Abstract
Abstract
The aim of this paper is to summarize the available evidence in the literature regarding the effects generated by exposure to an enriched environment (EE) on the modulation of epigenetic processes in the central nervous system under adverse environmental conditions. Searches were conducted in three databases: PubMed/Medline (1053 articles), Scopus (121 articles), and Embase (52 articles), which were subjected to eligibility criteria. Of the 1226 articles found, 173 duplicates were removed. After evaluating titles/abstracts, 904 studies were excluded, resulting in 49 articles, of which 14 were included in this systematic review. EE was performed using different inanimate objects. Adverse environmental conditions included CUMS, sepsis, nicotine exposure, PCP exposure, early stress, WAS, high fructose intake, TBI, and sevoflurane exposure. Regarding microRNA expression, after exposure to EE, an increase in the expression of miR-221 and miR-483 was observed in the prefrontal cortex, and a reduction in the expression of miR-92a-3p and miR-134 in the hippocampus. Regarding histone modifications, in the hippocampus, there was a reduction of HAT, HDAC/HDAC4, H3 (acetyl K14), H4 (acetyl K15), H3K4me3, K3k27me3, and HDAC2/3/5. In the cortex, there was a reduction of HDAC2, and in the prefrontal cortex, there was an increase in acetylated H3. Regarding DNA modifications, there was a reduction of DNMT in the hippocampus. This systematic review concludes that the benefits of EE on the brain and behavior of animals are directly related to different epigenetic mechanisms, reflecting in cell growth and neuroplasticity. EE may be a non-pharmacological and easy-to-apply alternative to prevent symptoms in disorders affecting brain tissue.
Graphical abstract
Keywords: Enriched environment, Epigenetics, Histones, MicroRNA, Nervous system
Introduction
Epigenetics was initially described by Waddington in the 1940s, specifically using the term "epigenetic landscape" to refer to the events that lead to the unfolding of the genetic program, which influences the resulting phenotype through the interaction between genes and the environment (Wang et al. 2011; Feinberg and Levchenko 2023).The mechanisms involved in epigenetic regulation are related to gene expression but do not involve changes to the DNA sequence; these molecular processes include DNA methylation, histone modifications, and the expression of non-coding RNAs (Fitz-James and Cavalli 2022).
DNA methylation can be succinctly described as the covalent addition of a methyl group to one of the nucleotides in the DNA molecule, most commonly associated with the methylation of the CpG dinucleotide (cytosine followed by guanine). This modification plays a crucial role in transcriptional regulation and phenotypic inheritance, as it can prevent the binding of transcription factors while recruiting proteins that compact chromatin. Consequently, this makes the DNA inaccessible for transcriptional processes (Li and Tollefsbol 2021). Histones make up the nucleosome, the functional unit of chromatin, and can either increase or decrease its compaction, thereby playing a role in gene expression regulation. This process is mediated by numerous modifications occurring on the N-terminal tails of these histone proteins (such as acetylation, ubiquitination, and phosphorylation); acetylation is catalyzed by the enzyme acetyltransferase (HAT), while deacetylation occurs through the enzyme histone deacetylase (HDAC). These enzymes are well-documented in the literature and are important mechanisms of epigenetic inheritance (Fitz-James and Cavalli 2022; Yu et al. 2020).
The expression of non-coding RNAs is another fundamental process for modifying the epigenetic state, capable of regulating gene transcription in various ways; one such example is the synthesis of microRNAs (miRNAs), which are small RNAs (19–22 nucleotides in length) formed in the cell nucleus and transported to the cytoplasm. These miRNAs are responsible for blocking messenger RNA (mRNA) translation by binding to target genes, thereby preventing the expression of the corresponding protein (gene silencing) or inducing the degradation of the mRNA (McKibben and Dwivedi 2021; Carthew and Sontheimer 2009; Yan and Bu 2021). All these mechanisms characterize the expression of the phenotypic state and are involved in various biological processes that affect aspects of health and disease, having a profound relationship with the environment.
Currently, environmental enrichment can be considered a fundamental aspect of animal housing and management; it involves modifications that provide physical, social, and cognitive stimuli, holding importance equal to nutritional precautions. This enrichment allows animals to engage in species-specific activities, positively influencing their quality of life, both in healthy individuals and those with certain conditions, such as neurological diseases (Baumans 2005; Gomez et al. 2015).
There is a wide variety of disorders that impact the nervous system, ranging from neurodevelopmental disorders to neurodegenerative diseases that predominantly affect the elderly. Additionally, trauma and disorders secondary to other pathological conditions, such as infectious diseases that result in damage to nervous tissue, can also be mentioned (Global, regional, and national burden of disorders affecting the nervous system 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021 2024). These unfavorable conditions related to the nervous system are strongly influenced by the environment and gene regulation mediated by epigenetic factors (Nikolac Perkovic 2021; Ghosh and Saadat 2023).
Exposure to an enriched environment is associated with various benefits for the nervous system; there is evidence that an enriched environment can stimulate myelination and repair of nervous tissue by increasing the secretion of molecules that regulate the development and myelination of nerve cells. This has significant importance in various neuropathological conditions (Gao et al. 2022). Studies with animals have also demonstrated benefits in social behavior, increased synaptic density in the hippocampus, elevated expression of dopamine receptor D2, promotion of neuronal recovery, changes in extracellular concentrations of glutamate and GABA, and enhanced learning flexibility. All these effects are mediated by sensory, cognitive, and social stimuli generated by an enriched environment, which trigger adaptive responses in the nervous system that impact various neurological contexts (Han et al. 2022). The benefits of exposure to an enriched environment also include the reversal of symptoms in pre-established pathological conditions, such as sepsis, by preventing cognitive losses associated with neurodegeneration. This leads to improved brain function, as evidenced by a reduction in long-term memory impairment (Córneo et al. 2022).
Given the significant influence of the environment on modifying the epigenetic profile, the stimuli provided by environmental enrichment can alter the phenotypic state resulting from changes in gene expression. However, the underlying mechanisms are not yet fully elucidated, particularly in adverse conditions such as pathologies and injuries. One major issue is the lack of standardized protocols. Clarifying the relationship between environment and epigenetic factors could be crucial for developing new therapeutic strategies based on enriched environments.
The present study aims to conduct a systematic review to understand the impacts of environmental enrichment on key epigenetic changes in the nervous system, analyzing DNA methylation, histone modifications, and non-coding RNA expression (miRNAs) under adverse conditions.
Methods
This systematic review was conducted following the PRISMA guideline (Page et al. 2020).
Eligibility Criteria
The selection of studies followed the eligibility criteria drawn up in accordance with PECOS (Population: rodents; Exposure: adverse environmental conditions; Control: standardized environment associated with adverse conditions; Outcomes: epigenetic processes [expression of microRNAs, modifications related to histones and DNA]; Studies Design: animal studies). No limitations on publication time and language were established in the selection of studies. Original studies were selected, while dissertations or theses, study protocols, review articles, gray literature, editorials, summaries, and articles that did not evaluate rodents, brain tissues, adverse environmental conditions, or epigenetic processes, or that did not present a control group exposed to the standardized environment associated with adverse environmental conditions, were excluded.
Information Sources and Search Strategy
Three electronic databases were used to search for articles (PubMed, Scopus, and Embase). The research was carried out in June 2024. The following search equation was used: ((Enriched environment) OR (Environmental enrichment)) AND (((((((((Epigenetic Processes) OR (Epigenetic Process)) OR (Histones)) OR (Histone)) OR (MicroRNAs)) OR (Micro RNA)) OR (MicroRNA)) OR (miRNA)) OR (miRNAs)). Adaptations were made for electronic databases. No filters were used to search for articles. The search for studies was carried out in pairs, with discrepancies resolved by a third evaluator.
Selection Process
Study selection was carried out with the help of Endnote X20 software (Clarivate Analytics, Philadelphia, USA). Duplicates were removed, and then studies were screened by reading titles and abstracts. Subsequently, the full texts were evaluated. This process was carried out by two researchers, and discrepancies were resolved by a third evaluator.
Data Collection Process
Data collection took place independently by two evaluators (MSSF and GCJS). Discrepancies were resolved by a third evaluator.
Data Items (outcomes)
Data were extracted about the study (author and year), animal characteristics (species, sex, age), and information on the number of animals per cage; environmental enrichment protocol, housing dimensions (length, width, and depth or height, in centimeters or meters), and exposure time of the environmental enrichment in weeks were collected. Finally, we also added information about the brain tissue used, type of adverse environmental condition, and environmental modifications.
Methodological Quality Assessment
The SYRCLE’s strategy was used to assess the methodological quality of the animal studies. The tool consisted of ten questions that evaluate methodological criteria: Q1—Was the allocation sequence adequately generated and applied? Q2—Were the groups similar at baseline or were they adjusted for confounders in the analysis? Q3—Was the allocation to the different groups adequately concealed? Q4—Were the animals randomly housed during the experiment? Q5—Were the caregivers and/or investigators blinded from knowledge of which intervention each animal received during the experiment? Q6—Were animals selected at random for outcome assessment? Q7—Was the outcome assessor-blinded? Q8—Were incomplete outcome data adequately addressed? Q9—Are reports of the study free of selective outcome reporting? Q10—Was the study free of other problems that could result in a high risk of bias? Questions were answered with options of 'Yes,' 'No,' or 'Not clear.' When the answer was 'Yes,' a score was given; when the answer was 'No' or 'Not clear,' no score was given. The overall scores for each article were calculated on a scale of 0–10 points, with the quality of each study being classified as high (8–10), moderate (5–7), or low (< 5). The quality outcomes are described in Table 1.
Table 1.
Methodological Quality Assessment
| Author, Year | Q1 | Q2 | Q3 | Q4 | Q5 | Q6 | Q7 | Q8 | Q9 | Q10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Córneo et al., 2022 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Gomez et al., 2015 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Ji et al., 2023 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Koseki et al., 2011 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Lin et al., 2020 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Mckibben et al., 2021 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Min et al., 2022 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Orock et al., 2021 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Seo et al., 2021 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Shen et al., 2019 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Wang et al. ,2016 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Wu et al., 2016 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Xin W et al., 2018 | Y | Y | Y | Y | N | Y | N | U | Y | U |
| Yu et al., 2020 | Y | Y | Y | Y | N | Y | N | U | Y | U |
Q1. Was the allocation sequence adequately generated and applied? Q2 Were the groups similar at baseline or were they adjusted for confounders in the analysis? Q3 Was the allocation to the different groups adequately concealed during? Q4 Were the animals randomly housed during the experiment? Q5 Were the caregivers and/or investigators blinded from knowledge which intervention each animal received during the experiment? Q6 Were animals selected at random for outcome assessment? Q7 Was the outcome assessor-blinded? Q8 Were incomplete outcome data adequately addressed? Q9 Are reports of the study free of selective outcome reporting? Q10 Was the study apparently free of other problems that could result in high risk of bias? Y Yes; N No; U Unclear
Results
Methodological Quality Assessment
The quality of the included studies is shown in Table 1. All studies showed adequate and randomized allocation with randomly selected rodents. Furthermore, incomplete results were handled appropriately, free from selective results and bias. Because these are studies with adverse conditions and environmental enrichment, it is not possible to consider the investigation and analysis of the results blind. In general, all studies presented good quality criteria.
Search Results
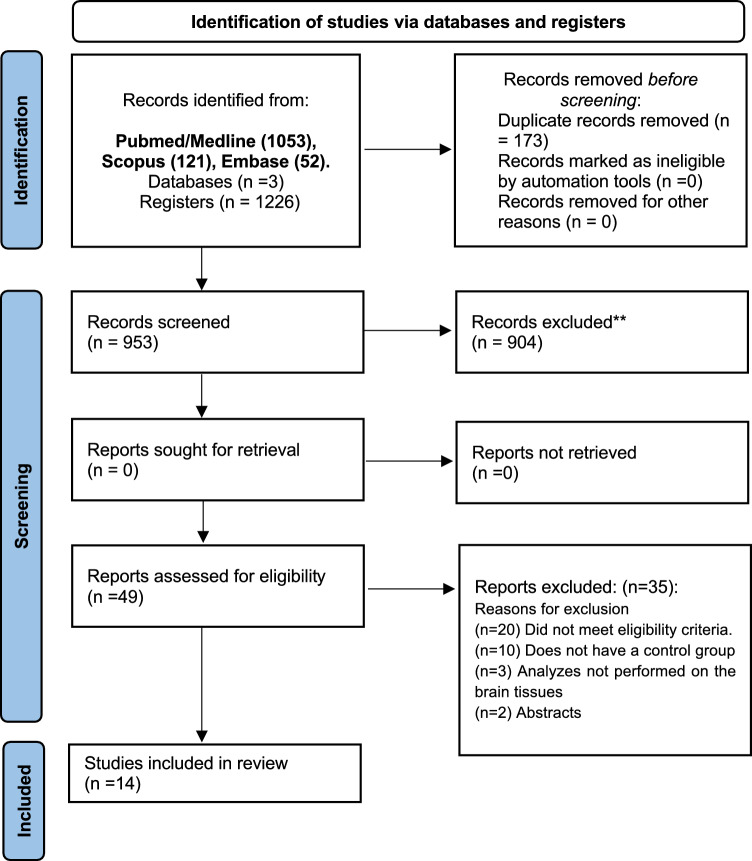
In total, 1226 articles were found in the databases [PubMed/Medline (n = 1053), Scopus (n = 121), and Embase (n = 52)]. Subsequently, 173 duplicates were removed, leaving 953 for screening. After screening, 904 articles were excluded, leaving 49 for analysis based on the eligibility criteria. Finally, 14 articles were included (Fig. 1).
Fig. 1.
PRISMA 2020 flow diagram for new systematic reviews which included searches of databases and registers only
Study Characteristics
The description of the included studies and environmental enrichment protocols is detailed in Table 2. The selected studies were published between the years 2011 and 2023, with the majority being published from 2020 onward (n = 9, 64.2%). Among the species of animals used in the study were observed the species: Sprague Dawley (n = 4) (Gomez et al. 2015; Wu et al. 2016; Shen et al. 2019; Ji and Zhao 2023), C57BL/6 (n = 4) (McKibben and Dwivedi 2021; Lin et al. 2020; Seo et al. 2021; Min et al. 2022), Kunming (n = 2) (Wang et al. 2016; Wang et al. 2018), Wistar (n = 1) (Córneo et al. 2022), ICR (n = 1) (Koseki et al. 2012), Fischer-344 (n = 1) (Orock et al. 2021), and Holtzman (n = 1) (McKibben and Dwivedi 2021). The animals used were aged between 21 days (Gomez et al. 2015) and 10 weeks (Shen et al. 2019). These rodents in their development period pass through the period of gestation and lactation (Gomez, et al. 2015) until the young age (Shen et al. 2019), and most of the protocols used only male animals (n = 10).
Table 2.
Sample characteristics and environmental enrichment protocol
| Author, year | Species, age | Sex | n per cage | Environmental Enrichment Protocol and Housing dimensions | Exposure time to environmental enrichment | |
|---|---|---|---|---|---|---|
| Type of environmental enrichment and inanimate objects | (Length, Width, Height) | |||||
| Córneo et al., 2022 | Wistar, 60 days old | M | 10 | Physical and Social Enrichment; Running wheels, toys, small house | 80 × 45 × 22 cm | 45 days |
| Gomez et al., 2015 | Sprague Dawley, 21 days | M | 10–15 | Physical and Social Enrichment; Plastic objects | 120 × 60 × 45 cm | – |
| Ji et al., 2023 | Sprague Dawley, 6 wks old | M | 10 | Physical and Social Enrichment; Running wheels, plastic-colored toys, houses, tunnels, and stairs | 100 × 80 × 80 cm | 3 wks |
| Koseki et al., 2011 | ICR mice, 3 wks old | M | 8 | Physical and Social Enrichment; Running wheels, toys, tunnels, and hiding places | 50 × 70 × 20 cm | 4 wks |
| Lin et al., 2020 | C57BL/6 mice, 6–7 wks | M | 6 | Physical and Social Enrichment; Running wheels, wooden toys, tunnels, igloos, huts, and retreats | 90 × 70 × 30 cm3 | 6 days |
| Mckibben et al., 2021 | Holtzman Rats, C56BL/6; | M/F | – | Physical and Social Enrichment; Toys, tubes, shreddable cotton, paper objects, and manzanita wood | – | 69 days |
| Min et al., 2022 | C57BL/6 mice, 8 wks | M | 3–5 | Physical and Social Enrichment; Running wheels, toys, tunnels, and shed | 43.18 × 22.86 × 19.05 cm | 3, 7, and 14 days |
| Orock et al., 2021 | Fischer-344 mice, 5wks | F | 8 | Physical and Social Enrichment; Toys, ramps with ample bedding, tunnels, and food enrichment | 78 × 52 × 100 cm | 1–7 Days |
| Seo et al.2, 021 | C57BL/6j mice; 8 wks | F | 6–8 | Physical and Social Enrichment; Running wheels, tunnels, small balls, and shaped blocks | 26 × 42 × 18 cm3 | 5 wks |
| Shen et al., 2019 | Sprague Dawley, 10 wks | M | 8 | Physical and Social Enrichment; Running wheels, toys, tunnels, house, and differently shaped objects | 100 × 80 × 80 cm | 3 wks |
| Wang et al., 2016 | Kunming mice, 8 wks | M | 25 | Physical and Social Enrichment; Running wheels and toys | 60 × 33 × 28 cm | 4 wks |
| Wu et al., 2016 | Sprague Dawley, 7 wks | F | 36 | Physical and Social Enrichment; Plastic toys and nesting material | 10.5 × 19 × 18 inch | 4 wks |
| Xin W et al., 2018 | Kunming mice, 6–8 wks | M | 30 | Physical and Social Enrichment; Running wheels and toys | 60 × 36 × 28 cm | 4 wks |
| Yu et al., 2020 | Rats, 6 wks old | M | 13 | Physical and Social Enrichment; Running wheels, toys, tunnels, shelters, ladder trapeze, and balls | 83 × 83 × 83 cm | 2 wks |
cm centimeters; wks weeks; M Male; F Female
Environmental enrichment was performed using different inanimate objects, with the majority being toys (n = 11) (Yu et al. 2020; McKibben and Dwivedi 2021; Córneo et al. 2022; Wu et al. 2016; Ji and Zhao 2023; Lin et al. 2020; Min et al. 2022; Wang et al. 2016, 2018; Koseki et al. 2012; Orock et al. 2021), running wheels (n = 10) (Yu et al. 2020; Córneo et al. 2022; Shen et al. 2019; Ji and Zhao 2023; Lin et al. 2020; Min et al. 2022; Wang et al. 2016, 2018; Koseki et al. 2012; Orock et al. 2021), tunnels (n = 8) (Yu et al. 2020; Shen et al. 2019; Ji and Zhao 2023; Lin et al. 2020; Seo et al. 2021; Min et al. 2022; Koseki et al. 2012; Orock et al. 2021), houses (n = 2) (Córneo et al. 2022; Ji and Zhao 2023), and balls (n = 2) (Yu et al. 2020; Seo et al. 2021). Other identified objects were plastic objects, stars, hiding places, huts, retreats, tubes, shreddable cotton, paper objects, manzanita wood, shed, ramps, food, shaped blocks, nesting materials, shelters, ladders, and trapeze. Additionally, the size of the boxes used to accommodate the animals varied with heights ranging from 26 (Seo, et al. 2021) to 120 cm (Gomez, et al. 2015), widths from 22.86 (Min et al. 2022) to 83 cm (Yu et al. 2020), and lengths from 18 (Seo et al. 2021) to 100 cm (Orock et al. 2021). The duration of environmental enrichment varied among the protocols, lasting from 1 day (Orock et al. 2021) to 69 days (McKibben and Dwivedi 2021), with most studies using protocols that lasted between 3 and 4 weeks (Wu et al. 2016; Shen et al. 2019; Ji and Zhao 2023; Wang et al. 2016, 2018; Koseki et al. 2012). All included studies carried out an environmental enrichment protocol based mainly on physical and social stimuli according to the standardization defined by Hosey and collaborators (Hosey et al. 2013).
The description of the adverse exposure condition, evaluated brain tissue, and epigenetic changes is detailed in Table 3. Various exposure protocols were used: CUMS (n = 3) (Shen et al. 2019; Ji and Zhao 2023; Seo, et al. 2021), stroke (n = 2) (Lin et al. 2020; Wang et al. 2016), sepsis (n = 1) (Córneo et al. 2022), nicotine exposure (n = 1) (Gomez, et al. 2015), PCP exposure (n = 1) (Koseki et al. 2012), early life stress (n = 1) (McKibben and Dwivedi 2021), POCD (n = 1) (Min et al. 2022), WAS (n = 1) (Orock et al. 2021), high fructose intake (n = 1) (Wu et al. 2016), TBI (n = 1) (Wang et al. 2018), and sevoflurane exposure (n = 1) (Yu et al. 2020). The expression of epigenetic changes occurred in various brain tissues such as the hippocampus (n = 8) (Yu et al. 2020; Córneo et al. 2022; Wu et al. 2016; Shen et al. 2019; Ji and Zhao 2023; Seo, et al. 2021; Min et al. 2022; Wang et al. 2016), prefrontal cortex (n = 3) (Gomez, et al. 2015; Wang et al. 2018; Koseki et al. 2012), cortex (n = 2) (Lin et al. 2020; Min et al. 2022), amygdala (n = 1) (Orock et al. 2021), and forebrain (n = 1) (Wang et al. 2016).
Table 3.
Impacts of environmental enrichment on epigenetic modifications in brain tissues under adverse conditions
| Author, Year | Adverse condition | Brain tissue | miR-expression | Histone modifications | DNA alterations | Summary of the effects of EE on epigenetic processes under adverse conditions |
|---|---|---|---|---|---|---|
| Córneo et al., 2022 | Diseases and trauma (Sepsis) | Hippocampus | – | ↔ HAT, ↓ HDAC | ↓ DNMT | EE decreased HDAC and DNMT activity, and protects against neuroinflammation and cognitive damage |
| Gomez et al., , 2015 | Substance exposure (Nicotine) | Prefrontal Cortex | ↑ miR-221, ↑ miR-483 | – | – | EE upregulated miR-221 and miR-483 and attenuates nicotine-induced increases in pERK1/2 and pCREB |
| Ji et al., 2023 | Psychological stress (CUMS) | Hippocampus | ↓ miR-92a-3p | – | – | EE decreased miR-92a-3p and protects damage to behavior and cognition through KLF2 pathway |
| Koseki et al., 2011 | Substance exposure (PCP) | Prefrontal Cortex | – | ↑ H3 acetylated at Lys, ↓ HDAC5, ↔ HDAC1 | – | EE prevented behavioral damage by reversing the reduction in histone H3-positive cells and the increase in HDAC5 protein expression |
| Lin et al., 2020 | Diseases and trauma (Stroke) | Cortex | – | ↓ HDAC2 | – | EE reversed upregulation of HDAC2 and promoted functional recovery after the damage |
| Mckibben et al., 2021 | Psychological stress (Early Life Stress) | Hypothalamus | ↑ miR-29b-1-5p, ↑ 301b-3p, ↑ -3065-5p | – | – | EE reversed downregulation of miR-29b-1-5p, miR-301b-3p, and miR-3065-5p, and improved behavior through MAPK signaling pathway |
| Min et al., 2022 | Diseases and trauma (POCD) | Cortex, Hippocampus | – | Cortex: ↔ H3 (acetyl K14), ↔ H4 (acetyl K15), ↔ HDAC. Hippocampus: ↑ H3 (acetyl K14), ↑ H4 (acetyl K15), ↓ HDAC | – | EE upregulated H3 (acetyl K14), H4 (acetyl K15), reduced HDAC in hippocampus, and attenuated post-surgical effects through Neuroglia 1 pathway |
| Orock et al., 2021 | Psychological stress (WAS) | Amygdala | – | ↑ H3K9 acetylation at GR promoter, ↓ H3K9 acetylation at CRH promoter | – | EE modulated H3K9 acetylation at GR and CRH promoter, and prevented changes in expression of glucocorticoid receptor and corticotrophin-releasing hormone |
| Seo et al., 2021 | Psychological stress (CUS) | Hippocampus | – | ↑ Acetylated H3, ↓ HDAC5, ↑ H3K4me3, ↓ H3K27me3 | – | EE prevented cognitive damage through acetylation of H3 and H3K4me3, and reducing the level of HDAC5 and H3K27me3 |
| Shen et al., 2019 | Psychological sress (CUMS) | Hippocampus | ↓ miR-134 | – | – | EE upregulated miR-134 and reverses several damages through SIRT1 and BDNF pathway |
| Wang et al., 2016 | Diseases and trauma (Stroke) | Forebrain, Hippocampus | – | Forebrain and Hippocampus: ↑ Acetylated H3, ↔ Acetylated H4; | – | EE upregulated H3 acetylation in forebrain and hippocampus, and attenuated cognitive damage through cholinergic circuits |
| Wu et al., 2016 | Dietary conditions (High fructose) | Hippocampus | – | ↓ Nuclear HDAC4, ↔ HDAC5 | – | EE decreased nuclear HDAC4 levels and prevented cognitive function through BDNF pathway |
| Xin W et al., 2018 | Diseases and trauma (TBI) | Prefrontal cortex | – | ↑ H3 acetylation, ↔ H4 acetylation | – | EE unregulated H3 acetylation at ChAT gene promoter, and attenuated cognitive damage through CREB and cholinergic pathways |
| Yu et al., 2020 | Substance exposure (Sevoflurane) | Hippocampus | – | ↑ Acetylated H3, ↑ Acetylated H4, ↓ HDAC2; ↓ HDAC3 | – | EE upregulated H3 and H4 acetylation, reduced HDAC2 and HDAC3 levels, and attenuated cognitive damage through BDNF pathway |
CUS Chronic unpredictable stress; CUMS Chronic unpredictable mild stress; DNMT DNA methyltransferase; HAT histone acetylase; H3K4me Trimethylation of H3K27me; H3 Trimethylation of H3K; 4H3K4 histone H3 lysine 4; HDAC histone deacetylase; HDAC1 histone deacetylase 1; HDAC1 histone deacetylase 2; HDAC3 histone deacetylase 3; HDAC5 histone deacetylase 5; mir microRNA; POCD Postoperative cognitive dysfunction; TBI traumatic brain injury; WAS Water avoidance stress. ↑ Significant Increased; ↓ Significant Decrease; ↔ Non-significant differences
Environmental Enrichment and Expression of Micrornas
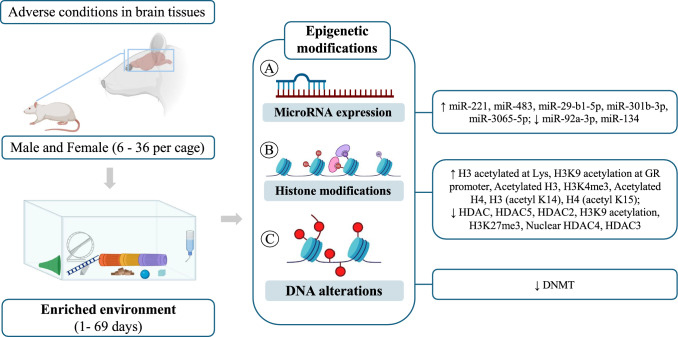
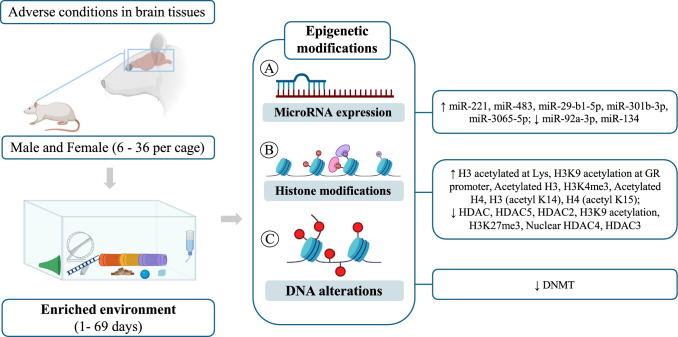
Four studies evaluated the expression of microRNAs, with two in the hippocampus (miR-92a-3P, miR-134) (Shen et al. 2019; Ji and Zhao 2023), one in the prefrontal cortex (miR-221, miR-483) (Gomez, et al. 2015), and one in the hypothalamus (miR-207, miR-219–1, miR-212) (McKibben and Dwivedi 2021). After the period of environmental enrichment, an increase in the expression of miR-221 and miR-483 was observed in the prefrontal cortex (Gomez et al. 2015), and a reduction in the expression of miR-92a-3P and miR-134 was observed in the hippocampus (Gomez, et al. 2015). No differences were observed in the expression of miR-207, miR-219–1, and miR-212 in the hypothalamus (McKibben and Dwivedi 2021).
Impact of Environmental Enrichment on the Levels of Enzymes Related to Histone and DNA Modifications
The presence of histone modifications was observed in ten articles. After environmental enrichment, in the hippocampus, there was a reduction of HAT (Córneo et al. 2022), HDAC/HDAC4 (Córneo et al. 2022; Wu et al. 2016; Min et al. 2022) in conditions of sepsis, POCD, and high fructose intake. On the other hand, an increase in H3 (acetyl K14), H4 (acetyl K15) (Min et al. 2022), H3K4me3, K3k27me3 (Seo et al. 2021), and HDAC2/3/5 (Yu et al. 2020; Seo et al. 2021) was observed under conditions of POCD, CUMS, and sevoflurane exposure.
In the cortex, there was a reduction of HDAC2 in stroke condition (Lin et al. 2020); in the prefrontal cortex, there was an increase in H3 acetylated at Lys in PCP exposure (Koseki et al. 2012); and in the amygdala, there was a reduction in H3K9 acetylation at the CRH promoter and an increase in H3K9 acetylation at the GR promoter in WAS condition (Orock et al. 2021). There were no changes to the other areas and markers. Only one study observed changes in DNA (Córneo et al. 2022). Córneo et al. (2022) observed that after environmental enrichment and with the sepsis model, there was a reduction in DNMT in the hippocampus of Wistar rats (Fig. 2).
Fig. 2.
DNA methyltransferase; HAT: histone acetylase; H3K4me: Trimethylation of H3K27me; H3: Trimethylation of H3K4H3K4: histone H3 lysine 4; HDAC: histone deacetylase; HDAC1: histone deacetylase 1; HDAC1: histone deacetylase 2; HDAC3: histone deacetylase 3; HDAC5: histone deacetylase 5; mir: microRNA
Discussion
The present systematic review aimed to evaluate the impacts of environmental enrichment on the modulation of epigenetic processes in different specific regions under adverse conditions. In this sense, we demonstrated that benefits produced by EE in the brain and animal behavior are directly related to the modulation of epigenetic mechanisms, particularly the modulation of histones toward a more acetylated profile, which results in a chromatin that is more permissive to the transcription machinery of genes associated with cell growth, development and maturation, as well as neuroplasticity in adverse environmental conditions.
The term 'epigenetics' is currently used to describe the study of mechanisms that cause heritable changes in gene expression and cellular phenotype, without any alteration to the sequence of nucleotide bases in DNA. These mechanisms, 'above genetics,' are responsible for modulating genomic structure and activity in response to external and internal cellular signals, thereby playing an essential role in the regulation of diverse cellular processes, including cell development and differentiation (Haig 2004; DeAngelis et al. 2008; Gonzalo 1985). At present, there are three principal epigenetic mechanisms that have been subjected to substantial molecular investigation: DNA methylation, post-translational modifications of histones, and gene silencing through non-coding RNAs, mainly microRNAs (Kim et al. 2009).
Briefly, during the process of DNA methylation, a methyl-CH3 radical is incorporated into a CpG dinucleotide through the activity of DNA methyltransferases (DNMTs), which typically results in the silencing of the associated gene (Lovrečić et al. 2013; Labbé et al. 2016). In contrast, post-translational histone modifications encompass a range of modifications occurring in the N-terminal tail of chromatin proteins. Histones can be modified through enzymatic processes of acetylation and methylation, which are more fully described in the literature, or through ubiquitination and phosphorylation. These modifications regulate chromatin packing and, as a result, affect the transcriptional activity of the chromatin itself (Qureshi and Mehler 2018; Kouzarides 2007). Finally, the process of gene silencing through microRNAs has been described more recently and consists of a combination of steps involving translational repression and degradation of the target mRNA (Jonas and Izaurralde 2015).
Given its dynamic nature, the epigenetic profile, along with its modifications, orchestrates gene expression and function at botch cellular and tissue levels. Furthermore, it mediates the interactions between genes and the external environment, from the development through to the cellular aging. Consequently, over the past decades, there has been a growing interest in the impact of epigenetic modifications across various fields of neurobiology, aiming to elucidate gaps in our understanding of topics such as learning and memory, as well as onset and progression of several psychiatric and neurological disorders (Praag et al. 2000; Anier and Kalda 2012).
As an example of external factors, environmental enrichment (EE) was first described by Donald Hebb (1949) and refers to simple strategies for manipulating standard laboratory housing conditions, with the aim of promoting neurorehabilitation by optimizing the diversity, quality, and intensity of environmental stimuli (Ball et al. 2019; Kempermann 2019). In general, animals maintained in EE are provided with larger living spaces, a variety of toys and other stimulating items (such as sound, light, and colors), periodic changes in food and water, and increased opportunities for physical activity and social interaction (Yu et al. 2014). Given the well-established impact of environmental conditions during childhood and adolescence on adult neuroplasticity, some researchers have recently sought to elucidate the role of epigenetic mechanisms in modulating behavioral, cognitive, and neurological performance in response to positive and/or negative stimuli (Lehmann and Herkenham 2011; Volkers and Scherder 2011).
Thus, the aim of this study was therefore to carry out the first systematic review of the influence of epigenetic mechanisms on the well-described impact of different EE protocols on the brains of rodents exposed to favorable or unfavorable environmental conditions. In a translational context, the elucidation of these mechanisms can contribute to the discovery of potential molecular targets through simple methods of non-pharmacological interventions for different pathologies and neuropsychological disorders.
Among the 14 studies selected here, 10 showed that the benefits promoted by EE protocols in young rodents’ brains exposed to different adverse conditions involve post-translational modifications of histones, usually acetylation or deacetylation process.
Consistent with existing literature, Lin et al. (2020) demonstrated that the upregulation of histone deacetylase 2 (HDAC2), a negative regulator of neuroplasticity, following a stroke in C57BL/6 mice, could be reversed with a 6-day environmental enrichment protocol. The reduction in HDAC2 in response to EE also resulted in the expression of neurotrophins and proteins associated with neuroplasticity, thereby contributing to the functional recovery of the cortex. Importantly, the HDAC2 knockdown mimicked the benefits promoted by EE, which reinforces that epigenetic modification of histones through HDAC2 is a critical step in recovery after damage. Similarly, Wang et al. (2016a, b) revealed that cognitive recovery of Kunming mice under EE conditions after stroke involves the maintenance of acetylation homeostasis in cholinergic pathways. In this study, 28 days of exposure to an EE partially reversed the reduction in acetylated histone levels in cholinergic cells of the forebrain and hippocampus in mice with post-damage cognitive impairment. Notably, there was a significant increase in Ac-H3 levels in both regions, impacting the cascade of acetylcholine neurotransmitter, which is essential for cognitive function. Later, they demonstrated that EE's ability to alleviate working memory impairment symptoms after traumatic brain injury also involves increasing Ac-H3 levels in the cholinergic system in the prefrontal cortex region of these mice (Wang et al. 2018). Similarly, C57BL/6 mice that were maintained on preoperative EE demonstrated resistance to post-surgical cognitive dysfunction, with the preservation of neuroglia 1 and an improvement in memory and learning. These benefits of EE were associated with the maintenance of homeostatic levels of HDAC activity and Ac-H3 and Ac-H4 in the hippocampus of these animals (Min et al. 2022).
Furthermore, 2 of the 10 studies selected for review on the role of histone modifications in the benefits of EE were conducted in models of mental and behavioral disorders. Seo et al. (2021) demonstrated that keeping C57BL/6j neonates in EE with racing wheels, tunnels, balls, and blocks prevents chronic stress and depressive behavior when they become adults. They demonstrated that these effects of postnatal EE are due to an increase in the level of Ac-H3 of the p11 gene in the hippocampus, a reduction in HDAC5 activity, as well as the prevention of stress-induced changes in the trimethylated state of histone H3 lysine 4 (H3K4) and H3K27. A study conducted on female Fischer-344 rats demonstrated that the stress-relieving effects of the short-term EE protocol in the brain-intestinal axis are due to the maintenance of histone H3 lysine 9 (H3K9) acetylation in specific promoters of the central nucleus of the amygdala.
The preservation of the epigenetic state resulted in a reduction in visceral and somatic nociception over the long term (Orock et al. 2021). In addition, Koseki et al. (2012) reported that a 4-week exposure to EE, using a variety of inanimate objects during adolescence, can prevent the development of schizophrenic behaviors in ICR mice. This effect was associated with increased levels of histone H3K9 acetylation and reduced HDAC5 activity in the prefrontal cortex of animals exposed to phencyclidine (PCP), a non-competitive antagonist of the N-methyl-d-Aspartate (NMDA) receptor.
Given the extensive literature on the impact of maternal drug use or nutrition on fetal neurodevelopment, Yu et al. (2020) investigated the role of histone modifications in the offspring of rats exposed to 2.5% sevoflurane. Additionally, they examined whether 2 days of EE could mitigate the epigenetic changes and cognitive impairment associated with this exposure. In this study, EE with plastic tunnels, wheels, and balls, among other toys, was effective in alleviating behavioral impairment in the offspring of rats exposed to 6 h of 2.5% sevoflurane. These effects were associated with a significant increase in the level of Ac-H3 and Ac-H4, a reduction in HDAC2 and HDAC3, as well as cognitive improvement associated with BDNF signaling in the pups' hippocampus. Similarly, EE with plastic toys and nesting material was observed to reduce the activity of nuclear HDAC4 in binding to promoters II and IV of the bdnf gene, which resulted in improved learning and memory in the offspring of females fed a high-glucose diet during gestation and lactation (Wu et al. 2016).
In the present systematic review, a total of four studies were selected regarding the potential of EE to improve neurobiological aspects through the modulation of microRNAs. An analysis of the hippocampus of Sprague Dawley rats with depressive behavior induced by mild stress revealed that the 3-week EE intervention protocol was effective in reducing the level of miR-92a-3p, which has been previously described as elevated in patients with depression and anxiety associated with substance abuse (Ji and Zhao 2023; Chen et al. 2021). In another study, the same protocol was also found to improve symptoms of chronic stress, depressive behavior, and cognitive deficits through the regulation of the SIRT1/mirR-134 signaling pathway by reducing the level of miR-134 and thus increasing the availability of its target SIRT1, which is necessary for greater BDNF expression and neuroplasticity (Shen et al. 2019).
Furthermore, rats in EE also showed a lower sensitivity to motor damage induced by nicotine abuse through an increase in 6 different microRNAs, mainly miR-221 and miR-483, compared to those kept in impoverished or standard environments. Interestingly, the differential expression screening conducted by McKibben and Dwivedi (2021) revealed that early life stress increases susceptibility to depressive behavior in adults by modulating the microRNA profile. In the analysis of the hypothalamus of rodents subjected to maternal separation, 29 microRNAs were upregulated while 21 were downregulated. In the group of downregulated miRNAs, the level of 3 specific miRNAs (miR-29b-1-5p, -301b-3p, and -3065-5p) exhibited a notable increase following the EE intervention, aligning with the control without stress. As expected, the modulation of these miRNAs also altered the expression of the respective target genes, such as MAPK6 and MMP19.
Among the articles reviewed in this study, only one examined the correlation between EE and DNA methylation. In this context, Córneo et al. (2022) demonstrated that intervention with EE protects against cognitive impairment induced by neuroinflammation through the repression of the hypermethylated state induced by both HDACs and DNMTs in a mouse model of sepsis. In summary, the studies discussed here demonstrate that the cognitive benefits of EE are linked to its capacity to modulate key biological processes in the animal brain. In different models, EE stimulation was able to protect the brain against adverse conditions at a "pre-transcriptional" level by preserving the acetylation profile. This was achieved by reversing the deacetylated or hypermethylated state of histones and DNA, respectively, or by promoting the activity of histone acetyltransferases. In addition, stimulation with EE was also effective in regulating these processes at the “post-transcriptional” level, by modulating the activity of microRNAs and, consequently, their respective target genes. The modulation of these mechanisms revealed the potential of EE to control the production and/or availability of neurotrophins and genes involved in the process of neurogenesis and neuroplasticity.
Limitations
Some limitations can be observed in the present review. Firstly, the heterogeneity of the epigenetic markers used in the included studies makes it difficult to observe the modulation of a specific potential target in the brain in response to environmental enrichment under adverse environmental conditions. This would require a more in-depth investigation not just of a single marker, but of an entire signaling pathway that may be activated/inactivated. Secondly, only one study included analyzes the impacts of environmental enrichment on DNMT activity. This enzyme participates in regulatory mechanisms, including molecular interactions, post-translational modifications, alternative splicing, and gene duplication or loss. Furthermore, the molecular functions of DNMTs are not limited to gene silencing and may also include transcriptional activation and transcriptional post-regulation. Finally, resolving these limitations can help in the discovery and establishment of new knowledge within molecular genetics and neuroscience, opening an effective field of possibility for translational studies with human beings with diverse chemical, pharmacological, pathological, and lifestyle-related exposures.
Future Directions
The evidence presented in this systematic review demonstrates a new frontier of scientific knowledge, pointing to environmental enrichment as a tool capable of modulating epigenetic processes mainly responsible for controlling gene expression in the central nervous system. These mechanisms may help future research to discover fundamental signaling pathways for the regulation of cognitive, behavioral, and neuroplasticity processes and can be applied in favorable and adverse environmental contexts. Next, it is necessary to develop translational studies that apply this non-pharmacological tool in pathologies that affect the central nervous system, including multiple sclerosis, Parkinson's, and Alzheimer's, which are known to be effectively regulated by epigenetic processes. Finally, environmental enrichment protocols should be standardized to avoid heterogeneity of methodology and results.
Conclusions
In summary, we have shown that the benefits of EE on the animal brain and behavior are directly related to different epigenetic mechanisms, particularly the modulation of histones toward a more acetylated profile, which results in a chromatin that is more permissive to the transcription machinery of genes involved in cell growth and neuroplasticity. In this sense, interventions based on positive environmental stimuli, such as EE, are emerging as non-pharmacological, low-cost, and simple-to-apply alternatives for preventing or mitigating symptoms in various disorders affecting the brain. This was the first systematic review on the subject, contributing to a detailed analysis of each EE protocol, the animal model studied, and the main effects achieved, finally advancing the standardization of the strategy for clinical application.
Author Contribution
Conceptualization, M.S.S.F, and R.F.S; methodology, M.S.S.F, C.J.L.; Formal analysis, M.R.C, and M.S.S.F; data curation, G.C.J.S, and M.S.S.F, writing—original draft preparation, M.S.S.F, F.O.S., G.B., F.H.Y., J.M.d.C., C.J.L., R.F.S writing—review and editing, M.S.S.F, G.B, L.P.A,M.R.C.,G.C.J.S, J.M.d.C, F.H.Y., C.J.L. and R.F.S, supervision: M.S.S.F, G.B, L.P.A, and R.F.S. All authors have read and agreed to the published version of the manuscript.
Funding
This research was conducted without external funding.
Data Availability
The data presented in this study are available upon request to the author Matheus Santos de Sousa Fernandes.
Declarations
Conflict of interest
The authors declare no competing interests.
Animal Ethics and Consent to Participate declarations
Not applicable.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Luca Paolo Ardigò and Fabrício Oliveira Souto have contributed equally to this work.
References
- Anier K, Kalda A (2012) Epigenetics in the central nervous system. Curr Geriatrics Reports 1:190–198 [Google Scholar]
- Ball NJ, Mercado E 3rd, Orduña I (2019) Enriched environments as a potential treatment for developmental disorders: a critical assessment. Front Psychol 10:466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baumans V (2005) Environmental enrichment for laboratory rodents and rabbits: requirements of rodents, rabbits, and research. Ilar J 46(2):162–170 [DOI] [PubMed] [Google Scholar]
- Carthew RW, Sontheimer EJ (2009) Origins and mechanisms of miRNAs and siRNAs. Cell 136(4):642–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen F et al (2021) Prognostic plasma exosomal microRNA biomarkers in patients with substance use disorders presenting comorbid with anxiety and depression. Sci Rep 11(1):6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Córneo E et al (2022) Enriched environment causes epigenetic changes in hippocampus and improves long-term cognitive function in sepsis. Sci Rep 12(1):11529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeAngelis JT, Farrington WJ, Tollefsbol TO (2008) An overview of epigenetic assays. Mol Biotechnol 38(2):179–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg AP, Levchenko A (2023) Epigenetics as a mediator of plasticity in cancer. Science 379(6632):eaaw3835. [DOI] [PMC free article] [PubMed]
- Fitz-James MH, Cavalli G (2022) Molecular mechanisms of transgenerational epigenetic inheritance. Nat Rev Genet 23(6):325–341 [DOI] [PubMed] [Google Scholar]
- Gao ZK et al (2022) Enriched environment effects on myelination of the central nervous system: role of glial cells. Neural Plast 2022:5766993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh P, Saadat A (2023) Neurodegeneration and epigenetics: a review. Neurologia (Engl Ed) 38(6):e62–e68 [DOI] [PubMed] [Google Scholar]
- Global, regional, and national burden of disorders affecting the nervous system, 1990–2021: a systematic analysis for the Global Burden of Disease Study 2021 (2024). Lancet Neurol 23(4):344–381. [DOI] [PMC free article] [PubMed]
- Gomez AM et al (2015) Prefrontal microRNA-221 mediates environmental enrichment-induced increase of locomotor sensitivity to nicotine. Int J Neuropsychopharmacol 19(1). [DOI] [PMC free article] [PubMed]
- Gonzalo S (2010) Epigenetic alterations in aging. J Appl Physiol (1985) 109(2):586–597. [DOI] [PMC free article] [PubMed]
- Haig D (2004) The (dual) origin of epigenetics. Cold Spring Harb Symp Quant Biol 69:67–70 [DOI] [PubMed] [Google Scholar]
- Han Y et al (2022) The role of enriched environment in neural development and repair. Front Cell Neurosci 16:890666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosey G, Melfi V, Pankhurst S (2013) Zoo animals: behaviour, management, and welfare. Oxford University Press, USA [Google Scholar]
- Ji X, Zhao Z (2023) Exposure to enriched environment ameliorated chronic unpredictable mild stress-induced depression-like symptoms in rats via regulating the miR-92a-3p/kruppel-like factor 2 (KLF2) pathway. Brain Res Bull 195:14–24 [DOI] [PubMed] [Google Scholar]
- Jonas S, Izaurralde E (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat Rev Genet 16(7):421–433 [DOI] [PubMed] [Google Scholar]
- Kempermann G (2019) Environmental enrichment, new neurons and the neurobiology of individuality. Nat Rev Neurosci 20(4):235–245 [DOI] [PubMed] [Google Scholar]
- Kim JK, Samaranayake M, Pradhan S (2009) Epigenetic mechanisms in mammals. Cell Mol Life Sci 66(4):596–612 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koseki T et al (2012) Exposure to enriched environments during adolescence prevents abnormal behaviours associated with histone deacetylation in phencyclidine-treated mice. Int J Neuropsychopharmacol 15(10):1489–1501 [DOI] [PubMed] [Google Scholar]
- Kouzarides T (2007) Chromatin modifications and their function. Cell 128(4):693–705 [DOI] [PubMed] [Google Scholar]
- Labbé C, Lorenzo-Betancor O, Ross OA (2016) Epigenetic regulation in Parkinson’s disease. Acta Neuropathol 132:515–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehmann ML, Herkenham M (2011) Environmental enrichment confers stress resiliency to social defeat through an infralimbic cortex-dependent neuroanatomical pathway. J Neurosci 31(16):6159–6173 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Tollefsbol TO (2021) DNA methylation methods: global DNA methylation and methylomic analyses. Methods 187:28–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YH et al (2020) HDAC2 (Histone deacetylase 2): a critical factor in environmental enrichment-mediated stroke recovery. J Neurochem 155(6):679–696 [DOI] [PubMed] [Google Scholar]
- Lovrečić L et al (2013) The role of epigenetics in neurodegenerative diseases. Neurodegener Dis 10:54744 [Google Scholar]
- McKibben LA, Dwivedi Y (2021) Early life and adult stress promote sex dependent changes in hypothalamic miRNAs and environmental enrichment prevents stress-induced miRNA and gene expression changes in rats. BMC Genomics 22(1):701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Min J et al (2022) Preoperative environment enrichment preserved neuroligin 1 expression possibly via epigenetic regulation to reduce postoperative cognitive dysfunction in mice. CNS Neurosci Ther 28(4):619–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikolac Perkovic M et al (2021) Epigenetics of Alzheimer's disease. Biomolecules 11(2). [DOI] [PMC free article] [PubMed]
- Orock A et al (2021) Environmental enrichment prevents stress-induced epigenetic changes in the expression of glucocorticoid receptor and corticotrophin releasing hormone in the central nucleus of the amygdala to inhibit visceral hypersensitivity. Exp Neurol 345:113841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page MJ, The PRISMA et al (2020) statement: an updated guideline for reporting systematic reviews. BMJ 2021:372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi IA, Mehler MF (2018) Epigenetic mechanisms underlying nervous system diseases. Handb Clin Neurol 147:43–58 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seo MK et al (2021) Early enriched environment prevents epigenetic p11 gene changes induced by adulthood stress in mice. Int J Mol Sci 22(4). [DOI] [PMC free article] [PubMed]
- Shen J et al (2019) The enriched environment ameliorates chronic unpredictable mild stress-induced depressive-like behaviors and cognitive impairment by activating the SIRT1/miR-134 signaling pathway in hippocampus. J Affect Disord 248:81–90 [DOI] [PubMed] [Google Scholar]
- van Praag H, Kempermann G, Gage FH (2000) Neural consequences of environmental enrichment. Nat Rev Neurosci 1(3):191–198 [DOI] [PubMed] [Google Scholar]
- Volkers KM, Scherder EJ (2011) Impoverished environment, cognition, aging and dementia. Rev Neurosci 22(3):259–266 [DOI] [PubMed] [Google Scholar]
- Wang J et al (2011) Quantifying the Waddington landscape and biological paths for development and differentiation. Proc Natl Acad Sci U S A 108(20):8257–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X et al (2016a) Enriched environment improves post-stroke cognitive impairment in mice by potential regulation of acetylation homeostasis in cholinergic circuits. Brain Res 1650:232–242 [DOI] [PubMed] [Google Scholar]
- Wu KL et al (2016b) Environmental stimulation rescues maternal high fructose intake-impaired learning and memory in female offspring: Its correlation with redistribution of histone deacetylase 4. Neurobiol Learn Mem 130:105–117 [DOI] [PubMed] [Google Scholar]
- Wang X et al (2018) Enriched environment improves working memory impairment of mice with traumatic brain injury by enhancing histone acetylation in the prefrontal cortex. PeerJ 6:e6113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan H, Bu P (2021) Non-coding RNA in cancer. Essays Biochem 65(4):625–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K et al (2014) Enriched environment induces angiogenesis and improves neural function outcomes in rat stroke model. J Neurol Sci 347(1–2):275–280 [DOI] [PubMed] [Google Scholar]
- Yu Z et al (2020) Enriched environment improves sevoflurane-induced cognitive impairment during late-pregnancy via hippocampal histone acetylation. Braz J Med Biol Res 53(10):e9861 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data presented in this study are available upon request to the author Matheus Santos de Sousa Fernandes.