both DNAs, indicating that the genomes are closely related. However, differences in the restriction profiles were apparent. Using maps predicted from the MHV-68 genome sequence, three regions of difference were identified. These are summarized in Fig. 1B.
FIG. 1.
Structure of the MHV-76 genome. (A) Restriction endonuclease profiles of MHV-68 and MHV-76 virion DNAs. Samples were analyzed on a 0.8% agarose gel. Fragments E and J are indicated in the HindIII profile of MHV-68. These are absent from the HindIII profile of MHV-76. Fragments containing the 40-bp repeat (upper pair) and the 100-bp repeat (lower pair) are connected by lines in the SacI profiles of MHV-68 and MHV-76. Molecular size markers and their sizes in kilobase pairs are indicated to the left. (B) Restriction endonuclease maps of the unique region of the MHV-68 genome, highlighting the three regions that differ in MHV-76. Variable copy numbers of the terminal repeat (not shown) flank the unique region, resulting in heterogeneous populations of fragments from the genome termini differing by increments of 1.2 kbp. None of the restriction endonucleases for which maps are shown cleave the terminal repeat. Three regions in which MHV-76 and MHV-68 differ are denoted by thicker horizontal lines. Two correspond to variations in the copy number of reiterations in the 40-bp repeat (at kbp 27) and 100-bp repeat (at kbp 100). The third represents a deletion at the left end. Minimal and maximal extents of the deletion as deduced from panel A are indicated by thicker and thinner lines, respectively. The location of the insert in MHV-68 cosmid A8 is indicated at the bottom, with terminal repeat sequences dashed, and the corresponding fragment nomenclature is included in the HindIII map.
Two regions correspond to noncoding tandem repeats containing variable copy numbers of 40- or 100-bp elements, at approximately kbp 27 and 100 in the genome, respectively. The former region was 0.4 kbp smaller in MHV-76 (approximately 10 copies fewer of the 40 bp element), and the latter region was 1.2 kbp larger (approximately 12 copies more of the 100-bp element). As examples, relevant bands are indicated for the SacI profile in Fig. 1A. Copy number variation in repeats has long been known to occur in herpesvirus genomes (for example, see reference 8).
The third region corresponds to the left end of the unique region. The profiles were consistent with the absence from the MHV-76 genome of a sequence which extends in the MHV-68 genome from a position between nucleotides 1 and 111 to a position between nucleotides 7962 and 10526. As examples, relevant bands (E and J) are indicated for the HindIII profile in Fig. 1A. The absence of sequences in HindIII E from MHV-76 and cosmid M1 (derived from MHV-76 DNA) was confirmed by Southern blot hybridization (Fig. 2A). The only other difference detected between the two genomes was the presence of an additional SphI site in the MHV-76 SphI fragment at nucleotides 32139 to 44301. This could in principle be the result of a single nucleotide difference.
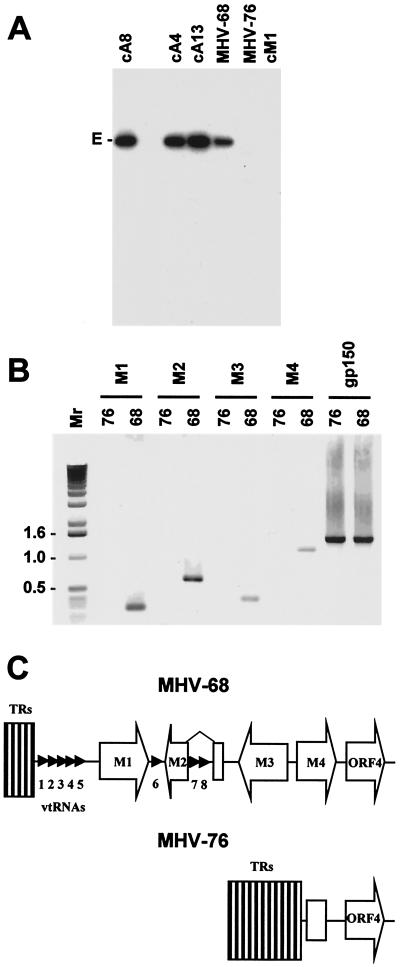
FIG. 2.
(A) Autoradiograph of a Southern blot of a 0.8% agarose gel showing the results of hybridizing radiolabeled HindIII E (isolated from a plasmid [11]) to the HindIII products of MHV-68 and MHV-76 DNAs and three MHV-68 cosmids (cA8, cA4, and cA13) and one MHV-76 cosmid (cM1) containing the left end of the genome. The HindIII E fragment (6,155 bp) is indicated. (B) PCR analysis of the MHV-68 and MHV-76 genomes. PCR amplification was performed on viral DNA templates using primers specific for M1, M2, M3, M4, and gp150 as indicated. Reaction products were analyzed on a 1% agarose gel. Molecular size markers and their sizes in kilobase pairs are indicated to the left. (C) Schematic diagram (not to scale) showing the structures of the left ends of the MHV-68 and MHV-76 genomes. The open arrows indicate protein-coding regions, and the small arrowheads indicate the vtRNA genes. The open rectangle in MHV-76 indicates residual M4 sequence. TRs, terminal repeats.
To characterize the sequence absent from MHV-76 accurately, the sequence of cosmid M1 was determined. In comparison with MHV-68, a 9,538-bp region corresponding to the last nucleotide of the terminal repeat and the first 9,537 bp of the unique region in MHV-68 was absent from the cosmid. The absence of this sequence is fully consistent with the genomic restriction profiles described above. Two minor differences were also observed between the 21,168-bp sequence of the cosmid insert and the relevant part of the MHV-68 sequence. One is trivial, mapping in a GC tract in the terminal repeat known to be variable in length (24). The other consists of a G residue at nucleotide 12280 in the coding region for MHV-68 ORF 6, which is present as an A residue in MHV-76, resulting in substitution of a serine for a glycine residue. This difference was confirmed by PCR and sequencing of the appropriate regions of the MHV-68 and MHV-76 DNAs (not shown).
We conclude that MHV-76 and MHV-68 are essentially identical, except for the absence from MHV-76 of 9,538 bp at the left end of the unique region. MHV-76 therefore represents a deletion mutant of MHV-68 or another highly related virus rather than a distinct viral species.
MHV-76 lacks MHV-68 genes M1, M2, M3, and M4 and vtRNAs.
To check that none of the sequences from the 9,538-bp deletion are present elsewhere in the genome, MHV-76 DNA was analyzed by PCR. MHV-76 and MHV-68 genomic DNAs were tested using primers specific for the M1, M2, M3, M4, and gp150 genes. The results are shown in Fig. 2B. All of the sequences assayed were amplified from MHV-68 DNA, but only the gp150 sequence was amplified from MHV-76 DNA.
These results are consistent with the findings described above. MHV-76 is thus a deletion mutant of MHV-68 that lacks M1, M2, M3, most of the 5′ portion of M4, and all eight vtRNA genes. This is depicted schematically in Fig. 2C.
MHV-76 growth in vitro is similar to that of MHV-68.
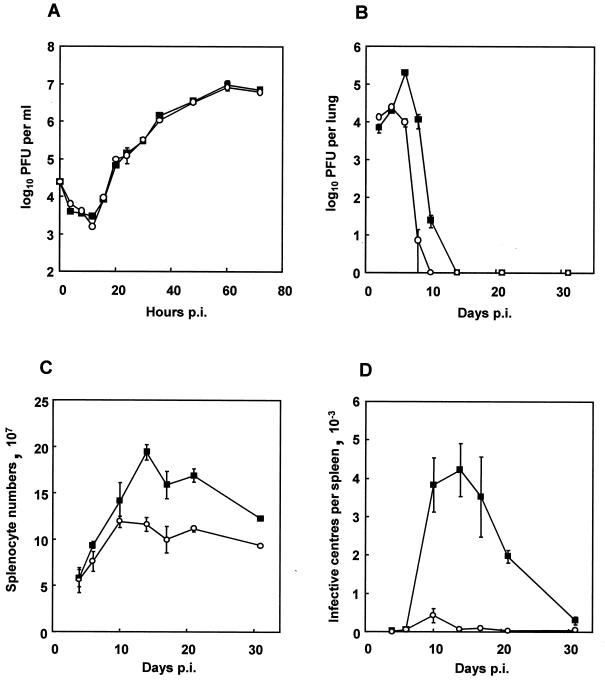
Figure 3A shows single-step growth curves determined for MHV-76 and MHV-68 in BHK-21 cells infected at an MOI of 5. As reported previously (33), MHV-68 replicated efficiently, attaining maximal titers at around 60 h p.i. MHV-76 replicated with the same kinetics and attained titers similar to those of MHV-68. Thus, there was no significant difference between MHV-68 and MHV-76 in their ability to replicate in vitro.
FIG. 3.
Biological characterization of MHV-76. (A) Single-step growth curves of MHV-68 (▪) and MHV-76 (○) on BHK-21 cells at an MOI of 5. Data are shown as mean log10 virus titer ± standard error and are representative of two separate experiments, each carried out in duplicate. (B) Viral replication in the lungs of BALB/c mice infected intranasally with 2 × 105 PFU of MHV-68 (▪) or MHV-76 (○). The mean log10 virus titer ± standard error for four mice per group is shown for each time point. (C) Numbers of spleen cells during intranasal infection of BALB/c mice with 2 × 105 PFU of MHV-68 (▪) or MHV-76 (○). The mean total number of splenocytes ± standard error for four mice per group is shown for each time point. (D) Latent virus in the spleens of BALB/c mice infected with 2 × 105 PFU of MHV-68 (▪) or MHV-76 (○) as determined by infective-center assay. Infectious virus titers (less than 50 PFU per spleen) were subtracted from the infective-center results. The mean number of infective centers per spleen ± standard error for four mice per group is shown for each time point.
MHV-76 is cleared more rapidly than MHV-68 from the lung.
The ability of MHV-76 to replicate productively in vivo was assessed. Mice were infected with MHV-76 or MHV-68, and the virus titer in the lungs was assessed at various times p.i. by plaque assay. Figure 3B shows that the titers of infectious MHV-68 peaked at day 6 p.i. and remained detectable through day 12 p.i. In contrast, MHV-76 titers peaked at day 4 p.i. and were undetectable at day 10 p.i. There was also a significant quantitative difference between the maximal titers achieved by each virus (MHV-68, 1.9 × 105 PFU/lung; MHV-76, 2.4 × 104 PFU/lung [P = 0.001]). These results indicate that MHV-76 replicates productively in vivo but is cleared more rapidly than MHV-68 from the site of primary viral replication.
MHV-76-induced splenomegaly is less marked than that induced by MHV-68.
To determine the extent of splenomegaly after MHV-76 infection, the total number of splenocytes in mice infected with MHV-76 or MHV-68 was determined. As seen in Fig. 3C, MHV-68 induced a characteristic increase in splenocyte number that peaked at 14 days p.i. MHV-76 infection also caused an increase in cellularity in the spleen, but the peak was significantly lower than that detected after MHV-68 infection (P = 0.01). In addition, maximal cell numbers were achieved slightly earlier, at between 10 and 14 days p.i.
MHV-76 establishes latency in the spleen but at much lower levels than MHV-68.
To investigate the ability of MHV-76 to establish latency in vivo, mice were infected and latent virus in the spleen was measured using an infective-center assay. The results are shown in Fig. 3D. MHV-68 infective centers were detected at around day 7 p.i. and peaked at day 14 p.i. In contrast, latent MHV-76 in the spleen peaked at day 10 p.i., and the amount of latent virus was very much less (day 10, P = 0.003; day 21, P < 0.001). MHV-76 infective centers were still detectable at 31 days p.i., at levels of approximately 5 per 107 splenocytes. This indicates that MHV-76 can establish latency in the spleen cell population but at much-reduced levels compared to MHV-68.
To check that MHV-76 did not have an altered cellular tropism in the spleen, we harvested spleens at 14 days p.i. and fractionated cells into CD19+ cells (B lymphocytes) and CD19− cells (non-B cells) using magnetic cell sorting as described previously (9). The purity of the separated fractions was tested by fluorescence-activated cell sorter analysis and found to be >95%. The two cell fractions as well as the total cell population were analyzed for the presence of latent virus using the infective-center assay. Greater than 90% of the infective centers present in the splenocytes were present within the CD19+-B-cell subpopulation (not shown). A small amount (<5%) of latent virus was also detected in the CD19− fractions. Thus, the distribution of the MHV-76 within different cell types in the spleen during splenomegaly appears to be similar to that of MHV-68 (35), in that the majority of infective centers were associated with B lymphocytes.
MHV-76 maintains long-term persistence in the spleen and lung.
The results of the infective-center assays described above demonstrated that MHV-76 was able to establish latency during the first 30 days of infection, albeit at a lower level than MHV-68. To ascertain whether MHV-76 is subsequently cleared or is able to persist, mice were infected with MHV-76 and MHV-68. After 5 months, organs from these mice were analyzed for the presence of virus. Three of the four mice were positive for MHV-76 infective centers in the spleen, at levels of approximately 1 to 2 latently infected cells per 107 splenocytes. No infectious virus was detected in this organ by plaque assay.
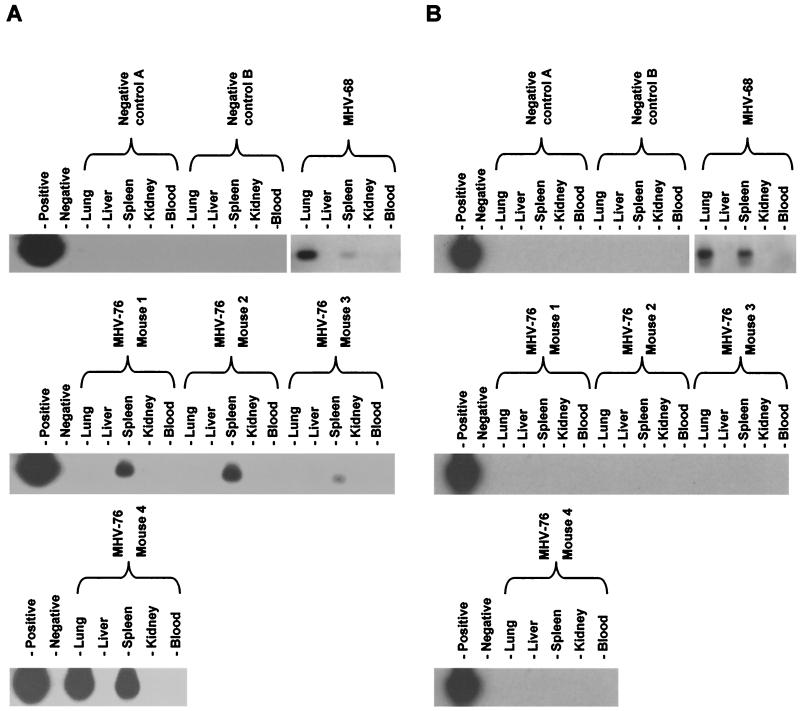
To analyze the presence of persisting virus in other organs, DNA was extracted from the livers, lungs, kidneys, spleens, and white blood cells of four MHV-76-infected mice at 5 months p.i. Organs from one mouse infected with MHV-68 were also harvested at 5 months p.i as a positive control, and those from two uninfected mice were harvested as negative controls. Extracted DNA was analyzed for the presence of the viral genome by PCR amplification using primers specific for MHV-68 gp150, followed by Southern blot hybridization with a gp150-specific probe to confirm the identity of the products. For each set of PCRs, samples containing purified viral DNA or no DNA were included as positive and negative controls, respectively. Using known amounts of cloned template, it was determined that this process was sensitive to 1 to 10 copies of the viral genome in 1 μg of high-molecular-weight DNA (not shown).
As shown in Fig. 4A, viral DNA was not detected in either of the uninfected mice. In agreement with our previous findings (32), viral genomes were detected in the spleens and lungs of the MHV-68-infected mouse but not in the blood or other organs. Viral DNA was found in spleens of all four mice infected with MHV-76 but in the lungs of only one of these mice. To address the possibility that MHV-76-infected mice may have been contaminated with MHV-68, we tested the same DNA samples by PCR using primers specific for the MHV-68 M3 gene, which is absent from MHV-76. Figure 4B shows that the M3 sequence was detected only in the MHV-68-infected mouse and not in any of the MHV-76-infected or uninfected mice. The inability to detect the M3 sequence confirmed that the MHV-76 viral stocks were free of MHV-68 and that the persisting virus in MHV-76-infected mice was definitely MHV-76.
FIG. 4.
Detection of MHV-76 DNA in the tissues of mice at 5 months p.i. Four BALB/c mice were infected with 2 × 105 PFU of MHV-76, and the lungs, livers, spleens, kidneys, and blood were harvested at 5 months p.i. Two uninfected mice were also harvested as negative controls, and one mouse infected 5 months previously with MHV-68 was harvested as a positive control. Viral DNA was extracted, and PCR amplification was performed on 1 μg of high-molecular-weight DNA. Samples containing MHV-68 viral DNA as a template (first lanes) or lacking DNA (second lanes) were used as positive and negative controls, respectively. (A) PCR amplification using primers specific for gp150. PCR products were analyzed by Southern blot hybridization with a 32P-labeled gp150 probe to confirm specificity. (B) PCR amplification using primers specific for M3 (a gene absent from MHV-76). PCR products were analyzed by Southern blot hybridization with a 32P-labeled M3 probe to confirm specificity.
These results show that MHV-76 is able to persist as efficiently as MHV-68 in the spleen but that it was not as efficient at persisting in lungs.
The inflammatory response is greater in the lung during MHV-76 infection than during MHV-68 infection.
The results described above show that there were clear differences between MHV-76 and MHV-68 in their ability to replicate productively in the lung and cause splenomegaly. This was probably due to differences in the host response to the virus, as there was no intrinsic difference in the ability of the viruses to replicate in vitro. As a preliminary response to this hypothesis, we undertook a histopathological study of organs after infection, examining the lungs and spleens of infected mice at various times p.i. Representative results are shown in Fig. 5.
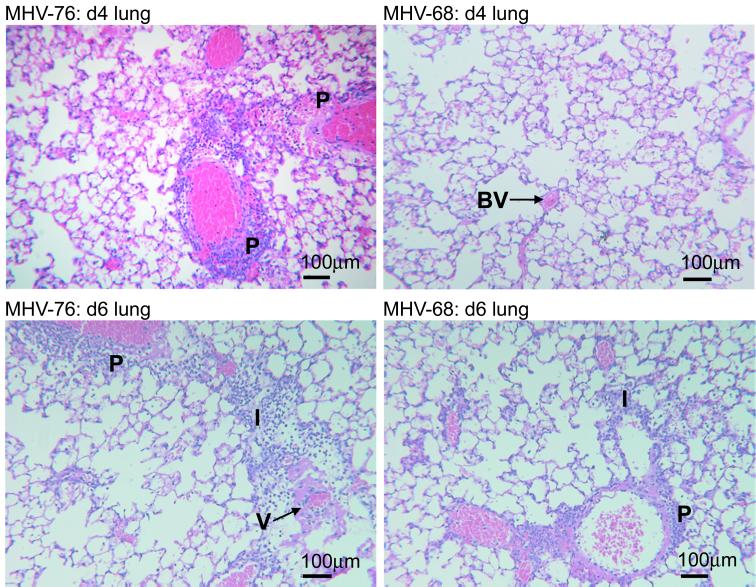
FIG. 5.
Histopathological changes in the lungs of mice infected with MHV-76 or MHV-68. Representative sections of lung and spleen stained with hematoxylin and eosin are shown for mice infected for the times indicated. Salient pathological features are highlighted as follows: P, perivascular infiltration; BV, blood vessel; I, interstitial infiltration; V, vasculitis.
At 4 days p.i. with MHV-76, there was a marked inflammatory response in the lungs, consisting of perivascular lymphoid infiltrates and edema. Subpleural lymphoid accumulations were also seen at this time p.i. (not shown). In contrast, no inflammatory response was observed at day 4 p.i. in the lungs of MHV-68-infected mice. By day 6 p.i., the inflammatory changes seen during MHV-76 infection were more severe and extensive than those on day 4, consisting of perivascular and interstitial lymphoid infiltration, thickened alveolar walls, and vasculitis. In contrast, the inflammatory changes were less pronounced in MHV-68 infection at day 6 p.i., consisting mostly of perivascular and interstitial lymphoid aggregations. At later times (days 8 and 10 p.i.), the inflammatory responses following MHV-76 and MHV-68 infections were similar (not shown).
These results demonstrate that the inflammatory changes seen in the infected lung occur earlier and are more substantial with MHV-76 than with MHV-68.
In the spleens of both MHV-68- and MHV-76-infected mice at day 10 p.i., there was evidence of follicular hyperplasia as revealed by germinal center formation and an increase in the marginal zone of the white pulp. However, the differences between the two groups, if any, were marginal and hard to quantify by histopathology alone (not shown).
Construction of rescuant viruses.
To determine definitively whether the differences observed between MHV-76 and -68 were due to deletion of genes at the left end of the genome or to unidentified mutations elsewhere in the genome, we constructed rescuants by cotransfecting MHV-76 genomic DNA with MHV-68 cosmid A8 into BHK-21 cells. Since rescuants in which the deletion was restored were likely to be less attenuated in vivo, the progeny were passaged in mice. Spleens were harvested, and virus was reactivated using an infective-center assay. Plaques were screened for the presence or absence of the M2 gene by PCR and assessed as representing pure viral clones on the basis of the presence or absence of M2 from all subsequent plaques.
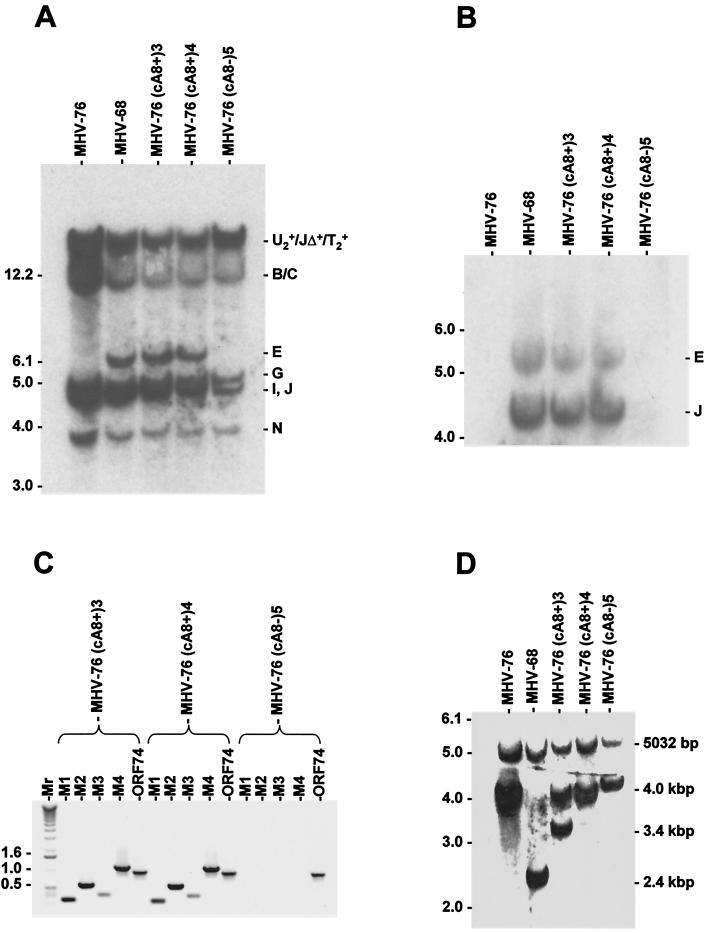
Three progeny clones were selected for further characterization. MHV-76(cA8+)3 and MHV-76(cA8+)4 were both positive for M2 and thus represented rescuants in which the deleted region had been restored. MHV-76(cA8-)5 was negative for M2 and thus represented unrestored MHV-76, which had been subjected to in vivo passage in precisely the same way as the rescuants. To check the genome structures, HindIII and EcoRI digests were analyzed by Southern blot hybridization using MHV-68 cosmid A8 as a probe. The results are shown in Fig. 6A. Cosmid A8 contains sequences from MHV-68 HindIII B, C, E, G, I, J, N, S, T2, U2, and Z2 (Fig. 1B). HindIII E, J, and U2 are absent from MHV-76, and the remaining part of HindIII J is present in a novel fragment containing the terminal repeats. This fragment (JΔ) and T2 should thus appear as a faint ladder of bands increasing to substantial sizes in increments of 1.2 kbp. As expected, rescuant viruses MHV-76(cA8+)3 and MHV-76(cA8+)4 yielded profiles identical to that of MHV-68, and HindIII E was absent from MHV-76 and MHV-76(cA8-)5. HindIII S and Z2 were too small to be present on the blot. Hybridization to fragments containing the terminal repeats (U2+, T2+, and JΔ+) appeared as high-molecular-weight products, and the laddering normally associated with such fragments was too weak to be detected. The same blot was reprobed with a fragment containing full-length M3 which crosses the boundary between HindIII E and J. Figure 6B shows that both fragments are present in MHV-68 and the two rescuants but absent from MHV-76 or MHV-76(cA8-)5. These data indicate that the sequence absent from MHV-76 was restored in MHV-76(cA8+)3 and MHV-76(cA8+)4.
FIG. 6.
Molecular characterization of rescuant viruses. (A) Autoradiograph of a Southern blot of a 0.8% agarose gel showing the results of hybridizing radiolabeled MHV-68 cosmid A8 to the HindIII products of DNAs isolated from BHK-21 cells infected with MHV-76, MHV-68, MHV-76(cA8+)3, MHV-76(cA8+)4, and MHV-76(cA8-)5. The sizes of molecular size markers (in kilobase pairs) are shown on the left, and HindIII fragments are shown on the right (see Fig. 1B for a map). (B) Autoradiograph of a Southern blot of a 0.8% agarose gel showing the results of hybridizing radiolabeled M3 probe (nucleotides 6060 to 7277 in the MHV-68 genome) to the HindIII products of DNA isolated from BHK-21 cells infected with MHV-76, MHV-68, MHV-76(cA8+)3, MHV-76(cA8+)4, or MHV-76(cA8-)5. The sizes of molecular size markers (in kilobase pairs) are shown on the left, and HindIII fragments are shown on the right (see Fig. 1B for a map). (C) PCR analysis of viral DNA from MHV-76(cA8+)3, MHV-76(cA8+)4, or MHV-76(cA8-)5 using primers specific for the MHV-68 M1, M2, M3, and M4 genes and ORF 74. Reaction products were analyzed on a 1% agarose gel. Molecular size markers and their sizes are shown to the left, with sizes in kilobase pairs. (D) Autoradiograph of a Southern blot of a 0.8% agarose gel showing the results of hybridizing a radiolabeled BamHI M probe (which contains the 100-bp repeat) to the HincII products of DNA isolated from BHK-21 cells infected with MHV-76, MHV-68, MHV-76(cA8+)3, MHV-76(cA8+)4, or MHV-76(cA8-)5. The sizes of molecular size markers (in kilobase pairs) are shown on the left. Fragments containing the 100-bp repeat, with approximate sizes, are indicated on the right, along with the adjacent 5,032-bp fragment present in each genome.
Sequence analysis of the region containing nucleotide 12280, which is a G residue in MHV-68 but an A residue in MHV-76, showed that the rescuants contain a G residue and MHV-76(cA8-)5 contains an A residue. This is consistent with rescue not only of the deleted sequences in MHV-76 by cosmid A8 but also of adjacent sequences.
We also analyzed viral DNA by PCR using primers specific for M1, M2, M3, and M4. The results are shown in Fig. 6C. ORF 74 primers amplified fragments of the anticipated size from all three viral DNA samples. Primers specific for M1, M2, M3, and M4 generated appropriately sized products from the rescuants but not from MHV-76(cA8-)5. PCR of the viral stocks produced the same results (not shown). This confirms that M1, M2, M3, and M4 were restored in the rescuants but not in MHV-76(cA8-)5.
Evidence that the rescuants were derived from MHV-76.
Variation in the copy number of reiterated sequences has been shown to occur in MHV-68 (1). Therefore, we utilized the copy number of the 100-bp repeat to assess the origin of the rescuants. Viral DNA was digested with HincII and analyzed by Southern blot hybridization using a radiolabeled BamM probe, which is predicted to hybridize to a fragment containing the 100-bp repeat and to an adjacent fragment of 5,032 bp. The results are shown in Fig. 6D. The MHV-68 fragment containing the 100-bp repeat was approximately 2.4 kbp in size, whereas that derived from MHV-76 was 4 kbp. A fragment of approximately 4 kbp was identified in both rescuants and MHV-76(cA8-)5, except that MHV-76(cA8+)3 also produced a fragment of 3.4 kbp. These results show that the copy number in MHV-68 DNA is approximately 22, as described previously (42). However, MHV-76 and three derived viruses contained a larger number (approximately 37 copies), and a subpopulation of MHV-76(cA8+)3 also contained an intermediate number (approximately 30). These results are consistent with derivation of the rescuants from rescued MHV-76 rather than from contaminating MHV-68. Since MHV-76(cA8+)3 contained variable numbers of this repeat domain, we did not include this rescuant in any further biological analyses.
The viruses are indistinguishable in vitro.
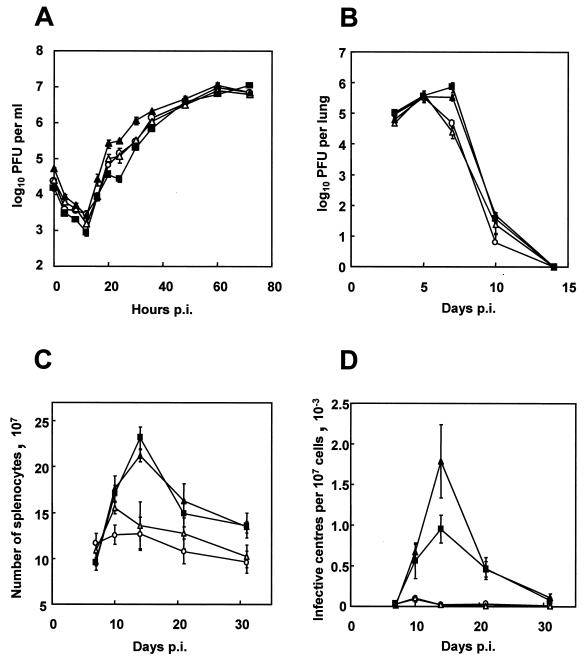
To investigate the effect of the deletion on viral replication in vitro, single-step growth curves of MHV-68, MHV-76, MHV-76(cA8+)4, and MHV-76(cA8-)5 were compared (Fig. 7A). All four viruses replicated with the same kinetics and attained similar maximal titers at around 60 h p.i. There was no indication that the deleted sequence had any effect on lytic replication in BHK-21 cells.
FIG. 7.
Biological characterization of MHV-68, MHV-76, and the rescuant viruses. Data points (mean value ± standard error) are shown in each graph for MHV-68 (▪), MHV-76 (○), MHV-76(cA8+)4 (▴), and MHV-76(cA8-)5 (▵). (A) Single-step growth curves comparing growth of viruses on BHK-21 cells at an MOI of 5. The data are representative of two separate experiments, and each experiment was done in duplicate. (B) Viral replication in the lungs of BALB/c mice infected intranasally with 2 × 105 PFU of virus. Data for four mice per group are shown at each time point. (C) Numbers of spleen cells during intranasal infection of BALB/c mice with 2 × 105 PFU of virus. Data for four mice per group are shown at each time point. (D) Latent virus in the spleens of BALB/c mice infected with 2 × 105 PFU of virus as determined by infective-center assay. Infectious virus titers (less than 50 PFU per spleen in every case) have been subtracted from the infective-center results. Data for four mice per group are shown at each time point.
A lack of M1, M2, M3, M4, and the vtRNA genes leads to attenuation in the lung.
Mice were infected with MHV-68, MHV-76, MHV-76(cA8+)4, or MHV-76(cA8-)5, and the virus titer in the lungs was assessed at various times p.i. by plaque assay. As shown in Fig. 7B, MHV-68 and MHV-76(cA8+)4 achieved peak titers at around 7 days p.i. and were cleared by 14 days p.i. MHV-76 and MHV-76(cA8-)5 achieved similar maximum titers, but the decrease in titer from this peak occurred earlier and was more rapid [MHV-68 versus MHV-76 at day 7 p.i., P = 0.037; MHV-76(cA8+)4 versus MHV-76 at day 7 p.i., P = 0.010]. These data show that the deletion leads to a significant attenuation of infection in the lungs.
Deletion of M1, M2, M3, M4, and the vtRNA genes severely attenuates virus pathogenicity in the spleen.
Mice were infected with MHV-68, MHV-76, MHV-76(cA8+)4, or MHV-76(cA8-)5, and the extent of ensuing splenomegaly was determined by counting the total number of splenocytes and the increase in latently infected cells as monitored by infective-center assay. The results are shown in Fig. 7C and D. After infection with MHV-68 or MHV-76(cA8+)4, there was a sharp increase in splenocyte number, peaking at around 14 days p.i. However, after infection with MHV-76 or MHV-76(cA8-)5, there was only a modest rise in splenocyte number, peaking earlier at around day 10 p.i. [MHV-68 versus MHV-76, P = 0.003; MHV-76(cA8+)4 versus MHV-76, P = 0.005].
In mice infected with MHV-68 and MHV-76(cA8+)4, infective centers rose sharply from day 7 p.i. to a peak at day 14 p.i. Infective centers from mice infected with MHV-76 and MHV-76(cA8-)5 were detectable in the spleen, but the peak occurred earlier (day 10 p.i.) and was significantly less [MHV-68 versus MHV-76, P = 0.002; MHV-76(cA8+)4 versus MHV-76, P = 0.008]. However, latently infected cells were detected in mice infected with MHV-76(cA8-)5 or MHV-76 even at the latest time point tested (32 days p.i.). These data show that the deleted sequence contributes significantly to splenomegaly and the acute rise in latently infected cells in the spleen but is not essential for the establishment of latency and persistence.
DISCUSSION
This work describes a primary analysis of a novel murine gammaherpesvirus, MHV-76. The genome of this virus was found to be essentially identical to that of MHV-68, with the single major difference being the absence of 9,538 bp at the left end of the unique region of the genome. MHV-76 replicated similarly in cell culture and had similar biological properties as MHV-68, but it appeared to be significantly less pathogenic in vivo in the context of a much more rapid and intense inflammatory response in the lungs. Recombinant viruses that had the deletion in MHV-76 rescued had biological properties similar to those of MHV-68.
MHV-68 was reported as having been isolated from the bank vole, C. glareolus (3), whereas MHV-76 was described as an independent isolate derived in the same study from the yellow-necked wood mouse, A. flavicollis. Our data strongly suggest that MHV-76 is a deletion mutant derived from MHV-68 or an extremely similar virus during passage. It is formally possible that this could have occurred in nature, since Clethrionomys and Apodemus species share similar habitats. This question is an important one and is currently being addressed by the investigation of free-living rodent samples from the United Kingdom.
MHV-76 lacks 9,538 bp containing the M1, M2, M3, M4, and vtRNA genes. We have shown that MHV-76 and MHV-68 differ dramatically in their pathogenic potential in mice. This could be due to the deletion or to unidentified mutations elsewhere in the genome. Consequently, we utilized rescuants in which the deleted region in MHV-76 had been restored by recombination with MHV-68 sequences. Replacement of the deleted sequence changed the phenotype of MHV-76 to one that was not significantly different from that of MHV-68. This shows definitively that the left end of the MHV-68 genome is nonessential for viral productive replication but plays a key role in pathogenesis. It is overwhelmingly likely that the key determinants are located in the deleted sequence. However, our results have not formally ruled out the possibility that the single nucleotide substitution at nucleotide 12280, resulting in replacement of a glycine by a serine residue in the ORF 6 protein, may play a role. We consider this unlikely, since a deleterious point mutation (unlike a deletion) can readily revert and is unlikely to survive under selection in mice. We are also encouraged to view this residue as noncritical, since it is not conserved in an independently isolated murine herpesvirus in which the ORF 6 protein is 91.3% identical to that of MHV-68 (it is an asparagine [S. Milligan, C. Chastel, S. Efstathiou, and A. J. Davison, unpublished data]).
The left end of the MHV-68 genome appears to have a propensity for undergoing recombination events in vitro (28). This could be due to the proximity of multiple terminal repeats or to the presence of highly transcribed vtRNA genes or a putative origin of replication (4). Naturally occurring deletions have been described for other gammaherpesvirus genomes. The strains of Epstein-Barr virus in the Raji, Daudi, and P3HR-1 cell lines all carry deletions of various sizes (15). For HVS, spontaneous deletions at the left end of the genome have been described (16). Indeed, the heterogeneity of the left end of the HVS genome has been held responsible for the oncogenic potential of the virus (19, 20). It is also of note that the left end of the HVS genome encodes seven small viral RNAs (2).
Unique genes at the left end of the genome have attracted considerable attention in other gammaherpesviruses as well as in MHV-68. In Kaposi's sarcoma-associated herpesvirus, this region contains the K1 gene, which shows unusual levels of divergence in different virus isolates (45) and has been associated with cellular transformation (17). In HVS, this region contains the STP gene, which is required for cellular immortalization and oncogenesis (16, 23), as well as the HVS small viral RNAs (2). In Epstein-Barr virus, this region encodes LMP-1, a classic oncoprotein that is essential for B-cell immortalization (15). For MHV-68, however, less is known about the function of the genes in this region and their contribution to viral pathogenesis. Indeed, only the function of M3 has been identified, as a soluble chemokine binding protein (25, 40) potentially subverting the inflammatory response. Deletion of M1 had no phenotypic effect on viral replication in vitro or in vivo (6, 28), although one study implicated this region of the genome in the regulation of latency (6). Also, deletion of vtRNA genes 1 to 4 had no effect on viral pathogenesis (28). Our studies confirm and extend these findings in demonstrating that genes M1, M2, M3, and M4 and all eight vtRNA genes are nonessential for lytic virus replication in vitro and in vivo and for the establishment and maintenance of latency in vivo.
Functions encoded at the left end of the MHV-68 genome are nevertheless involved in virus pathogenicity. The more rapid clearance from the lungs characteristic of MHV-76, combined with an enhanced inflammatory response, indicates a key role for this region in the avoidance of restrictions placed on viral productive replication, for example, via interferons or the immune response. The M1, M3, M4, and vtRNA genes are transcribed during lytic infection (29, 41) and thus potentially play roles in infection of the lung. Previous studies have analyzed MHV-68 mutants lacking M1 (6, 28) but did not assess viral replication in the lung after intranasal infection.
The dramatically smaller extent of splenomegaly and lower number of infective centers observed during MHV-76 infection constitute the first evidence that genes at the left end of the MHV-68 genome are specifically involved in splenomegaly and generation of latently infected B cells. It is not clear, however, precisely which of the known genes is responsible. M2 and the vtRNA genes are candidates, since they are transcribed during latent infection of splenocytes (14, 29). M2 is also expressed predominantly in B cells during splenomegaly (38). M1, M3, and M4 have not been shown definitively to be associated with latency (14, 41) but may still have a role in splenomegaly, since low levels of productive viral replication occur in the spleen during the establishment of latency. Chemokines play an important role in directing the migration of leukocytes to sites of inflammation (18). Thus, the chemokine-binding actions of M3 may play an important role in splenomegaly. One report has implicated M1 in the suppression of viral reactivation (6). However, the effect was observed only at later time points in peritoneal exudate cells after intraperitoneal infection. In addition, that study employed recombinant viruses containing the human cytomegalovirus immediate-early promoter, which may in itself affect MHV-68 replication. A suppressive effect of M1 was not observed in our experiments.
Both MHV-76 and MHV-68 persisted in the spleens of infected mice for at least 5 months p.i. Thus, the genetic elements necessary for this process are clearly not present in the left unique portion of the genome. However, the persistence of MHV-76 in the lungs was much less consistent, occurring in only one of four mice. Long-term persistence of virus in the lung is a consistent feature of MHV-68 infection (32). Thus, although the numbers are low, our data suggest that sequences missing in MHV-76 are important for long-term persistence in the lung. The more rapid and intense inflammatory response seen in the lung during MHV-76 infection may indicate that the virus is more effectively cleared from this site by the host response. In addition, there may be subtle variations in genome copy number in the spleen that are influenced by the locus absent in MHV-76. These questions await the results of quantitative experiments on DNA load over an extended time course.
In summary, we have demonstrated that a region of the genome containing M1, M2, M3, M4, and the vtRNA genes plays a key role in infection in mice by MHV-68, probably though evasion of the host innate defense mechanisms and immune response. Ongoing studies in this laboratory are expected to reveal the contributions of individual genes. MHV-76 is anticipated to be a useful virus from which to construct appropriate mutants.
ACKNOWLEDGMENTS
This work was funded by the Wellcome Trust, The Royal Society, and the MRC (United Kingdom). A.I.M. holds a Wellcome Trust Veterinary Clinical Scholarship. J.P.S. is a Royal Society University Research Fellow.
REFERENCES
- 1.Adler H, Messerle M, Wagner M, Koszinowski U H. Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J Virol. 2000;74:6964–6974. doi: 10.1128/jvi.74.15.6964-6974.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albrecht J C, Nicholas J, Biller D, Cameron K R, Biesinger B, Newman C, Wittmann S, Craxton M A, Coleman H, Fleckenstein B, et al. Primary structure of the herpesvirus saimiri genome. J Virol. 1992;66:5047–5058. doi: 10.1128/jvi.66.8.5047-5058.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Blaskovic D, Stancekova M, Svobodova J, Mistrikova J. Isolation of five strains of herpesviruses from two species of free living small rodents. Acta Virol. 1980;24:468. [PubMed] [Google Scholar]
- 4.Bowden R J, Simas J P, Davis A J, Efstathiou S. Murine gammaherpesvirus 68 encodes tRNA-like sequences which are expressed during latency. J Gen Virol. 1997;78:1675–1687. doi: 10.1099/0022-1317-78-7-1675. [DOI] [PubMed] [Google Scholar]
- 5.Cardin R D, Brooks J W, Sarawar S R, Doherty P C. Progressive loss of CD8+ T cell-mediated control of a gamma-herpesvirus in the absence of CD4+ T cells. J Exp Med. 1996;184:863–871. doi: 10.1084/jem.184.3.863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clambey E T, Virgin H W, Speck S H. Disruption of the murine gammaherpesvirus 68 M1 open reading frame leads to enhanced reactivation from latency. J Virol. 2000;74:1973–1984. doi: 10.1128/jvi.74.4.1973-1984.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cunningham C, Davison A J. A cosmid-based system for constructing mutants of herpes simplex virus type 1. Virology. 1993;197:116–124. doi: 10.1006/viro.1993.1572. [DOI] [PubMed] [Google Scholar]
- 8.Davison A J, Wilkie N M. Nucleotide sequences of the joint between the L and S segments of herpes simplex virus types 1 and 2. J Gen Virol. 1981;55:315–331. doi: 10.1099/0022-1317-55-2-315. [DOI] [PubMed] [Google Scholar]
- 9.Dutia B M, Stewart J P, Clayton R A, Dyson H, Nash A A. Kinetic and phenotypic changes in murine lymphocytes infected with murine gammaherpesvirus-68 in vitro. J Gen Virol. 1999;80:2729–2736. doi: 10.1099/0022-1317-80-10-2729. [DOI] [PubMed] [Google Scholar]
- 10.Efstathiou S, Ho Y M, Hall S, Styles C J, Scott S D, Gompels U A. Murine herpesvirus 68 is genetically related to the gammaherpesviruses Epstein-Barr virus and herpesvirus saimiri. J Gen Virol. 1990;71:1365–1372. doi: 10.1099/0022-1317-71-6-1365. [DOI] [PubMed] [Google Scholar]
- 11.Efstathiou S, Ho Y M, Minson A C. Cloning and molecular characterization of the murine herpesvirus 68 genome. J Gen Virol. 1990;71:1355–1364. doi: 10.1099/0022-1317-71-6-1355. [DOI] [PubMed] [Google Scholar]
- 12.Ehtisham S, Sunil-Chandra N P, Nash A A. Pathogenesis of murine gammaherpesvirus infection in mice deficient in CD4 and CD8 T cells. J Virol. 1993;67:5247–5252. doi: 10.1128/jvi.67.9.5247-5252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Flano E, Husain S M, Sample J T, Woodland D L, Blackman M A. Latent murine gamma-herpesvirus infection is established in activated B cells, dendritic cells, and macrophages. J Immunol. 2000;165:1074–1081. doi: 10.4049/jimmunol.165.2.1074. [DOI] [PubMed] [Google Scholar]
- 14.Husain S M, Usherwood E J, Dyson H, Coleclough C, Coppola M A, Woodland D L, Blackman M A, Stewart J P, Sample J T. Murine gammaherpesvirus M2 gene is latency-associated and its protein a target for CD8(+) T lymphocytes. Proc Natl Acad Sci USA. 1999;96:7508–7513. doi: 10.1073/pnas.96.13.7508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kieff E. Epstein-Barr virus and its replication. In: Fields B N, Knipe D M, Howley P M, editors. Fields virology. Vol. 2. Philadelphia, Pa: Lippincott-Raven; 1996. pp. 2343–2396. [Google Scholar]
- 16.Koomey J M, Mulder C, Burghoff R L, Fleckenstein B, Desrosiers R C. Deletion of DNA sequence in a nononcogenic variant of herpesvirus saimiri. J Virol. 1984;50:662–665. doi: 10.1128/jvi.50.2.662-665.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee H, Veazey R, Williams K, Li M, Guo J, Neipel F, Fleckenstein B, Lackner A, Desrosiers R C, Jung J U. Deregulation of cell growth by the K1 gene of Kaposi's sarcoma-associated herpesvirus. Nat Med. 1998;4:435–440. doi: 10.1038/nm0498-435. [DOI] [PubMed] [Google Scholar]
- 18.Luster A D. Chemokines—chemotactic cytokines that mediate inflammation. N Engl J Med. 1998;338:436–445. doi: 10.1056/NEJM199802123380706. [DOI] [PubMed] [Google Scholar]
- 19.Medveczky M M, Szomolanyi E, Hesselton R, DeGrand D, Geck P, Medveczky P G. Herpesvirus saimiri strains from three DNA subgroups have different oncogenic potentials in New Zealand White rabbits. J Virol. 1989;63:3601–3611. doi: 10.1128/jvi.63.9.3601-3611.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Medveczky P, Szomolanyi E, Desrosiers R C, Mulder C. Classification of herpesvirus saimiri into three groups based on extreme variation in a DNA region required for oncogenicity. J Virol. 1984;52:938–944. doi: 10.1128/jvi.52.3.938-944.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mistrikova J, Rajcani J, Mrmusova M, Oravcova I. Chronic infection of Balb/c mice with murine herpesvirus 72 is associated with neoplasm development. Acta Virol. 1996;40:297–301. [PubMed] [Google Scholar]
- 22.Mistrikova J, Remenova A, Lesso J, Stancekova M. Replication and persistence of murine herpesvirus 72 in lymphatic system and peripheral blood mononuclear cells of Balb/C mice. Acta Virol. 1994;38:151–156. [PubMed] [Google Scholar]
- 23.Murthy S C, Trimble J J, Desrosiers R C. Deletion mutants of herpesvirus saimiri define an open reading frame necessary for transformation. J Virol. 1989;63:3307–3314. doi: 10.1128/jvi.63.8.3307-3314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nash, A. A., B. M. Dutia, J. P. Stewart, and A. J. Davison. Natural history of murine gammaherpesvirus infection. Philos. Trans. R. Soc. Lond. B, in press. [DOI] [PMC free article] [PubMed]
- 25.Parry C M, Simas J P, Smith V P, Stewart C A, Minson A C, Efstathiou S, Alcami A. A broad spectrum secreted chemokine binding protein encoded by a herpesvirus. J Exp Med. 2000;191:573–578. doi: 10.1084/jem.191.3.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Raslova H, Mistrikova J, Kudelova M, Mishal Z, Sarasin A, Blangy D, Berebbi M. Immunophenotypic study of atypical lymphocytes generated in peripheral blood and spleen of nude mice after MHV-72 infection. Viral Immunol. 2000;13:313–327. doi: 10.1089/08828240050144644. [DOI] [PubMed] [Google Scholar]
- 27.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 28.Simas J P, Bowden R J, Paige V, Efstathiou S. Four tRNA-like sequences and a serpin homologue encoded by murine gammaherpesvirus 68 are dispensable for lytic replication in vitro and latency in vivo. J Gen Virol. 1998;79:149–153. doi: 10.1099/0022-1317-79-1-149. [DOI] [PubMed] [Google Scholar]
- 29.Simas J P, Swann D, Bowden R, Efstathiou S. Analysis of murine gammaherpesvirus-68 transcription during lytic and latent infection. J Gen Virol. 1999;80:75–82. doi: 10.1099/0022-1317-80-1-75. [DOI] [PubMed] [Google Scholar]
- 30.Staden R. Computer handling of DNA sequencing projects. In: Bishop M J, Rawlings C J, editors. Nucleic acid and protein sequence analysis: a practical approach. Oxford, United Kingdom: IRL Press; 1987. pp. 173–217. [Google Scholar]
- 31.Stewart J P. Of mice and men: murine gammaherpesvirus 68 as a model. EBV Rep. 1999;6:31–35. [Google Scholar]
- 32.Stewart J P, Usherwood E J, Ross A, Dyson H, Nash T. Lung epithelial cells are a major site of murine gammaherpesvirus persistence. J Exp Med. 1998;187:1941–1951. doi: 10.1084/jem.187.12.1941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sunil-Chandra N P. Studies on the pathogenesis of a murine gammaherpesvirus (MHV-68). Ph.D. thesis. Cambridge, United Kingdom: University of Cambridge; 1991. [Google Scholar]
- 34.Sunil-Chandra N P, Efstathiou S, Arno J, Nash A A. Virological and pathological features of mice infected with murine gamma-herpesvirus 68. J Gen Virol. 1992;73:2347–2356. doi: 10.1099/0022-1317-73-9-2347. [DOI] [PubMed] [Google Scholar]
- 35.Sunil-Chandra N P, Efstathiou S, Nash A A. Murine gammaherpesvirus 68 establishes a latent infection in mouse B lymphocytes in vivo. J Gen Virol. 1992;73:3275–3279. doi: 10.1099/0022-1317-73-12-3275. [DOI] [PubMed] [Google Scholar]
- 36.Tripp R A, Hamilton-Easton A M, Cardin R D, Nguyen P, Behm F G, Woodland D L, Doherty P C, Blackman M A. Pathogenesis of an infectious mononucleosis-like disease induced by a murine gamma-herpesvirus: role for a viral superantigen? J Exp Med. 1997;185:1641–1650. doi: 10.1084/jem.185.9.1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Usherwood E J, Ross A J, Allen D J, Nash A A. Murine gammaherpesvirus-induced splenomegaly: a critical role for CD4 T cells. J Gen Virol. 1996;77:627–630. doi: 10.1099/0022-1317-77-4-627. [DOI] [PubMed] [Google Scholar]
- 38.Usherwood E J, Roy D J, Ward K, Surman S L, Dutia B M, Blackman M A, Stewart J P, Woodland D L. Control of gammaherpesvirus latency by latent antigen-specific CD8(+) T cells. J Exp Med. 2000;192:943–952. doi: 10.1084/jem.192.7.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Usherwood E J, Stewart J P, Robertson K, Allen D J, Nash A A. Absence of splenic latency in murine gammaherpesvirus 68-infected B cell-deficient mice. J Gen Virol. 1996;77:2819–2825. doi: 10.1099/0022-1317-77-11-2819. [DOI] [PubMed] [Google Scholar]
- 40.van Berkel V, Barrett J, Tiffany H L, Fremont D H, Murphy P M, McFadden G, Speck S H, Virgin H W., IV Identification of a gammaherpesvirus selective chemokine binding protein that inhibits chemokine action. J Virol. 2000;74:6741–6747. doi: 10.1128/jvi.74.15.6741-6747.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Virgin H W, Presti R M, Li X Y, Liu C, Speck S H. Three distinct regions of the murine gammaherpesvirus 68 genome are transcriptionally active in latently infected mice. J Virol. 1999;73:2321–2332. doi: 10.1128/jvi.73.3.2321-2332.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Virgin H W, Latreille P, Wamsley P, Hallsworth K, Weck K E, Dal Canto A J, Speck S H. Complete sequence and genomic analysis of murine gammaherpesvirus 68. J Virol. 1997;71:5894–5904. doi: 10.1128/jvi.71.8.5894-5904.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Weck K E, Barkon M L, Yoo L I, Speck S H, Virgin H W. Mature B cells are required for acute splenic infection, but not for establishment of latency, by murine gammaherpesvirus 68. J Virol. 1996;70:6775–6780. doi: 10.1128/jvi.70.10.6775-6780.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weck K E, Kim S S, Virgin IV H W, Speck S H. Macrophages are the major reservoir of latent murine gammaherpesvirus 68 in peritoneal cells. J Virol. 1999;73:3273–3283. doi: 10.1128/jvi.73.4.3273-3283.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zong J C, Ciufo D M, Alcendor D J, Wan X, Nicholas J, Browning P J, Rady P L, Tyring S K, Orenstein J M, Rabkin C S, Su I J, Powell K F, Croxson M, Foreman K E, Nickoloff B J, Alkan S, Hayward G S. High-level variability in the ORF-K1 membrane protein gene at the left end of the Kaposi's sarcoma-associated herpesvirus genome defines four major virus subtypes and multiple variants or clades in different human populations. J Virol. 1999;73:4156–4170. doi: 10.1128/jvi.73.5.4156-4170.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]