Abstract
With devastating health and socioeconomic impact worldwide, much work is left to understand the Coronavirus Disease 2019 (COVID-19), with emphasis in the severely affected elderly population. Here, we present a proteomics study of lung tissue obtained from aged vs. young rhesus macaques (Macaca mulatta) and olive baboons (Papio Anubis) infected with severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Using age as a variable, we identified common proteomic profiles in the lungs of aged infected non-human primates (NHPs), including key regulators of immune function, as well as cell and tissue remodeling, and discuss the potential clinical relevance of such parameters. Further, we identified key differences in proteomic profiles between both NHP species, and compared those to what is known about SARS-CoV-2 in humans. Finally, we explored the translatability of these animal models in the context of aging and the human presentation of the COVID-19.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-024-01264-3.
Keywords: Aging, SARS-CoV-2, COVID-19, Lung proteomics, Non-human primates
Introduction
With an estimated 774 million cases or more reported and 0.90% mortality globally [1], the Coronavirus disease (COVID-19) pandemic is still an unresolved public health concern. Aged individuals, and especially, those with pre-existing co-morbidities, are at increased risk of respiratory complications and even death [2, 3]. Indeed, a 62-fold increase in mortality after severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection has been reported in the > 65-year-old age group compared to the < 55 age group [4, 5].
Despite abundant research to understand COVID-19 pathogenesis, we still lack information on specific tissue responses driven by the infection. Most published proteomic reports are from human clinical samples obtained post-mortem [6–8], with limited, if any, studies investigating key differences in lung-specific responses related to the age of the individuals.
To address this knowledge gap, here we used pre-existing lung samples from our established models of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection in non-human primates (NHPs) [9–12]. We sought to identify differences in the proteomes of two age groups (aged vs. young) in two NHP species, rhesus macaques (Macaca mulatta) and olive baboons (Papio anubis) during SARS-CoV-2 infection. We previously published that both species are susceptible to SARS-CoV-2, with baboons having more lung inflammation and longer SARS-CoV-2 viral shedding compared to macaques [9]. Their differential progression to COVID-19 makes them suitable as models to test vaccines and therapies. Global proteomics were conducted to obtain their proteomic profiles and relative quantification in the lungs at the time of necropsy (14 days post-infection), and results were then compared to published reports from human infections.
Materials and methods
Study approval
Pre-existing samples used in this study were from infected NHPs that were housed under Animal Biosafety Level 3 (ABSL3) facilities at the Southwest National Primate Research Center (SNPRC), where they were treated according to the standards recommended by AAALAC International and the NIH Guide for the Care and Use of Laboratory Animals. NHP studies were approved by the Institutional Animal Care and Use Committee (IACUC) at Texas Biomedical Research Institute (protocol# 1714 PC 6).
Animal study and tissue processing
We used a biorepository of pre-existing lung tissue samples from 3-year-old (young group, n = 6) and 17 to 22-year-old (aged group, n = 8) rhesus macaques and 2-year-old (young group, n = 6) and 10 to 20-year-old (aged group, n = 7) olive baboons; gender matched, infected with SARS-CoV-2 USA/WA1 2020 strain through multiple routes (ocular, intratracheal and intranasal) with a dose of 1.05 × 106 PFU/per animal. Lung tissues were collected at necropsy after 14 days post-infection as described [9]. Briefly, approximately 0.5 cm3 tissues pieces were obtained at necropsy and snap-frozen with dry ice and stored at -80 °C until processing. Tissues were thawed in ice and homogenized in a total of 2 mL of 1X DPBS containing 1% SDS and a protease inhibitor cocktail (Roche) using Precellys Lysing Kit (CKMix50 – 7 mL). After homogenization, samples were filtered to remove large debris, and were inactivated at 60 °C for 1 h [9].
Proteomic analyses
Lung homogenate samples were mixed with 10% SDS/50 mM triethyl-ammonium bicarbonate (TEAB) in the presence of protease and phosphatase inhibitors (Halt; Thermo Scientific). Aliquots corresponding to 100 µg protein (EZQ™ Protein Quantitation Kit; Thermo Fisher) were reduced with tris(2-carboxyethyl)phosphine hydrochloride (TCEP), alkylated in the dark with iodoacetamide and applied to S-Traps (mini; Protifi) for tryptic digestion (sequencing grade; Promega) in 50 mM TEAB. Peptides were eluted from the S-Traps with 0.2% formic acid in 50% aqueous acetonitrile and quantified using Pierce™ Quantitative Fluorometric Peptide Assay (Thermo Scientific).
DIA-MS (Data Independent Acquisition Mass Spectrometry) was conducted on an Orbitrap Fusion Lumos (Thermo Scientific) mass spectrometer. On-line HPLC separation used an RSLC NANO HPLC system (Thermo Scientific/Dionex: column, PicoFrit™ (New Objective; 75 μm i.d.) packed to 15 cm with C18 adsorbent (Vydac; 218MS 5 μm, 300 Å); mobile phase A, 0.5% acetic acid (HAc)/0.005% trifluoroacetic acid (TFA) in water; mobile phase B, 90% acetonitrile/0.5% HAc/0.005% TFA/9.5% water; gradient 3 to 42% B in 120 min; flow rate, 0.4 μl/min. A pool was made of all of the samples, and 2-µg peptide aliquots were analyzed using gas-phase fractionation and 4-m/z windows (30 k resolution for precursor and product ion scans, all in the orbitrap) to create a DIA chromatogram library [13] by searching against a Prosit-generated predicted spectral library (Gessulat 2019) [14] based on either the reference protein sequence database for UniProt_Papio_anubis_9555_20220312 (44,734 sequences; 24,127,678 residues) or UniProt_Macaca_mulatta_9544_20220315 (44,390 sequences; 28,513,169 residues), as appropriate for the experiment. A database of common contaminants (without any bovine serum protein entries; 124 sequences; 62,564 residues) was also used. Experimental samples were randomized for sample preparation and analysis. Injections of 2 µg of peptides were employed. MS data for experimental samples were acquired in the Orbitrap using 8-m/z windows (staggered; 30 k resolution for precursor and product ion scans) and searched against the chromatogram library. Carbamidomethylation of cysteine was considered as a fixed modification for generation of the chromatogram library and for searching the MS data from the experimental samples. Scaffold DIA (v3.2.1; Proteome Software) was used for all DIA-MS data processing.
Data and statistical analyses
Protein identifiers were transformed to gene symbols by: 1- Annotated gene name, 2- Sequence homology. Significant DAPs were selected based on the following thresholds: Log2 Fold Change < -0.5 or > 0.5, p-value < 0.05. IPA (v90348151; QIAGEN) was used to perform pathway analysis, pathway comparison between species, and regulator prediction analyses. R v4.2.2 was used to generate Volcano plots (EnhancedVolcano package), with supporting packages Bioconductor and ggplot2. Heatmaps were generated in NG-CHM Heat Map Viewer v2.22.2 [15, 16].
Results and discussion
Comparative proteomics of SARS-CoV-2 infected lung tissues from aged vs. young rhesus macaques and olive baboons – shared DAPs
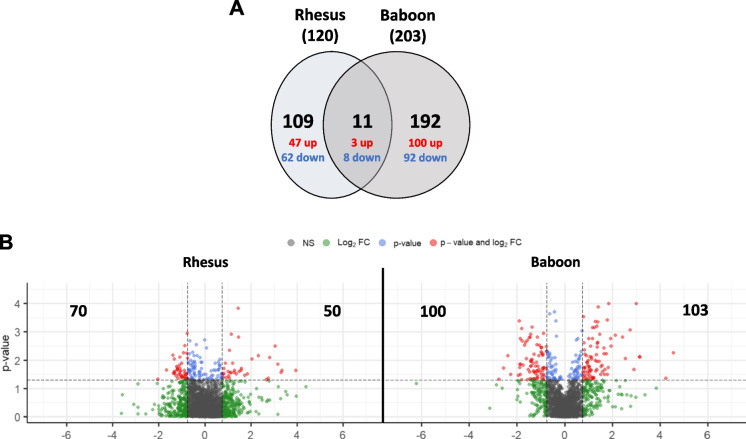
We performed untargeted proteomics by DIA-MS in pre-existing samples of lung tissue of SARS-CoV-2 infected rhesus macaques and olive baboons [9] for the identification and relative quantification of proteins in infected NHPs. We first determined significant Differentially Abundant Proteins (DAPs) (log2 fold change ≤ -0.5 or ≥ 0.5, p-value < 0.05) in both aged vs. young rhesus macaques (RM) and olive baboons (OB), with rhesus macaques having 120 and olive baboons 203 total DAPs (Fig. 1, Tables 1 and 2). Of those, 11 DAPs were shared between both species, and 109 and 192 DAPs were exclusive to rhesus and baboons, respectively (Fig. 1A).
Fig. 1.
Differentially expressed proteins (DAPs) in aged vs. young rhesus macaques and aged vs. young olive baboons. A Venn diagram showing the number of shared and unique DAPs in each NHP species; B Volcano plots showing the distribution of DAPs. The x-axis shows log2-transformed protein fold changes, the y-axis shows -log10-transformed p-values; a total of 120 DAPs and 203 DAPs were identified for aged rhesus macaque and olive baboon groups, respectively
Table 1.
List of DAPs for rhesus macaques (aged vs. young). Accession number, protein name, gene name, p-value and log2 Fold Change are provided. Proteins are ordered by increasing p-value. Top 5 DAPs are highlighted in bold
| Accession Number | Protein name | Gene name | p-value | Log2 Aged Rhesus |
|---|---|---|---|---|
| F7EDT3 | Calpain 5 | CAPN5 | 0.00015 | 1.45 |
| F6TSU2 | Protein-tyrosine-phosphatase receptor J | PTPRJ | 0.0011 | -0.771 |
| F7H1V9 | Pentraxin | APCS | 0.0012 | 1.14 |
| F6UQB6 | Heme-binding protein 1 | HEBP1 | 0.0015 | 1.46 |
| F6V089 | t-SNARE coiled-coil homology domain-containing protein | PPP1R8 | 0.0021 | -0.644 |
| A0A1D5Q9N1 | Radixin | RDX | 0.0031 | -0.875 |
| F7ESZ3 | Biglycan | BGN | 0.0032 | 3.03 |
| A0A1D5R6M8 | Immunoglobulin heavy constant mu | IGHM | 0.0045 | 1.06 |
| A0A5F7ZIS1 | RALY heterogeneous nuclear ribonucleoprotein | RALY | 0.0052 | -0.533 |
| F6YXJ2 | Deoxyribonuclease II | DNASE2 | 0.0054 | -0.936 |
| F6R2V4 | BAG cochaperone 3 | BAG3 | 0.0057 | -0.799 |
| F7C9U4 | CD93 molecule | CD93 | 0.0059 | -0.669 |
| H9FSW2 | Argonaute RISC component 1 | AGO1 | 0.0066 | -1.38 |
| F6RJC0 | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase | PLCB3 | 0.007 | 2.33 |
| F6SFP9 | D-aminoacyl-tRNA deacylase | DTD1 | 0.0079 | -1.1 |
| A0A1D5QRS2 | Histone H2A | H2AC12 | 0.0082 | 2.81 |
| F6ZAW8 | Hippocalcin like 1 | HPCAL1 | 0.0085 | -0.755 |
| G7MHG1 | Complement C1q subcomponent subunit B | C1QB | 0.0089 | 2.05 |
| F7GPV6 | Multifunctional protein ADE2 | PAICS | 0.0092 | -1.34 |
| A0A5F7ZHW5 | Lectin, mannose binding 2 | LMAN2 | 0.0095 | 0.64 |
| F7FMM5 | TROVE domain-containing protein | RO60 | 0.01 | -0.605 |
| A0A5F7ZQM9 | Proteasome 20S subunit beta 8 | PSMB8 | 0.012 | -0.726 |
| A0A1D5Q716 | Uncharacterized protein | MCAM | 0.012 | -0.519 |
| A0A5F7Z998 | Vitronectin | VTN | 0.012 | 0.589 |
| A0A1D5Q3K1 | NAD(P)H-hydrate epimerase | NAXE | 0.013 | -1.02 |
| F7EHH7 | Protein-tyrosine-phosphatase | PTPRG | 0.013 | -0.71 |
| A0A5F8A5N3 | Uncharacterized protein | NAPG | 0.013 | 0.893 |
| F7E8L2 | Phosphoenolpyruvate carboxykinase (GTP) | PCK2 | 0.013 | 1.21 |
| P00002 | Cytochrome c | CYCS | 0.014 | -0.56 |
| F7HSJ5 | Histone H2A | H2AC21 | 0.014 | 3.15 |
| A0A5F7ZEM7 | Early endosome antigen 1 | EEA1 | 0.015 | -1.11 |
| A0A1D5QTI7 | 60S ribosomal protein L17 | RPL17 | 0.015 | -0.605 |
| F7DJM1 | SEC22 vesicle-trafficking protein homolog B | SEC22B | 0.015 | 0.62 |
| F6UZQ6 | Reticulon | RTN4 | 0.015 | 0.637 |
| F7H9U1 | Latent transforming growth factor beta binding protein 4 | LTBP4 | 0.017 | -1.24 |
| G7N7C5 | Citrate synthase | CS | 0.018 | -0.523 |
| A0A1D5Q3X0 | Cysteinyl-tRNA synthetase | CARS1 | 0.018 | 0.642 |
| A0A1D5R2U5 | IK cytokine | IK | 0.019 | -1.2 |
| A0A1D5RGB7 | EWS RNA binding protein 1 | EWSR1 | 0.02 | -1.1 |
| F6YK65 | C3/C5 convertase | CFB | 0.02 | 0.827 |
| F6QAX2 | Tubulin beta chain | TUBB3 | 0.02 | 1.58 |
| F6W1K5 | UBX domain protein 1 | UBXN1 | 0.021 | -1.2 |
| A0A5F7ZJ61 | Transforming acidic coiled coil containing protein 2 | TACC2 | 0.021 | -0.671 |
| A0A5F7ZWV4 | Secernin 1 | SCRN1 | 0.021 | 0.576 |
| A0A1D5RGN2 | Cell adhesion molecule 1 | CADM1 | 0.022 | -1.66 |
| A0A1D5Q4R1 | Protein-tyrosine-phosphatase | PTPRM | 0.022 | -1.22 |
| A0A1D5R9J9 | Nucleoside diphosphate kinase | NME3 | 0.022 | -0.721 |
| F7CA88 | Ribonuclease inhibitor | LOC720791 | 0.022 | -0.558 |
| A0A5F7Z9N3 | 60S ribosomal protein L4 | RPL4 | 0.022 | 0.587 |
| F7H400 | Kinectin 1 | KTN1 | 0.023 | -0.898 |
| A0A5F8APU1 | U6 snRNA-associated Sm-like protein LSm4 | LSM4 | 0.023 | -0.697 |
| A0A5F7ZVB9 | Inter-alpha-trypsin inhibitor heavy chain 2 | ITIH2 | 0.023 | 1.28 |
| F6UZ20 | Fibrinogen gamma chain | FGG | 0.023 | 1.57 |
| A0A1D5R0R7 | Histone H2B | H2BC5 | 0.023 | 3.33 |
| F7HAP4 | Histone H2A | H2AC6 | 0.023 | 3.94 |
| F7CDG7 | U1 small nuclear ribonucleoprotein 70 kDa | SNRNP70 | 0.024 | -1.05 |
| A0A1D5RJP5 | Arylsulfatase B | ARSB | 0.024 | -0.66 |
| F6TCB7 | Bis(5'-adenosyl)-triphosphatase | FHIT | 0.025 | -1.33 |
| A0A5F7ZHK9 | H15 domain-containing protein | H1-2 | 0.025 | 1.01 |
| F6QXG0 | Angiopoietin-2 | ANGPT2 | 0.025 | 1.49 |
| A0A5F7ZA28 | Uncharacterized protein | FUBP3 | 0.026 | -1.1 |
| A0A5F7ZC04 | Transcription elongation factor SPT5 | SUPT5H | 0.026 | -0.651 |
| A0A1D5R7P5 | Inositol-1-monophosphatase | IMPA1 | 0.027 | -1.34 |
| A0A1D5QIK3 | SAFB like transcription modulator | SLTM | 0.027 | -0.724 |
| A0A5F8ANH4 | Coagulation factor II | F2 | 0.027 | 1.11 |
| A0A1D5QZ18 | Complement component 8 subunit beta | C8B | 0.027 | 1.26 |
| A0A5F8AKB8 | Histone H2B | H2BC17 | 0.027 | 3.3 |
| A0A1D5R536 | Kinectin 1 | KTN1 | 0.028 | -0.864 |
| F6UZ60 | Fibrinogen alpha chain | FGA | 0.028 | 1.12 |
| F7HAB9 | Golgi associated, gamma adaptin ear containing, ARF binding protein 1 | GGA1 | 0.029 | -1.37 |
| F6WIQ0 | Uncharacterized protein | SET | 0.029 | -1.32 |
| A0A5F7ZB67 | Pyruvate kinase | PKLR | 0.029 | 0.721 |
| A0A1D5R9K8 | Microfibril associated protein 4 | MFAP4 | 0.029 | 0.828 |
| A0A1D5QPA5 | KH-type splicing regulatory protein | KHSRP | 0.03 | -0.814 |
| I0FHZ5 | Phosphoinositide phospholipase C | PLCD1 | 0.03 | 0.748 |
| A0A5F7ZAZ2 | C1q domain-containing protein | C1QC | 0.03 | 1.37 |
| A0A5F7ZNS5 | Uncharacterized protein | ACOT2 | 0.03 | 1.85 |
| A0A1D5QVX8 | ATP-dependent (S)-NAD(P)H-hydrate dehydratase | NAXD | 0.031 | -1.14 |
| A0A5F8A3T7 | Ig-like domain-containing protein | 0.031 | 0.79 | |
| F7AL41 | Formin-binding protein 1-like | FNBP1L | 0.032 | -0.837 |
| F7GRA6 | Complement C8 gamma chain | C8G | 0.032 | 0.7 |
| A0A1D5RJK6 | Uncharacterized protein | FUBP1 | 0.033 | -1.07 |
| H9END9 | Tropomyosin alpha-4 chain isoform 2 | TPM4 | 0.033 | -0.624 |
| A0A5F7ZEZ9 | Fibrinogen beta chain | FGB | 0.034 | 1.72 |
| A0A5F7ZFX7 | Fructose-bisphosphatase | FBP2 | 0.035 | -1.25 |
| A0A5F7ZJW9 | Uncharacterized protein | MANF | 0.035 | -0.838 |
| F6YBP7 | Adipsin | CFD | 0.035 | 1.03 |
| H9Z3B8 | Copper transport protein ATOX1 | ATOX1 | 0.036 | -0.942 |
| A0A1D5R0N5 | Uncharacterized protein | MYL6 | 0.036 | 0.572 |
| A0A1D5Q833 | PTPRF interacting protein alpha 1 | PPFIA1 | 0.037 | -1.02 |
| A0A1D5QJR4 | Host cell factor C1 | HCFC1 | 0.039 | -1.04 |
| F7GXX7 | Small ubiquitin-related modifier | SUMO1P1 | 0.039 | -0.968 |
| A0A1D5QFS4 | Scaffold attachment factor B | SAFB | 0.039 | -0.889 |
| F7APH2 | Heterogeneous nuclear ribonucleoprotein D0 isoform c | HNRNPD | 0.039 | -0.556 |
| F6T0W5 | Tetranectin | CLEC3B | 0.04 | 0.803 |
| F6QQP0 | Uncharacterized protein | 0.041 | 0.737 | |
| A0A1D5QMN6 | Serpin family B member 6 | SERPINB6 | 0.042 | -0.577 |
| F6SJD2 | CXXC motif containing zinc binding protein | CZIB | 0.042 | 0.678 |
| A0A5F7ZI29 | Uncharacterized protein | JCHAIN | 0.042 | 0.731 |
| F6TQ14 | Carbonic anhydrase | CA2 | 0.042 | 0.92 |
| F7F874 | Thiamine pyrophosphokinase | TPK1 | 0.043 | -1.18 |
| F7FF62 | Proteasome activator complex subunit 1 isoform 1 | PSME1 | 0.043 | -0.76 |
| A0A5F7ZKJ3 | Pyruvate dehydrogenase phosphatase regulatory subunit | PDPR | 0.043 | 1.06 |
| F7HNR1 | Anterior gradient 2, protein disulphide isomerase family member | AGR2 | 0.043 | 2.74 |
| H9FXA8 | Proteasome subunit alpha type | PSMA7 | 0.044 | -0.848 |
| A0A1D5QYN2 | Glutathione reductase | GSR | 0.044 | -0.611 |
| I0FNJ1 | Epididymal secretory protein E1 | NPC2 | 0.045 | -0.777 |
| F7FLX2 | GDP-4-keto-6-deoxy-D-mannose-3,5-epimerase-4-reductase | GFUS | 0.045 | 0.503 |
| F7F6K7 | Junctional cadherin 5 associated | JCAD | 0.046 | -2.05 |
| A0A5F8ATC0 | Eukaryotic translation initiation factor 2 subunit beta | EIF2S2 | 0.046 | -0.827 |
| A0A1D5QSC4 | Epidermal growth factor receptor pathway substrate 15 | EPS15 | 0.046 | -0.703 |
| A0A5F8A837 | Uncharacterized protein | SUGT1 | 0.046 | -0.563 |
| F7F2A5 | Metavinculin | VCL | 0.046 | -0.5 |
| F7HER2 | Actinin alpha 3 | ACTN3 | 0.048 | -0.771 |
| G7MLE5 | Phosphoglycerate mutase | PGAM2 | 0.048 | -0.542 |
| F7E3N5 | Phosphatase domain containing paladin 1 | PALD1 | 0.048 | -0.524 |
| A0A5F8ATN0 | Histone H2A | H2AZ2 | 0.048 | 2.66 |
| A0A5F8AJ37 | RAB8B, member RAS oncogene family | RAB8B | 0.049 | -0.55 |
| F7H4P3 | Proteasome subunit beta | PSMB4 | 0.05 | -0.804 |
| A0A5F7ZMU2 | Uncharacterized protein | CFHR5 | 0.05 | 0.766 |
Table 2.
List of DAPs for olive baboons (aged vs. young). Accession number, protein name, gene name, p-value and log2 Fold Change are provided. Proteins are ordered by increasing p-value. Top 5 DAPs are highlighted in bold
| Accession number | Protein name | Gene name | p-value | Log2 aged baboon |
|---|---|---|---|---|
| A0A096NQN2 | Pentraxin | APCS | < 0.0001 | 2.99 |
| A0A096NEV9 | Ig-like domain-containing protein | IGHA | < 0.0001 | 1.84 |
| A0A2I3M4W7 | Dematin actin binding protein | DMTN | 0.00013 | 1.41 |
| A0A2I3MAI1 | Transketolase | TKT | 0.00023 | -0.63 |
| A0A096N1Y1 | Clusterin | CLU | 0.00029 | 0.787 |
| A0A2I3MM55 | Joining chain of multimeric IgA and IgM | JCHAIN | 0.00038 | 1.78 |
| A0A2I3MW37 | Paraspeckle component 1 | PSPC1 | 0.0004 | -1.91 |
| A0A096P228 | Complement C4-A | C4A | 0.00042 | 1.29 |
| A0A096NJK4 | Serpin family A member 3 | SERPINA3 | 0.00043 | 1.38 |
| A0A2I3MWE6 | Ig-like domain-containing protein | IGHM | 0.00062 | 1.35 |
| A0A2I3LD62 | Transcription elongation regulator 1 | TCERG1 | 0.00068 | -1.37 |
| A0A2I3N2B9 | IGv domain-containing protein | 0.00071 | 1.25 | |
| A0A096NIN4 | Cell division cycle 5 like | CDC5L | 0.00071 | 1.17 |
| A0A096MM00 | Non-POU domain containing octamer binding | NONO | 0.00075 | -1.8 |
| A0A096NN03 | HDGF like 3 | HDGFL3 | 0.00084 | 2.74 |
| A0A2I3NEJ3 | Laminin subunit beta 2 | LAMB2 | 0.0009 | 0.731 |
| A0A096NUT9 | RALY heterogeneous nuclear ribonucleoprotein | RALY | 0.0011 | -0.892 |
| A0A2I3LWK4 | Immunoglobulin heavy variable 3–72 | IGHV3-72 | 0.0012 | 1.46 |
| A0A0A0MUA3 | Heterogeneous nuclear ribonucleoprotein M | HNRNPM | 0.0012 | -1.43 |
| A0A096MPI8 | Filaggrin family member 2 | FLG2 | 0.0013 | 2.12 |
| A0A096NSI5 | Calretinin | CALB2 | 0.0015 | 2.43 |
| A0A096N317 | Splicing factor, proline- and glutamine-rich | SFPQ | 0.0015 | -1.15 |
| A0A2I3MJZ3 | Actinin alpha 2 | ACTN2 | 0.0015 | -1.44 |
| A0A2I3MDY2 | Myocardin related transcription factor B | MRTFB | 0.0016 | -1.48 |
| A0A0A0MU60 | Aldehyde dehydrogenase family 16 member A1 | ALDH16A1 | 0.0017 | 1.93 |
| A0A2I3NGY5 | Ig-like domain-containing protein | 0.0017 | 1.16 | |
| A0A2I3NC97 | Ig-like domain-containing protein | 0.0017 | 1 | |
| A0A2I3MP06 | Tropomyosin 4 | TPM4 | 0.0017 | 0.55 |
| A0A2I3M158 | Ubiquitin recognition factor in ER associated degradation 1 | UFD1 | 0.0017 | -1.87 |
| A0A0A0MU58 | Actinin alpha 4 | ACTN4 | 0.002 | -1.56 |
| A0A2I3LZX6 | Uncharacterized protein | HNRNPH2 | 0.0021 | -1.79 |
| P05770 | Apolipoprotein E | APOE | 0.0023 | 1.39 |
| A0A2I3M2P0 | Heterogeneous nuclear ribonucleoprotein H1 | HNRNPH1 | 0.0023 | -1.65 |
| A0A096N1T1 | Histidine rich glycoprotein | HRG | 0.0024 | 0.801 |
| A0A2I3MT71 | Complement C1r subcomponent like | C1RL | 0.0025 | 1.36 |
| A0A2I3LTI2 | General vesicular transport factor p115 | USO1 | 0.0028 | -1.17 |
| A0A096NKK0 | Serpin family B member 9 | SERPINB9 | 0.0029 | -1.07 |
| A0A2I3MTT7 | Ribosomal protein L22 | RPL22 | 0.0029 | -1.69 |
| A0A096NK84 | Cysteine rich protein 1 | CRIP1 | 0.0032 | 1.95 |
| A0A2I3LU29 | PC4 and SFRS1 interacting protein 1 | PSIP1 | 0.0032 | 1.09 |
| A0A096P0K3 | Spectrin alpha, non-erythrocytic 1 | SPTAN1 | 0.0033 | -0.771 |
| A0A2I3MTE9 | Ig-like domain-containing protein | 0.0035 | 1.36 | |
| A0A2I3M360 | Inter-alpha-trypsin inhibitor heavy chain 5 | ITIH5 | 0.0036 | -0.971 |
| A0A2I3LFB9 | Zinc finger protein 207 | ZNF207 | 0.0036 | -0.985 |
| A0A2I3M0F1 | Lymphocyte cytosolic protein 2 | LCP2 | 0.0037 | 1.65 |
| A0A2I3MCZ4 | Ig-like domain-containing protein | 0.0044 | 0.832 | |
| A0A2I3LP09 | Shisa family member 7 | SHISA7 | 0.0048 | -0.728 |
| A0A0A0MU78 | Shisa family member 7 | SHISA7 | 0.0048 | -0.749 |
| A0A096N709 | Transaldolase | TALDO1 | 0.0048 | -0.771 |
| A0A096N345 | Hemopexin | HPX | 0.0049 | 0.689 |
| A0A2I3LXY2 | Non-specific serine/threonine protein kinase | SLK | 0.005 | -1 |
| A0A2I3LHU4 | HLA class II histocompatibility antigen, DR beta 3 chain | 0.0054 | 4.55 | |
| A0A096N4D7 | Ro60, Y RNA binding protein | RO60 | 0.0057 | -0.627 |
| A0A2I3LMG7 | Spectrin alpha, non-erythrocytic 1 | SPTAN1 | 0.0057 | -0.978 |
| A0A096NFT8 | MARCKS like 1 | MARCKSL1 | 0.0059 | 1.75 |
| A0A2I3N4Y0 | Calumenin | CALU | 0.006 | 1.65 |
| A0A2I3LNH3 | KH-type splicing regulatory protein | KHSRP | 0.006 | -0.719 |
| A0A096P4D3 | Multimerin 2 | MMRN2 | 0.0061 | 0.851 |
| A0A2I3LJS5 | Myosin heavy chain 10 | MYH10 | 0.0062 | -0.923 |
| A0A2I3NAA8 | Apoptosis inducing factor mitochondria associated 1 | AIFM1 | 0.0062 | -1.3 |
| A0A096N7V3 | Complement component 4 binding protein alpha | C4BPA | 0.0063 | 1.33 |
| A0A2I3M1T5 | Adenosine deaminase RNA specific | ADAR | 0.0066 | -0.934 |
| A0A2I3MSR7 | Tropomyosin 3 | TPM3 | 0.0067 | 0.647 |
| A0A2I3MIX0 | DNA damage-binding protein 1 | DDB1 | 0.0067 | -0.627 |
| A0A096MYE5 | Myosin regulatory light chain 12A | MYL12A | 0.0068 | 0.914 |
| A0A096NL32 | Enoyl-CoA delta isomerase 1 | ECI1 | 0.0068 | -2.39 |
| A0A2I3MZY8 | Aldo–keto reductase family 1 member C1 homolog | AKR1C1 | 0.0069 | -1.09 |
| A0A2I3MSB6 | Cytochrome b-c1 complex subunit 7 | UQCRB | 0.0075 | 3.13 |
| A0A2I3LPQ6 | CXADR Ig-like cell adhesion molecule | CXADR | 0.0076 | 1.06 |
| A0A096MWL2 | Syndecan binding protein | SDCBP | 0.0076 | 0.622 |
| A0A2I3M6Q6 | Nuclear mitotic apparatus protein 1 | NUMA1 | 0.0077 | -0.665 |
| A0A2I3LE24 | PTPRF interacting protein alpha 1 | PPFIA1 | 0.0077 | -0.696 |
| A0A2I3LH89 | Ig-like domain-containing protein | 0.0078 | 1.12 | |
| A0A096P0S3 | V-type proton ATPase subunit G | ATP6V1G1 | 0.0079 | 3.14 |
| A0A096NBS2 | Selenocysteine lyase | SCLY | 0.0085 | -0.526 |
| A0A096NZE2 | Small RNA binding exonuclease protection factor La | SSB | 0.0086 | -1.61 |
| A0A096NFU9 | Ig-like domain-containing protein | 0.0088 | 2.54 | |
| A0A096NV22 | Endoplasmic reticulum oxidoreductase 1 alpha | ERO1A | 0.009 | 0.597 |
| A0A096NRJ1 | CD59 glycoprotein | 0.009 | 0.56 | |
| A0A096P5I3 | Keratin 27 | KRT27 | 0.0092 | 1.13 |
| A0A2I3LH21 | Dynein cytoplasmic 1 intermediate chain 2 | DYNC1I2 | 0.0096 | -1.36 |
| A0A2I3M314 | QKI, KH domain containing RNA binding | QKI | 0.0098 | -1.41 |
| A0A2I3N4K0 | G3BP stress granule assembly factor 1 | G3BP1 | 0.01 | 1.28 |
| A0A096N6A0 | Myristoylated alanine rich protein kinase C substrate | MARCKS | 0.01 | 0.988 |
| A0A2I3N200 | Heterogeneous nuclear ribonucleoprotein D | HNRNPD | 0.01 | -0.604 |
| A0A096P2E8 | Nuclear cap-binding protein subunit 3 | NCBP3 | 0.011 | 1.44 |
| A0A096NM11 | Ig-like domain-containing protein | 0.011 | 1.35 | |
| A0A2I3MSK0 | Nucleoporin 153 | NUP153 | 0.011 | -1.4 |
| A0A2I3LID9 | RNA binding protein fox-1 homolog 1 | RBFOX1 | 0.011 | -1.42 |
| B0VYX8 | Cytochrome c oxidase subunit 5A, mitochondrial | COX5A | 0.012 | 0.938 |
| A0A2I3LRD7 | Laminin subunit alpha 3 | LAMA3 | 0.012 | 0.93 |
| A0A096NT92 | Galectin-3-binding protein | LGALS3BP | 0.012 | 0.553 |
| A0A096N5V4 | Chitinase-3-like protein 1 | CHI3L1 | 0.012 | -1.2 |
| A0A2I3MZJ5 | Talin 1 | TLN1 | 0.013 | -0.643 |
| A0A2I3M4M4 | Cell division cycle and apoptosis regulator 1 | CCAR1 | 0.013 | -1.34 |
| A0A2I3MIF7 | Filamin-C | FLNC | 0.014 | -1.84 |
| A0A0A0MWY7 | Splicing factor U2AF subunit | U2AF2 | 0.014 | -1.89 |
| A0A096MQP5 | Ig-like domain-containing protein | 0.015 | 1.57 | |
| A0A096NVY7 | BH3-interacting domain death agonist | BID | 0.015 | 0.966 |
| A0A096NEY1 | Choline-specific glycerophosphodiester phosphodiesterase | ENPP6 | 0.015 | -1.75 |
| A0A096NKT3 | Desmoplakin | DSP | 0.016 | 0.618 |
| A0A096MNQ3 | C-X-C motif chemokine | PPBP | 0.016 | -0.575 |
| A0A2I3MIV3 | Actinin alpha 1 | ACTN1 | 0.016 | -1.13 |
| A0A0A0MXC4 | Transcription elongation factor SPT5 | SUPT5H | 0.016 | -1.23 |
| A0A096MPU5 | Serpin B6-like | SERPINB6 | 0.016 | -1.4 |
| A0A2I3LKR9 | Septin | SEPTIN11 | 0.016 | -1.43 |
| A0A2I3M614 | Keratin, type I cuticular Ha3-I | KRT33A | 0.017 | 1.53 |
| A0A0A0MWZ3 | Calponin | CNN1 | 0.017 | 1.03 |
| A0A2I3LNI7 | Acidic nuclear phosphoprotein 32 family member A | ANP32A | 0.017 | 1.02 |
| A0A096N402 | Cathepsin D | CTSD | 0.017 | 0.545 |
| A0A096N9Y0 | Clathrin interactor 1 | CLINT1 | 0.017 | -0.729 |
| A0A2I3M3R5 | Prefoldin subunit 4 | PFDN4 | 0.018 | 1.82 |
| A0A096NUK6 | Myosin light chain 9 | MYL9 | 0.018 | 0.841 |
| A0A2I3MGL1 | Solute carrier family 12 member 2 | SLC12A2 | 0.018 | 0.783 |
| A0A2I3NB00 | EPS8 like 2 | EPS8L2 | 0.018 | -0.57 |
| A0A0A0MUI0 | Tripartite motif containing 28 | TRIM28 | 0.018 | -0.869 |
| A0A2I3N473 | 60S ribosomal protein L8 | RPL8 | 0.018 | -1.88 |
| A0A2I3MQY3 | Immunoglobulin kappa variable 4–1 | IGKV4-1 | 0.019 | 0.84 |
| A0A096N1J7 | Apolipoprotein D | APOD | 0.019 | 0.617 |
| A0A096NFB0 | All-trans-retinol dehydrogenase [NAD( +)] ADH1B | ADH7 | 0.019 | -0.898 |
| A0A096MWK3 | Filamin A | FLNA | 0.019 | -0.938 |
| A0A096N6E3 | Hyaluronan and proteoglycan link protein 1 | HAPLN1 | 0.019 | -2.56 |
| A0A2I3LE14 | Keratin 10 | KRT10 | 0.02 | 1.03 |
| A0A096N0H9 | Adducin 2 | ADD2 | 0.02 | 0.514 |
| A0A096N1Y4 | PKSO | ADH7 | 0.02 | -0.795 |
| A0A2I3MZ49 | Serine and arginine rich splicing factor 5 | SRSF5 | 0.021 | 2.9 |
| A0A2I3MDH7 | Serine and arginine rich splicing factor 10 | SRSF10 | 0.021 | 1.68 |
| A0A096N0E7 | Marginal zone B and B1 cell specific protein | MZB1 | 0.021 | 1.35 |
| A0A096N6Z6 | Poly(rC) binding protein 1 | PCBP1 | 0.021 | -1.09 |
| A0A096NA79 | Transcription factor Sp1 | SP1 | 0.021 | -1.52 |
| A0A096MU45 | Ig-like domain-containing protein | 0.022 | 1.1 | |
| A0A2I3MM66 |
Protein S100 TYMP |
S100A11 | 0.022 | 0.506 |
| A0A096MN47 | Keratin 77 | KRT77 | 0.023 | 1.08 |
| A0A2I3LS05 | 2-phospho-D-glycerate hydro-lyase | ENO2 | 0.023 | -0.942 |
| A0A096MVE7 | RWD domain containing 1 | RWDD1 | 0.024 | 1.3 |
| A0A2I3N9B6 | Host cell factor C1 | HCFC1 | 0.024 | -0.707 |
| A0A2I3LXU6 | Ribosomal protein | RPL10A | 0.024 | -1.16 |
| A0A2I3MXU2 | Synaptotagmin binding cytoplasmic RNA interacting protein | SYNCRIP | 0.024 | -1.19 |
| A0A2I3M825 | Zinc finger RNA binding protein | ZFR | 0.024 | -1.31 |
| A0A2I3M8L5 | Synaptopodin 2 | SYNPO2 | 0.025 | 1.58 |
| A0A2I3M1I3 | Latent transforming growth factor beta binding protein 1 | LTBP1 | 0.025 | 0.972 |
| A0A096MV46 | Immunoglobulin kappa variable 6D-41 (non-functional) | IGKV6D-41 | 0.025 | 0.764 |
| A0A096NR90 | Ig-like domain-containing protein | 0.026 | 1.04 | |
| A0A2I3MML7 | Ig-like domain-containing protein | 0.026 | 0.842 | |
| A0A2I3LKS6 | Tight junction protein 1 | TJP1 | 0.026 | -0.559 |
| A0A096P3B1 | Aldo–keto reductase family 1 member C1 homolog | AKR1C1 | 0.026 | -1.05 |
| A0A2I3MLZ3 | Seryl-tRNA synthetase | SARS1 | 0.026 | -1.3 |
| A0A096N2S9 | F-spondin | SPON1 | 0.026 | -1.47 |
| A0A096MYJ5 | Copine 3 | CPNE3 | 0.027 | 0.539 |
| A0A096NAR9 | Spectrin beta chain | SPTBN1 | 0.028 | -0.624 |
| A0A096N5A4 | HtrA serine peptidase 2 | HTRA2 | 0.029 | 0.528 |
| A0A2I3LNW4 | Tight junction protein 2 | TJP2 | 0.029 | -0.729 |
| A0A096MNW7 | Adipocyte-type fatty acid-binding protein | FABP4 | 0.029 | -1.23 |
| A0A2I3N8Y4 | 3-beta-hydroxysterol Delta (14)-reductase | LBR | 0.031 | 1.79 |
| A0A2I3LFF0 | Keratin 17 | KRT17 | 0.031 | 1.67 |
| A0A096P1P8 | Frataxin, mitochondrial | FXN | 0.031 | 0.969 |
| A0A2I3LJL3 | Non-specific protein-tyrosine kinase | PTK2 | 0.031 | -0.525 |
| A0A2I3MM96 | Heat shock protein family A (Hsp70) member 5 | HSPA5 | 0.031 | -1.87 |
| A9L8U0 | Hemoglobin subunit theta 1 | HBQ1 | 0.032 | 1.51 |
| A0A2I3N512 | 4-trimethylaminobutyraldehyde dehydrogenase | ALDH9A1 | 0.032 | 0.727 |
| A0A2I3LIT0 | GLI pathogenesis related 2 | GLIPR2 | 0.032 | -0.513 |
| A0A096N1H4 | Splicing factor 3b subunit 1 | SF3B1 | 0.032 | -0.9 |
| A0A0A0MWU1 | Bromodomain containing 4 | BRD4 | 0.032 | -0.965 |
| A0A096N5N8 | Uncharacterized protein | HNRNPA3 | 0.032 | -2.26 |
| A0A096P131 | Acidic nuclear phosphoprotein 32 family member B | ANP32B | 0.034 | 0.83 |
| A0A096P5P4 | Junction plakoglobin | JUP | 0.034 | 0.652 |
| A0A096ML84 | Uncharacterized protein | HNRNPA3 | 0.034 | -1.94 |
| A9X1C4 | Huntingtin interacting protein K | HYPK | 0.035 | 1.06 |
| A0A096NPT5 | Heterogeneous nuclear ribonucleoprotein A3 | HNRNPA3 | 0.036 | -1.15 |
| A0A096MXZ5 | Serine and arginine rich splicing factor 9 | SRSF9 | 0.037 | 0.767 |
| A0A096NZG7 | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase | BST1 | 0.037 | 0.509 |
| A0A2I3M5I3 | Small nuclear ribonucleoprotein U5 subunit 200 | SNRNP200 | 0.037 | -0.749 |
| A0A2I3LIJ2 | Septin | SEPTIN7 | 0.037 | -0.873 |
| A0A096N4V8 | Keratin, type II cytoskeletal 6C | KRT5 | 0.039 | 1.18 |
| A0A2I3M017 | Parvalbumin | PVALB | 0.039 | 0.543 |
| A0A096MV19 | Thyroid hormone receptor associated protein 3 | THRAP3 | 0.039 | -0.742 |
| A0A096P4S7 | Serine/arginine-rich splicing factor 1 | SRSF1 | 0.04 | 0.871 |
| A0A2I3N6J2 | Tensin 1 | TNS1 | 0.041 | -0.64 |
| A0A096MQV4 | Lamin A/C | LMNA | 0.041 | -1.16 |
| A0A2I3MZ19 | RNA binding protein, mRNA processing factor | RBPMS | 0.042 | 1.23 |
| A0A096NDH2 | CSD domain-containing protein | YBX1 | 0.042 | 1.16 |
| A0A2I3LXT9 | PDS5 cohesin associated factor B | PDS5B | 0.042 | -0.97 |
| A0A096N233 | Taxilin alpha | TXLNA | 0.043 | 4.23 |
| A0A096NUU1 | Charged multivesicular body protein 4B | CHMP4B | 0.043 | 1 |
| A0A2I3MLK1 | Pinin | PNN | 0.044 | -1.18 |
| A0A2I3M0D0 | Zinc finger CCCH-type containing 11A | ZC3H11A | 0.044 | -1.45 |
| A0A2I3N574 | RRM domain-containing protein | RBMX | 0.045 | 1.21 |
| A0A2I3M706 | Ig-like domain-containing protein | 0.045 | 0.556 | |
| A0A096P534 | Phenazine biosynthesis like protein domain containing | PBLD | 0.045 | -1.28 |
| A0A096NPS7 | All-trans-retinol dehydrogenase [NAD( +)] ADH1B | ADH7 | 0.045 | -1.32 |
| A0A2I3LFE6 | Four and a half LIM domains 1 | FHL1 | 0.046 | -1.39 |
| A0A096NWE1 | Crystallin beta-gamma domain containing 3 | CRYBG3 | 0.046 | -2.76 |
| A4K2N4 | Syndecan | SDC4 | 0.047 | 1.13 |
| A0A2I3LK27 | Moesin | MSN | 0.047 | -0.711 |
| A0A096NZQ0 | Ferredoxin 1 | FDX1 | 0.048 | 0.929 |
| A0A2I3MLU1 | CDKN2A interacting protein | CDKN2AIP | 0.048 | -1.8 |
| A0A096ML57 | Acyl-CoA dehydrogenase family member 8 | ACAD8 | 0.049 | 0.877 |
| A0A096NAU0 | Cytokeratin-1 | KRT1 | 0.049 | 0.8 |
| A0A2I3N6R9 | Heterogeneous nuclear ribonucleoprotein R | HNRNPR | 0.049 | -0.897 |
| A0A2I3LZ61 | Vitelline membrane outer layer 1 homolog | VMO1 | 0.049 | -0.906 |
| A0A096N6J5 | Calcium-activated chloride channel regulator 1 | CLCA1 | 0.049 | -1.57 |
| A0A2I3LNR5 | Amyloid beta precursor like protein 2 | APLP2 | 0.05 | 1.59 |
| A0A2I3MNR7 | Helix-destabilizing protein | HNRNPA1 | 0.05 | -1.35 |
Of the 11 shared DAPs, three were significantly upregulated in both NHP species when comparing aged vs. young animals (Fig. 1A, Tables 1 and 2) (listed here as human protein, followed by gene name equivalent and log2 fold changes in both NHPs). Serum amyloid P-component or SAP [APCS (RM 1.14/OB 2.99)] is a pentraxin associated with amyloid deposits with roles in innate and acute immune responses. Increased SAP levels could potentially be linked to disease severity since other serum amyloid-associated proteins (e.g. Serum amyloid A-component) are known markers of COVID-19 severity [17, 18]. This was followed by the upregulation of Immunoglobulin heavy constant mu [IGHM (RM 1.06/OB 1.35)] and Immunoglobulin J chain [JCHAIN (RM 0.73/OB 1.78)], both components of immunoglobulin M (IgM), directly associated with SARS-CoV-2 infection and regulation of the host complement system. High IgM levels have been associated with an altered immune response and represent a risk factor for the development of recurrent lung infections, such as SARS-CoV-2 [19].
The remaining eight DAPs were significantly downregulated in both NHP species (aged vs. young) during early stages of SARS-CoV-2 infection (Fig. 1A, Tables 1 and 2): Host cell factor 1 [HCFC1 (RM -1.04/OB -0.707)] is crucial to regulate the host cell cycle and is recognized by SARS-CoV-2 [20], although no direct correlation with COVID-19 progression has been described. Liprin-alpha-1 [PPFIA1 (RM -1.02 /OB -0.696)], a LAR family transmembrane protein-tyrosine phosphatase involved in cell–matrix interactions [21], was previously identified as a potential gene candidate of Acute Lung Injury (ALI) risk [22], which is the most severe form of COVID-19, and has also been linked to age-associated changes [23]. Next is far upstream element-binding protein 2 [KHSRP or FUBP2 (RM -0.814/OB -0.719)], involved in mRNA biogenesis, stability and trafficking, and regulation of innate and adaptive immune responses, with decreased host antiviral defense in their absence [24, 25], which could be associated with the increased disease pathology and susceptibility observed in aged animals. Further, we observed downregulation of transcription elongation factor SPT5 [SUPT5H (RMs -0.651/OB -1.23)], a component of the DRB sensitivity-induced complex (DSIF) that regulates mRNA processing and facilitates rapid induction of pro-inflammatory genes through NF-κB activation [26, 27]. SPT5 downregulation would result in decreased immune response, which could favor SARS-CoV-2 replication.
Two RNA-binding proteins (RBPs) were also downregulated in both NHP species. RBP Raly [RALY (RM -0.533/OB -0.892)] targets ssRNAs and acts as an antiviral, although the SARS-CoV-2 genome was found to be depleted of interacting motifs with RALY, suggesting that the viral genome might have evolved to avoid interactions with this host defense protein [28]. Still, this might be a double-edged sword, with viral evasion and lower basal levels to respond against infection. On the other hand, RBP RO60 [RO60 (RM -0.605/OB -0.627)] binds to non-coding misfolded Y RNAs, and regulates some pro-inflammatory genes in autoimmune disorders such as Guillain-Barré syndrome and rheumatoid arthritis. Autoantibodies against RO60 are found in COVID-19 patients, suggestive of some viral-host interaction [29].
The last two proteins downregulated in both rhesus macaques and olive baboons (aged vs. young) were: heterogeneous nuclear ribonucleoprotein D0 [HNRNPD (RM -0.556/OB -0.604)], which binds to heterogenous nuclear RNA (hnRNA) and is thought to prevent age-related neurodegenerative diseases [30], has been found under expressed in COVID-19 patients, further supporting interactions of SARS-CoV-2 with host factors [31]. Finally, Serpin B6 [SERPINB6 (RM -0.577/OB -1.4)] is a serine protease inhibitor and its downregulation is associated with increased SARS-CoV-2 uptake and replication [32], which could explain the higher COVID-19 pathology observed in aged infected animals.
Lastly, we found a few other DAPs (aged vs. young) that were shared by rhesus macaques and olive baboons but only significant (log2 fold change ≤ -0.5 or ≥ 0.5 and p-value < 0.05) in one of these two NHP species. Among the ones over expressed, we found Desmoplakin [DSP (RM 0.444/OB 0.618)] and Junction plakoglobin [JUP (RM 0.0392/OB 0.652)], significantly upregulated in aged olive baboons. These are structural proteins of the cardiac desmosome and intermediate filaments. DSP has been described as a major player in aging lung diseases [33], and is potentially associated with viral myocarditis [34], while levels of JUP were altered in COVID-19 recovered patients with long-term cardiac dysfunction [35]. Also associated with the potential to cause cardiac disease, Tropomyosin alpha-4 chain [TPM4 (RM 0.0628/OB 0.55)] is a common autoantigen in humans identified as part of the COVID-19 autoimmunogenicity repertoire [36]. We also found Clusterin [CLU (RM 0.307/OB 0.787)], which will be discussed in the next section.
Among the ones under expressed, Latent-transforming growth factor beta-binding protein 4 [LTBP4 (RMs -1.24/OB—0.218)] is a regulator of TGFβ signaling, and it has recently emerged as a driver of age-related organ pathologies in a mitochondrial-dependent manner [37]. It was previously found decreased in COVID-19 lung autopsy specimens [38]. Further, Far Upstream element-Binding Protein 1 [FUBP1 (RM -1.07/OB -0.441)] is a master regulator of cell function with oncogenic associations as a dual-agent [39]. Downregulated FUBP1 is associated with higher risk of COVID-19 due to its antiviral function, which may be contributing to increased disease severity in aged NHPs [40]. Next, Alpha-actinin-1 [ACTN1 (RM -0.223/OB -1.13)], a cytoskeleton protein implicated in inflammatory and degenerative autoimmune diseases, is shown to interact with SARS-CoV-2 proteins. Indeed, upregulation of related protein Alpha-actinin-4 results in a protective response to COVID-19 [41], suggesting that downregulation of Alpha-actinin-1 could be associated with disease susceptibility. Finally, Bromodomain-containing protein 4 [BRD4 (RM -0.176/OB -0.965)], an epigenetic regulator with multifaceted roles in aging-related diseases [42], has been associated with the cardiac cytokine storm during SARS-CoV-2 infection [43].
Overall, these results suggest the dysregulation of several immune processes in aged NHPs during SARS-CoV-2 infection, and provide clues of potential host factors that might be associated with increased susceptibility as we age.
Top DAPs in SARS-CoV-2 infected lung tissues from aged vs. young rhesus macaques and olive baboons
We next explored significant species-specific DAPs in aged vs. young SARS-CoV-2 infected rhesus macaques (Table 1, Suppl. Fig S1A) and aged vs. young SARS-CoV-2 infected olive baboons (Table 2, Suppl. Fig S1B) (p-value < 0.05, absolute log2 fold change > 0.5) (Fig. 1).
The top five DAPs based on p-value for rhesus macaques were (gene name, log2 fold change, Accession #) (Table 1, Suppl. Fig S1A): calpain 5 (CAPN5, 1.45, F7EDT3), Protein-Tyrosine phosphatase receptor J (PTPRJ,—0.771, F6TSU2), pentraxin (APCS, 1.14, F7H1V9), heme-binding protein 1 (HEBP1, 1.46, F6UQB6) and t-SNARE coiled-coil homology domain-containing protein (PPP1R8, -0.644, F6V089). Calpains are calcium-activated proteases involved in apoptosis, cell proliferation and motility, and overactivation has been previously associated with aging diseases and pathological lung conditions [44, 45]. SARS-CoV-2 appears to activate calpains, resulting in increased cell death, and their inhibition has been studied as a means of stopping COVID-19 related cytokine storm, inflammation, and pulmonary fibrosis [46–48]. Thus, upregulation of calpain 5 could be involved in the increased disease severity observed in aged NHPs. PTPRJ contributes to protein dephosphorylation, with roles in cell adhesion, migration, proliferation and differentiation, as well as regulation of a variety of immune cells. Downregulation of PTPRJ, as observed here, has been associated with numerous diseases, such as idiopathic pulmonary fibrosis [49]. Indeed, pulmonary fibrosis is one of the long-term consequences of COVID-19 [50]. Thus, under expression of PTPRJ in aged rhesus macaques could indicate an increased potential for developing post-COVID conditions. PTPRJ has also been shown to interact with SARS-CoV-2 ORF3a [51]. Regarding immune effector pentraxin or serum amyloid P component, which was over expressed in aged rhesus macaques, elevated levels of pentraxin 3 have been associated with COVID-19 severity and mortality, and it has been proposed as a promising biomarker for long COVID [52]. Next, HEBP1 has high affinity for heme and porphyrins modulating mitochondrial dynamics, and has been linked to synaptic vulnerability during aging [53]. Its metabolism is interfered by SARS-CoV-2 [54]. Finally, no association has been shown between PPP1R8 and COVID-19.
The top five DAPs based on p-value for olive baboons were (Gene name, Log2 Fold Change, Accession #) (Table 2, Suppl. Fig. S1B): pentraxin (APCS, 2.99, A0A096NQN2), also a top upregulated DAP in RMs (see above), immunoglobulin heavy constant alpha (IGHA, 1.84, A0A096NEV9), dematin (DMTN, 1.41, A0A2I3M4W7), Transketolase (TKT, -0.63, A0A2I3MAI1), and clusterin (CLU, 0.787, A0A096N1Y1). The over expression of IGHA in aged baboon lungs found here is consistent with the basal increase seen during aging in humans [55]. Although IgA plays an important role during early SARS-CoV-2 neutralization [56], hyperactivation can result in IgA-mediated diseases and has been linked to post-COVID conditions [57], suggesting again that risk of developing long COVID is increased as we age. Dematin, a cytoskeleton-associated protein with functions in formation, bundling and stabilization of F-actin, was also over expressed in aged baboons. Interestingly, there was increased phosphorylation of dematin in patients with post-COVID-19 interstitial lung changes associated with upregulation of proinflammatory immune signatures [58], suggesting an altered immune response during aging. Conversely, transketolase, and enzyme that connects the pentose phosphate pathway to glycolysis, was under expressed in aged infected baboons, although reports indicate that SARS-CoV-2 infection upregulates transketolase levels [59]. Lastly, clusterin is a glycoprotein that functions as a stress-activated and ATP-independent molecular chaperone. It is predicted to be a strong interactor of ACE2 receptor, and was found increased in the lungs of coronavirus-infected individuals [60], in agreement with our observations in aged baboons. Interestingly, it seems that cellular localization of clusterin (intra- or extracellular) leads to divergent effects on epithelial cell regeneration and lung repair during fibrosis, with both beneficial and detrimental roles [61], although its role during COVID-19 is yet to be understood.
Overall, several of the factors found differentially expressed in aged NHPs seem to play a role in the development of post-COVID diseases, in agreement with a greater risk of persisting symptoms associated with COVID-19 among older people (65 + years) [62].
Pathway analysis in aged NHPs
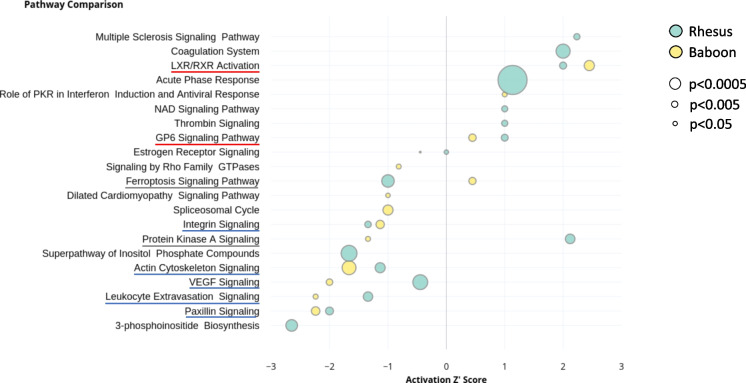
We further explored pathways relevant to SARS-CoV-2 infection focusing in aged NHPs. We identified nine common pathways enriched in both NHP species studied (Fig. 2 and Table 3). Based on the calculated IPA Z-scores, the ones with reduced activation were Paxillin signaling, Leukocyte Extravasation signaling, VEGF signaling, Actin cytoskeleton signaling, and Integrin signaling. The ones with increased activation were GP6 signaling pathway and LXR/RXR activation. We also identified two divergent pathways: Protein kinase A signaling (activation increased in rhesus macaques but decreased in olive baboons) and Ferroptosis signaling (activation decreased in rhesus macaques but increased in olive baboons) pathways.
Fig. 2.
Comparison of top 21 canonical pathways in aged rhesus macaques and aged olive baboons. Rhesus macaques (cyan) and olive baboons (yellow) activation Z-scores (an inferred measurement of the pathway activation stated based on measured levels and IPA knowledge) is shown by bubble position on the X-axis, pathway names in the Y-axis, where p-values are represented by bubble size. Highly overlapping pathways were removed from analysis and only the most comprehensive pathway is displayed. Enriched pathways in both rhesus macaques and olive baboons are underlined in red (increased activation), blue (reduced activation), or grey (divergent)
Table 3.
Comparison of canonical pathways in aged rhesus macaques and aged olive baboons. List of all pathways reported in IPA, with pathway activation z-score and -log p-values provided for the ones detected in old rhesus macaques and old olive baboons. Pathways are ordered by activation trend and are considered significantly enriched if -log (p-value) > 1.3 (equivalent to p-value < 0.05). Enriched pathways in both NHP species are highlighted in bold
| Canonical pathways | Rhesus z-score | Baboon z-score | Rhesus -log p-value | Baboon -log p-value |
|---|---|---|---|---|
| LXR/RXR Activation | 2.0000 | 2.4490 | 2.0803 | 3.0287 |
| Paxillin Signaling | -2.0000 | -2.2360 | 2.2914 | 2.5223 |
| Leukocyte Extravasation Signaling | -1.3420 | -2.2360 | 2.7958 | 1.4927 |
| Protein Kinase A Signaling | 2.1210 | -1.3420 | 2.8470 | 1.4524 |
| Actin Cytoskeleton Signaling | -1.1340 | -1.6670 | 2.9773 | 4.0654 |
| D-myo-inositol (1,4,5,6)-Tetrakisphosphate Biosynthesis | -2.6460 | N/A | 3.7825 | 0.0000 |
| D-myo-inositol (3,4,5,6)-tetrakisphosphate Biosynthesis | -2.6460 | N/A | 3.7825 | 0.0000 |
| 3-phosphoinositide Degradation | -2.6460 | N/A | 3.6098 | 0.0000 |
| 3-phosphoinositide Biosynthesis | -2.6460 | N/A | 3.4238 | 0.0000 |
| Integrin Signaling | -1.3420 | -1.1340 | 1.9137 | 2.4694 |
| VEGF Signaling | -0.4470 | -2.0000 | 4.3537 | 1.8903 |
| Multiple Sclerosis Signaling Pathway | 2.2360 | N/A | 1.8347 | 0.0000 |
| Intrinsic Prothrombin Activation Pathway | 2.0000 | N/A | 3.8150 | 0.4957 |
| Coagulation System | 2.0000 | N/A | 4.1297 | 0.5618 |
| Superpathway of Inositol Phosphate Compounds | -1.6670 | N/A | 4.6609 | 0.0000 |
| D-myo-inositol-5-phosphate Metabolism | -1.6670 | N/A | N/A | N/A |
| Ferroptosis Signaling Pathway | -1.0000 | 0.4470 | 3.6630 | 2.1370 |
| GP6 Signaling Pathway | 1.0000 | 0.4470 | 2.0326 | 2.2068 |
| Acute Phase Response Signaling | 1.1340 | N/A | 8.5300 | N/A |
| MicroRNA Biogenesis Signaling Pathway | 0.0000 | -1.1340 | N/A | N/A |
| Spliceosomal Cycle | N/A | -1.0000 | 0.0000 | 2.9871 |
| EIF2 Signaling | 0.0000 | -1.0000 | N/A | N/A |
| Thrombin Signaling | 1.0000 | N/A | 1.8118 | 0.4724 |
| Dilated Cardiomyopathy Signaling Pathway | N/A | -1.0000 | 0.5990 | 1.3127 |
| Complement System | 1.0000 | N/A | N/A | N/A |
| NAD Signaling Pathway | 1.0000 | N/A | 1.7796 | 0.3971 |
| Role of PKR in Interferon Induction and Antiviral Response | N/A | 1.0000 | 0.6606 | 1.4429 |
| RHOA Signaling | N/A | -0.8160 | N/A | N/A |
| Signaling by Rho Family GTPases | N/A | -0.8160 | 0.2883 | 1.4431 |
| ILK Signaling | 0.0000 | -0.7070 | N/A | N/A |
| Apoptosis Signaling | N/A | -0.4470 | N/A | N/A |
| Estrogen Receptor Signaling | 0.0000 | -0.4470 | 1.3055 | 0.4987 |
| Asparagine Degradation I | N/A | N/A | 1.8914 | 0.0000 |
| Agranulocyte Adhesion and Diapedesis | N/A | N/A | 0.8124 | 1.3602 |
| D-myo-inositol (1,4,5)-Trisphosphate Biosynthesis | N/A | N/A | 1.9170 | 0.0000 |
| Virus Entry via Endocytic Pathways | N/A | N/A | 0.7534 | 2.3407 |
| IL-33 Signaling Pathway | N/A | N/A | 2.1435 | 0.0000 |
| Sertoli Cell-Sertoli Cell Junction Signaling | N/A | N/A | N/A | N/A |
| Protein Ubiquitination Pathway | N/A | N/A | 2.0631 | 0.3427 |
| Myo-inositol Biosynthesis | N/A | N/A | 1.4976 | 0.0000 |
| Production of Nitric Oxide and Reactive Oxygen Species in Macrophages | N/A | N/A | 1.4523 | 0.5971 |
| Role of Tissue Factor in Cancer | N/A | N/A | 3.0278 | 0.0000 |
| Pentose Phosphate Pathway | N/A | N/A | 0.0000 | 2.3660 |
| Gluconeogenesis I | N/A | N/A | N/A | N/A |
| Tumoricidal Function of Hepatic Natural Killer Cells | N/A | N/A | N/A | N/A |
| Granzyme B Signaling | N/A | N/A | N/A | N/A |
| UVA-Induced MAPK Signaling | N/A | N/A | 2.4270 | 0.0000 |
| FAT10 Signaling Pathway | N/A | N/A | N/A | N/A |
| Remodeling of Epithelial Adherens Junctions | N/A | N/A | N/A | N/A |
| Extrinsic Prothrombin Activation Pathway | N/A | N/A | N/A | N/A |
| Regulation of Cellular Mechanics by Calpain Protease | N/A | N/A | N/A | N/A |
| Noradrenaline and Adrenaline Degradation | N/A | N/A | 0.0000 | 1.3499 |
| Thyroid Hormone Biosynthesis | N/A | N/A | 0.0000 | 1.7413 |
| GDP-L-fucose Biosynthesis I (from GDP-D-mannose) | N/A | N/A | 1.8914 | 0.0000 |
| TCA Cycle II (Eukaryotic) | N/A | N/A | 2.0203 | 0.0000 |
| CSDE1 Signaling Pathway | N/A | N/A | N/A | N/A |
| Isoleucine Degradation I | N/A | N/A | 0.0000 | 1.9398 |
| Thiamin Salvage III | N/A | N/A | 2.1910 | 0.0000 |
| FXR/RXR Activation | N/A | N/A | N/A | N/A |
| Apelin Cardiomyocyte Signaling Pathway | N/A | N/A | N/A | N/A |
| Regulation of eIF4 and p70S6K Signaling | N/A | N/A | 1.5330 | 0.0000 |
| Antigen Presentation Pathway | N/A | N/A | 1.5826 | 0.0000 |
| NADH Repair | N/A | N/A | N/A | N/A |
| Inhibition of ARE-Mediated mRNA Degradation Pathway | N/A | N/A | N/A | N/A |
| L-carnitine Biosynthesis | N/A | N/A | 0.0000 | 1.5672 |
| 2-ketoglutarate Dehydrogenase Complex | N/A | N/A | 1.5931 | 0.0000 |
| BAG2 Signaling Pathway | N/A | N/A | N/A | N/A |
| Fatty Acid β-oxidation III (Unsaturated, Odd Number) | N/A | N/A | 0.0000 | 1.4443 |
| Pentose Phosphate Pathway (Non-oxidative Branch) | N/A | N/A | N/A | N/A |
| Chronic Myeloid Leukemia Signaling | 0.0000 | N/A | N/A | N/A |
| Coronavirus Replication Pathway | N/A | N/A | N/A | N/A |
| Germ Cell-Sertoli Cell Junction Signaling | N/A | N/A | N/A | N/A |
| Tight Junction Signaling | N/A | N/A | N/A | N/A |
| Clathrin-mediated Endocytosis Signaling | N/A | N/A | N/A | N/A |
| Phagosome Maturation | N/A | N/A | N/A | N/A |
| Insulin Secretion Signaling Pathway | 0.0000 | N/A | N/A | N/A |
| Valine Degradation I | N/A | N/A | N/A | N/A |
| Gap Junction Signaling | N/A | N/A | N/A | N/A |
| Inosine-5'-phosphate Biosynthesis II | N/A | N/A | N/A | N/A |
| Ethanol Degradation II | N/A | N/A | 0.0000 | 1.3938 |
| Glycolysis I | N/A | N/A | N/A | N/A |
| Rapoport-Luebering Glycolytic Shunt | N/A | N/A | N/A | N/A |
| Glutathione Redox Reactions II | N/A | N/A | N/A | N/A |
| 14–3-3-mediated Signaling | N/A | N/A | N/A | N/A |
| Death Receptor Signaling | N/A | 0.0000 | N/A | N/A |
| PPARα/RXRα Activation | N/A | N/A | N/A | N/A |
| Iron homeostasis signaling pathway | N/A | N/A | N/A | N/A |
Most of these pathways have been previously associated with COVID-19. Indeed, disruption of the VEGF-related pathways was previously seen during both acute and long COVID-19 [63], in agreement with our observations. Further, disruption of the host actin cytoskeleton, as seen here, is tightly connected to pathological processes during SARS-CoV-2 infection, where the virus highjacks cytoskeletal functions leading to increased viral loads, dissemination, and immune dysfunction [64]. Conversely, while integrin activation is essential for SARS-CoV-2 infection [65], we found reduced integrin signaling in aged NHPs. Indeed, it has been shown that inflammatory cytokines activate certain integrins, promoting cellular adherence to counter-receptors such as ICAMs resulting in phagocytosis and cytotoxic killing [66]. Low integrin signaling in the elderly could potentially be associated to decreased killing and thus, increased susceptibility to infections. In addition, activation of the GP6 pathway was observed in whole blood from COVID-19 patients and its upregulation constituted a molecular signature for COVID-19 progression and severity [67, 68], indicating a potential link with the increased pathology observed in aged NHPs. Next, the liver X receptor/retinoid X receptor or LXR/RXR activation pathway, which plays a key role in the regulation of cholesterol, lipid metabolism, and inflammation, was found enriched here as well as in a multi-omics study that linked this pathway to COVID-19 severity [69]. Interestingly, a higher HDL-cholesterol measured before infection was associated with a lower risk of death during COVID-19 in older humans [70]. Lastly, activation of the ferroptosis pathway by SARS-CoV-2 might be associated with COVID-19 cardiovascular complications [71], although we only found increased activation in aged olive baboons (but decreased in rhesus macaques). This could be related to the higher disease pathology observed in baboons compared to rhesus [72].
Other statistically significant pathways were identified in only one of the two NHP species studied and are presented in Suppl. Table S1 (rhesus macaques) and Suppl. Table S2 (olive baboons). In aged rhesus macaques, the top three altered pathways based on p-value were: the Acute Phase Response signaling, the Complement system, and the Extrinsic Prothrombin Activation Pathway. The Acute Phase Response signaling, a rapid inflammatory response that provides protection against microorganisms, was found activated and suggests an overactive immune response in aged NHPs, similar to our observations in elderly humans [73]. The Complement system was also activated, specifically, the Alternative Pathway, which may play a major role in SARS-CoV-2 pathogenesis [74]. Upregulation of both acute phase response signaling and the complement system were previously seen in COVID-19 patients, linking it to IL-6 production, increased inflammation and tissue damage [75, 76]. Finally, the Extrinsic Prothrombin Activation Pathway had a z-score = 0, but with increased levels of fibrin, alpha-thrombin and fibrinogen, as well as coagulation factor II, indicating increased risk of COVID-19-related coagulopathy in aged rhesus macaques, similar to humans [77]. Inhibition of the complement pathway as a therapeutic strategy during COVID-19 resulted in reduced acute-phase reaction and thrombin activity, further linking these three pathways to disease severity [78]. Complete pathway results for rhesus macaques can be found in Suppl. Table S1.
In aged olive baboons, the top three statistically significant enriched pathways based on p-value (with assigned z-score) were: the Actin cytoskeleton signaling, the Integrin-linked kinase (ILK) signaling pathway, and the LXR/RXR signaling pathways. The Actin cytoskeleton signaling was inhibited in both NHP species, as mentioned earlier, suggesting reduced focal adhesion assembly and actin polymerization, like what we observed previously in elderly humans [73]. ILK signaling (closely related to actin cytoskeleton signaling) was also inhibited in aged olive baboons; and the LXR/RXR signaling pathway (activated in both species) suggests an increase in lipogenesis, cholesterol efflux, and transport. This finding agrees with several studies that identified a dysregulation of the lipid transport system as a key signature of COVID-19 [69, 79]. And contrary to what we observed here, inhibition of the LXR/RXR pathway was associated with prolonged viral RNA shedding during COVID-19, often associated with carriers with increased contagion risk such as the elderly. A potential explanation is that we might not have captured this effect since we only studied lung tissues from acute SARS-CoV-2 infection in NHPs. Complete pathway results for olive baboons can be found in Suppl. Table S2.
Clinical relevance and study limitations
Acute respiratory distress in rhesus macaques and olive baboons partially recapitulates the progression of SARS-CoV-2 infection in humans, making them suitable animal models to test vaccines and therapies [9]. Indeed, our published studies of SARS-CoV-2 infection in NHP models indicate that both young and old rhesus macaques develop clinical signs of viral infection, mild-to-moderate pneumonitis, and extra-pulmonary pathologies [9]. However, independent of their age, rhesus macaques can clear SARS-CoV-2 viral particles to undetectable levels within two weeks. Conversely, baboons have prolonged viral RNA shedding and substantially more lung inflammation compared with macaques, with higher lung inflammation in aged vs. young baboons [9].
Human proteomic studies provide a platform to uncover new molecular pathways associated with COVID-19 severity, identifying the involvement of complement factors, the coagulation system, inflammation modulators, and pro-inflammatory factors upstream and downstream of IL-6 [80, 81]. Indeed, a proteomics study identified inflammatory response modulator S100A8/A9 as being highly expressed in fatal COVID-19 cases [82]. Another study reported blood clot formation, severe extracellular matrix restructuring, and impaired tissue repair signaling in end-stage COVID-19 lung biopsies [6]. Other groups performed multiorgan proteomics analyses and revealed shared (e.g. RIPK1 or BRD4, the latter also showing altered expression in this study) and tissue-specific factors associated with SARS-CoV-2 infection, suggesting the need of organ-specific therapeutic interventions [83, 84]. However, most of these studies are performed in blood or serum (systemic changes) or are limited to post-mortem lung samples (tissue-specific responses), which are not easily available, with very few addressing COVID-19 lung-specific responses in aging individuals. NHP models that recapitulate COVID-19 pathology and progression represent a suitable alternative to address this gap in knowledge. Indeed, a COVID-19 infection model in cynomolgus macaques was able to recapitulate moderate COVID-19 symptoms [85].
In our current study we determined tissue-specific responses to SARS-CoV-2 infection using lung tissues from both young and aged NHPs (Fig. 3). Aged infected NHPs had significantly increased DAPs associated with regulation of the cytokine storm and the complement system (e.g. SAP, IGHM, JCHAIN, Clusterin). We also found increased DAPs associated with regulation of heart functions and the cardiovascular system (e.g. DSP, JUP, TPM4), specifically controlling blood flux. Indeed, aging plasma in healthy individuals was found enriched for heart and aorta specific proteins, revealing age-specific changes, which could potentially contribute to COVID-19 responses [86]. Other studies demonstrated that aged vascular endothelial cells (ECs) are highly susceptible to SARS-CoV-2 infection and subsequent endothelial dysfunction, resulting in fatal pneumonia with thrombosis in aged mice, suggesting age-associated EC responses in severe human COVID-19 [87, 88]. In this regard, L-arginine has been shown to enhance cardiac rehabilitation after myocardial infarction, whose risk is increased after COVID-19 [89, 90]. Proteins involved in clearance of cellular debris and apoptosis (e.g. Clusterin) were also more abundant in aged infected NHPs, as well as proteins involved in accelerating viral evasion and replication in host cells (e.g. RALY, RO60).
Fig. 3.
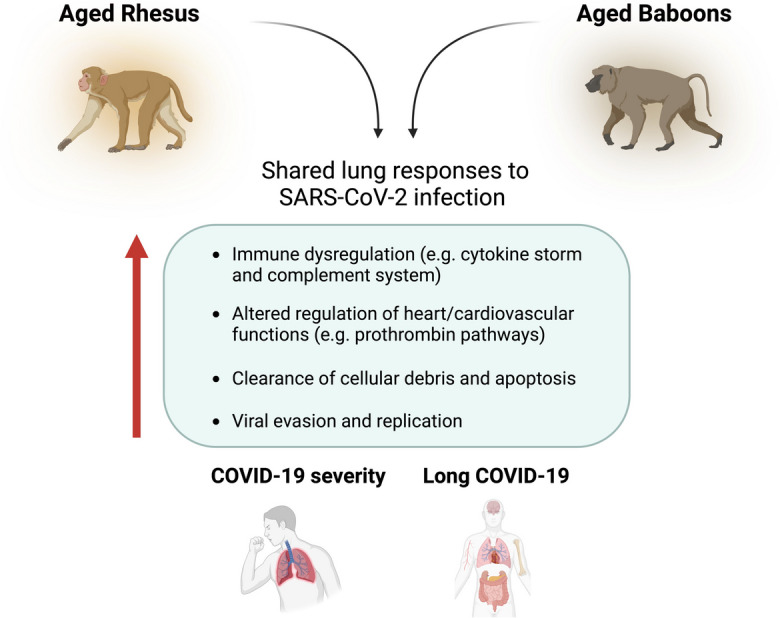
Summary figure of shared lung responses to SARS-CoV-2 infection in aged rhesus macaques and olive baboons. Figure created with BioRender.com
Proteins and pathways previously seen to be involved in long COVID (e.g. Pentraxin, IGHA, VEGF signaling) were also enriched in aged NHPs (Fig. 3). These findings suggest that aged NHPs can serve as a good model to study long-term health consequences and outcomes for long COVID-19, a poorly understood condition, especially in the elderly population [62]. Indeed, in the elderly, long COVID-19 seems to make existing chronic diseases worse, where elders with disorders such as heart failure, lung disease, or dementia, among others, develop more serious symptoms after recovering from COVID-19 [62].
Our results in the current study were limited by the number of available NHP lung tissue samples, which might not be representative of the overall NHP responses. In addition, samples from uninfected controls were not available, thus limiting our interpretation of the effect of SARS-CoV-2 infection (compared to uninfected NHPs), as well as the differences between young and aged NHPs at the baseline level (no infection). Indeed, lower pre-infection levels of HDL-cholesterol and vitamin D deficiency was associated with greater risk of adverse COVID-19 outcomes in elderly people [70, 91], as well as antecedent use of RASIs (renin–angiotensin system inhibitors) was associated with lower all-cause mortality in elderly hypertensive COVID-19 patients [92], which we were not able to determine here since pre-infection samples were not available. Further, the study had a single observation, at 14 days post-infection (necropsy timepoint), which did not allow us to study the proteome dynamics as disease progresses. Also, we used pre-existing tissue biopsies, previously snap-frozen, of approximately 0.5 cm3. The small size of these biopsies meant we may not have captured all local responses to infection. Indeed, a deep spatial proteomics study of human lung autopsy specimens confirmed region-specific dysregulation of protein expression in key pulmonary structures, including alveolar epithelia, bronchial epithelia, and blood vessels, among others [7].
Despite these limitations, important findings were uncovered that support the use of these models as discovery agents for respiratory infectious diseases, especially in the context of aging where limited human samples are available. This and future studies can provide clues to delineate the mechanisms that explain why COVID-19 severity is increased in the elderly.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
We would like to thank Dr. Luis D. Giavedoni and the Southwest National Primate Research Center (SNPRC) Biorepository services for providing the NHP samples studied. We thank Dr. Luis Martinez-Sobrido and his laboratory personnel for providing the SARS-CoV-2 viral stock used in the original study to infect the NHP from which the samples were obtained. MS analyses were conducted in the Institutional Mass Spectrometry Laboratory of UT Health San Antonio, with support from UT Health San Antonio for the laboratory and the University of Texas System for purchase of the Orbitrap Fusion Lumos mass spectrometer. We thank Mr. Pardo and Ms. Molleur for their MS technical support.
Author contributions
A.G.-V., A.A.-G. and A.A., sample processing and data analysis. S.T.W., DIA-MS analysis. D.K.S. provided samples. A.G.-V., J.B.T. study conceptualization, experimental design. A.G.-V., A.A.-G., N.M.C, and J.B.T wrote the manuscript. B.I.R., L.S.S., J.T., and J.B.T. provided funding and critical comments. All authors read and approved the final version of this manuscript.
Funding
Studies supported by the National Institutes of Health/National Institute on Aging (NIH/NIA) 3P01-AG-051428-05S1 (Supplement) award to B.I.R., L.S.S., J.B.T. and J.T. A.G.-V., A.A.-G., and J.B.T. are part of the Interdisciplinary NextGen Tuberculosis Research Advancement Center (IN-TRAC) at Texas Biomed, which is supported by the NIH/National Institute of Allergy and Infectious Diseases (NIAID) under the award number P30-AI-168439. The content in this publication is solely the responsibility of the authors and does not necessarily represent the official views of the NIH.
Data availability
Data supporting findings are available in Supplementary Materials. Raw Mass Spectrometry files and the Data-Independent Acquisition file are publicly available at MassIVE (ProteomeXchange consortium), http://massive.ucsd.edu, under MassIVE identifier: MSV000094333; ProteomeXchange identifier: PXD050683 (for both rhesus macaque and olive baboon species).
Declarations
Conflict of interest
The authors declare no conflict of interest exists.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Andreu Garcia-Vilanova, Email: garciavilanovaandreu@gmail.com.
Anna Allué-Guardia, Email: aallueguardia@txbiomed.org.
Jordi B. Torrelles, Email: jtorrelles@txbiomed.org
References
- 1.WHO (WHO). "WHO Coronavirus (COVID-19) Dashboard - https://covid19.who.int/." WHO. (accessed 2023).
- 2.Chen Y, et al. Aging in COVID-19: Vulnerability, immunity and intervention. Ageing Res Rev. 2021;65:101205. 10.1016/j.arr.2020.101205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.O’Driscoll M, et al. Age-specific mortality and immunity patterns of SARS-CoV-2. Nature. 2021;590(7844):140–5. 10.1038/s41586-020-2918-0. [DOI] [PubMed] [Google Scholar]
- 4.Farshbafnadi M, Kamali Zonouzi S, Sabahi M, Dolatshahi M, Aarabi MH. Aging & COVID-19 susceptibility, disease severity, and clinical outcomes: The role of entangled risk factors. Exp Gerontol. 2021;154:111507. 10.1016/j.exger.2021.111507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yanez ND, Weiss NS, Romand JA, Treggiari MM. COVID-19 mortality risk for older men and women. BMC Public Health. 2020;20(1):1742. 10.1186/s12889-020-09826-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gindlhuber J, et al. Proteomic profiling of end-stage COVID-19 lung biopsies. Clin Proteomics. 2022;19(1):46. 10.1186/s12014-022-09386-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mao Y, et al. Deep spatial proteomics reveals region-specific features of severe COVID-19-related pulmonary injury. Cell Rep. 2024;43(2):113689. 10.1016/j.celrep.2024.113689. [DOI] [PubMed] [Google Scholar]
- 8.Nie X, et al. Multi-organ proteomic landscape of COVID-19 autopsies. Cell. 2021;184(3):775-791.e14. 10.1016/j.cell.2021.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Singh DK, et al. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat Microbiol. 2021;6(1):73–86. 10.1038/s41564-020-00841-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Trichel AM. Overview of Nonhuman Primate Models of SARS-CoV-2 Infection. Comp Med. 2021;71(5):411–32. 10.30802/AALAS-CM-20-000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singh DK, et al. Myeloid cell interferon responses correlate with clearance of SARS-CoV-2. Nat Commun. 2022;13(1):679. 10.1038/s41467-022-28315-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosa BA, et al. IFN signaling and neutrophil degranulation transcriptional signatures are induced during SARS-CoV-2 infection. Commun Biol. 2021;4(1):290. 10.1038/s42003-021-01829-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Searle BC, et al. Chromatogram libraries improve peptide detection and quantification by data independent acquisition mass spectrometry. Nat Commun. 2018;9(1):5128. 10.1038/s41467-018-07454-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gessulat S, et al. Prosit: proteome-wide prediction of peptide tandem mass spectra by deep learning. Nat Methods. 2019;16(6):509–18. 10.1038/s41592-019-0426-7. [DOI] [PubMed] [Google Scholar]
- 15.Broom BM, et al. A Galaxy Implementation of Next-Generation Clustered Heatmaps for Interactive Exploration of Molecular Profiling Data. Cancer Res. 2017;77(21):e23–6. 10.1158/0008-5472.CAN-17-0318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.M. C. Ryan et al. "Interactive clustered heat map builder: An easy web-based tool for creating sophisticated clustered heat maps." F1000Res, 2019;8. 10.12688/f1000research.20590.2. [DOI] [PMC free article] [PubMed]
- 17.Zinellu A, Paliogiannis P, Carru C, Mangoni AA. Serum amyloid A concentrations, COVID-19 severity and mortality: An updated systematic review and meta-analysis. Int J Infect Dis. 2021;105:668–74. 10.1016/j.ijid.2021.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Almusalami EM, Lockett A, Ferro A, Posner J. Serum amyloid A-A potential therapeutic target for hyper-inflammatory syndrome associated with COVID-19. Front Med (Lausanne). 2023;10:1135695. 10.3389/fmed.2023.1135695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.V. Gallo ,et al. "Clinical, immunological, and functional characterization of six patients with very high IgM levels." J Clin Med, 2020;9(3). 10.3390/jcm9030818. [DOI] [PMC free article] [PubMed]
- 20.Sobhy H. The potential functions of protein domains during COVID infection: An analysis and a review. COVID Rev. 2021;1:384–93. 10.3390/covid1010032. [Google Scholar]
- 21.Serra-Pages C, Medley QG, Tang M, Hart A, Streuli M. Liprins, a family of LAR transmembrane protein-tyrosine phosphatase-interacting proteins. J Biol Chem. 1998;273(25):15611–20. 10.1074/jbc.273.25.15611. [DOI] [PubMed] [Google Scholar]
- 22.Christie JD, et al. Genome wide association identifies PPFIA1 as a candidate gene for acute lung injury risk following major trauma. PLoS One. 2012;7(1): e28268. 10.1371/journal.pone.0028268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rodwell GE, et al. A transcriptional profile of aging in the human kidney. PLoS Biol. 2004;2(12):e427. 10.1371/journal.pbio.0020427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.K. B. Karunakaran, N. Balakrishnan, M. K. Ganapathiraju. "Interactome of SARS-CoV-2 / nCoV19 modulated host proteins with computationally predicted PPIs." Res Sq, 2020. 10.21203/rs.3.rs-28592/v1.
- 25.Labeau A, et al. Characterization and functional interrogation of the SARS-CoV-2 RNA interactome. Cell Rep. 2022;39(4):110744. 10.1016/j.celrep.2022.110744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Walker AP, Fodor E. Interplay between Influenza Virus and the Host RNA Polymerase II Transcriptional Machinery. Trends Microbiol. 2019;27(5):398–407. 10.1016/j.tim.2018.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Diamant G, Bahat A, Dikstein R. The elongation factor Spt5 facilitates transcription initiation for rapid induction of inflammatory-response genes. Nat Commun. 2016;7:11547. 10.1038/ncomms11547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.A. Lal, M. Galvao Ferrarini, A. J. Gruber, "Investigating the Human Host-ssRNA Virus Interaction Landscape Using the SMEAGOL Toolbox." Viruses, 2022;14(7). 10.3390/v14071436. [DOI] [PMC free article] [PubMed]
- 29.Fujii H, et al. High levels of anti-SSA/Ro antibodies in COVID-19 patients with severe respiratory failure: a case-based review : High levels of anti-SSA/Ro antibodies in COVID-19. Clin Rheumatol. 2020;39(11):3171–5. 10.1007/s10067-020-05359-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Q, et al. Nuclear speckle specific hnRNP D-like prevents age- and AD-related cognitive decline by modulating RNA splicing. Mol Neurodegener. 2021;16(1):66. 10.1186/s13024-021-00485-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang C, et al. Abnormal global alternative RNA splicing in COVID-19 patients. PLoS Genet. 2022;18(4):e1010137. 10.1371/journal.pgen.1010137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johansen MD, et al. Increased SARS-CoV-2 Infection, Protease, and Inflammatory Responses in Chronic Obstructive Pulmonary Disease Primary Bronchial Epithelial Cells Defined with Single-Cell RNA Sequencing. Am J Respir Crit Care Med. 2022;206(6):712–29. 10.1164/rccm.202108-1901OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.van Moorsel CHM. Desmoplakin: An Important Player in Aging Lung Disease. Am J Respir Crit Care Med. 2020;202(9):1201–2. 10.1164/rccm.202006-2457ED. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rezaei Bookani K, et al. A case series of desmoplakin cardiomyopathy: a mimic of viral myocarditis. Eur Heart J Case Rep. 2022;6(8):ytac341. 10.1093/ehjcr/ytac341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen Y, et al. 2022 “Immune response pattern across the asymptomatic, symptomatic and convalescent periods of COVID-19,.” Biochim Biophys Acta Proteins Proteom. 1870;2:140736. 10.1016/j.bbapap.2021.140736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang JY, Zhang W, Roehrl MW, Roehrl VB, Roehrl MH. An autoantigen profile of human A549 lung cells reveals viral and host etiologic molecular attributes of autoimmunity in COVID-19. J Autoimmun. 2021;120:102644. 10.1016/j.jaut.2021.102644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Neikirk K, et al. Latent transforming growth factor beta binding protein 4: A regulator of mitochondrial function in acute kidney injury. Aging Cell. 2023;22(12):e14019. 10.1111/acel.14019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.M. Fassan, et al. "Multi-design differential expression profiling of COVID-19 lung autopsy specimens reveals significantly deregulated inflammatory pathways and SFTPC impaired transcription." Cells, 2022;11(6). 10.3390/cells11061011. [DOI] [PMC free article] [PubMed]
- 39.Debaize L, Troadec MB. The master regulator FUBP1: its emerging role in normal cell function and malignant development. Cell Mol Life Sci. 2019;76(2):259–81. 10.1007/s00018-018-2933-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shi W, et al. Integration of risk variants from GWAS with SARS-CoV-2 RNA interactome prioritizes FUBP1 and RAB2A as risk genes for COVID-19. Sci Rep. 2023;13(1):19194. 10.1038/s41598-023-44705-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Y. Zhou, et al. "A comprehensive SARS-CoV-2-human protein-protein interactome network identifies pathobiology and host-targeting therapies for COVID-19." Res Sq, 2022. 10.21203/rs.3.rs-1354127/v2.
- 42.J. Sun, Y. Gui, S. Zhou, X. L. Zheng. "Unlocking the secrets of aging: Epigenetic reader BRD4 as the target to combatting aging-related diseases." J Adv Res, 2023. 10.1016/j.jare.2023.11.006. [DOI] [PMC free article] [PubMed]
- 43.Mills RJ, et al. BET inhibition blocks inflammation-induced cardiac dysfunction and SARS-CoV-2 infection. Cell. 2021;184(8):2167-2182.e22. 10.1016/j.cell.2021.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nixon RA. The calpains in aging and aging-related diseases. Ageing Res Rev. 2003;2(4):407–18. 10.1016/s1568-1637(03)00029-1. [DOI] [PubMed] [Google Scholar]
- 45.Kovacs L, Su Y. Redox-dependent Calpain signaling in airway and pulmonary vascular remodeling in COPD. Adv Exp Med Biol. 2017;967:139–60. 10.1007/978-3-319-63245-2_9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Juibari AD, Rezadoost MH, Soleimani M. The key role of Calpain in COVID-19 as a therapeutic strategy. Inflammopharmacology. 2022;30(5):1479–91. 10.1007/s10787-022-01002-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Inal J, Paizuldaeva A, Terziu E. Therapeutic use of calpeptin in COVID-19 infection. Clin Sci (Lond). 2022;136(20):1439–47. 10.1042/CS20220638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ma C, et al. Boceprevir, GC-376, and calpain inhibitors II, XII inhibit SARS-CoV-2 viral replication by targeting the viral main protease. Cell Res. 2020;30(8):678–92. 10.1038/s41422-020-0356-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.H. Li, et al. "The structure, function and regulation of protein tyrosine phosphatase receptor type J and its role in diseases." Cells, 2022;12(1). 10.3390/cells12010008. [DOI] [PMC free article] [PubMed]
- 50.Rodriguez-Galan I, Albaladejo-Blazquez N, Ruiz-Robledillo N, Pascual-Lledo JF, Ferrer-Cascales R, Gil-Carbonell J. Impact of COVID-19 on quality of life in survivors with pulmonary sequelae. Sci Rep. 2024;14(1):6926. 10.1038/s41598-024-57603-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Z, et al. Interactomes of SARS-CoV-2 and human coronaviruses reveal host factors potentially affecting pathogenesis. Embo J. 2021;40(17):e107776. 10.15252/embj.2021107776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Crupi L, Capra AP, Pantò G, Repici A, Calapai F, Squeri R, Ardizzone A, Esposito E. Serum Pentraxin 3 as promising biomarker for the long-lasting inflammatory response of COVID-19. Int J Mol Sci. 2023;24(18):14195. 10.3390/ijms241814195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Graham LC, et al. Regional molecular mapping of primate synapses during normal healthy aging. Cell Rep. 2019;27(4):1018-1026.e4. 10.1016/j.celrep.2019.03.096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.G. C. Lechuga, et al. "SARS-CoV-2 Proteins Bind to Hemoglobin and Its Metabolites." Int J Mol Sci, 2021;22(16). 10.3390/ijms22169035. [DOI] [PMC free article] [PubMed]
- 55.Batory G, Jancso A, Puskas E, Redei A, Lengyel E. Antibody and immunoglobulin levels in aged humans. Arch Gerontol Geriatr. 1984;3(2):175–88. 10.1016/0167-4943(84)90009-8. [DOI] [PubMed] [Google Scholar]
- 56.D. Sterlin, et al. "IgA dominates the early neutralizing antibody response to SARS-CoV-2." Sci Transl Med 2021;13(577). 10.1126/scitranslmed.abd2223. [DOI] [PMC free article] [PubMed]
- 57.Farooq H, Ur Rehman MA, Asmar A, Asif S, Mushtaq A, Qureshi MA. The pathogenesis of COVID-19-induced IgA nephropathy and IgA vasculitis: A systematic review. J Taibah Univ Med Sci. 2022;17(1):1–13. 10.1016/j.jtumed.2021.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.George PM, et al. A persistent neutrophil-associated immune signature characterizes post-COVID-19 pulmonary sequelae. Sci Transl Med. 2022;14(671):eabo5795. 10.1126/scitranslmed.abo5795. [DOI] [PubMed] [Google Scholar]
- 59.D. Bojkova, et al. "Targeting the pentose phosphate pathway for SARS-CoV-2 therapy." Metabolites,2021;11(10). 10.3390/metabo11100699. [DOI] [PMC free article] [PubMed]
- 60.Varela AA, Cheng S, Werren JH. Novel ACE2 protein interactions relevant to COVID-19 predicted by evolutionary rate correlations. PeerJ. 2021;9:e12159. 10.7717/peerj.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Habiel DM, et al. Divergent roles for clusterin in lung injury and repair. Sci Rep. 2017;7(1):15444. 10.1038/s41598-017-15670-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mansell V, Hall Dykgraaf S, Kidd M, Goodyear-Smith F. Long COVID and older people. Lancet Healthy Longev. 2022;3(12):e849–54. 10.1016/S2666-7568(22)00245-8. [DOI] [PubMed] [Google Scholar]
- 63.R. Talotta. "Impaired VEGF-A-Mediated neurovascular crosstalk induced by SARS-CoV-2 spike protein: A potential hypothesis explaining long COVID-19 symptoms and COVID-19 vaccine side effects?." Microorganisms, 2022;10(12). 10.3390/microorganisms10122452. [DOI] [PMC free article] [PubMed]
- 64.M. Aminpour, S. Hameroff, J. A. Tuszynski, "How COVID-19 hijacks the cytoskeleton: therapeutic implications." Life (Basel), 2022; 12(6). 10.3390/life12060814. [DOI] [PMC free article] [PubMed]
- 65.Liu J, Lu F, Chen Y, Plow E, Qin J. Integrin mediates cell entry of the SARS-CoV-2 virus independent of cellular receptor ACE2. J Biol Chem. 2022;298(3):101710. 10.1016/j.jbc.2022.101710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Mezu-Ndubuisi OJ, Maheshwari A. The role of integrins in inflammation and angiogenesis. Pediatr Res. 2021;89(7):1619–26. 10.1038/s41390-020-01177-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.D. L. Ng et al. "A diagnostic host response biosignature for COVID-19 from RNA profiling of nasal swabs and blood." Sci Adv, 2021; 7(6). 10.1126/sciadv.abe5984. [DOI] [PMC free article] [PubMed]
- 68.De R, Azad RK. Molecular signatures in the progression of COVID-19 severity. Sci Rep. 2022;12(1):22058. 10.1038/s41598-022-26657-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lipman D, Safo SE, Chekouo T. Multi-omic analysis reveals enriched pathways associated with COVID-19 and COVID-19 severity. PLoS One. 2022;17(4):e0267047. 10.1371/journal.pone.0267047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mostaza JM, et al. Pre-infection HDL-cholesterol levels and mortality among elderly patients infected with SARS-CoV-2. Atherosclerosis. 2022;341:13–9. 10.1016/j.atherosclerosis.2021.12.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Q, Chen Z, Zhou X, Li G, Zhang C, Yang Y. Ferroptosis and multi-organ complications in COVID-19: mechanisms and potential therapies. Front Genet. 2023;14:1187985. 10.3389/fgene.2023.1187985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Singh DG, Singh SR, Cole B, Alfson J, Clemmons KJ, Gazi E, Gonzalez M, Escobedo O, Lee R, Chatterjee T-H, Goez-Gazi A, Sharan Y, Thippeshappa R, Gough R, Alvarez M, Blakley C, Ferdin A, Bartley J, Staples C, Parodi H, Callery P, Mannino J, Klaffke A, Escareno B, PlattII P, Hodara RN, Scordo V, Oyejide JM, Ajithdoss A, Copin DK, Baum R, Kyratsous A, Alvarez C, Rosas X, Ahmed B, Goodroe M, Dutton A, Hall-Ursone J, Frost S, Voges PA, Ross AK, Sayers CN, Chen K, Hallam C, Khader C, Mitreva SA, Anderson M, Martinez-Sobrido TJC, Patterson L, Turner JL, Torrelles J, Dick JB Jr, Brasky EJ, Schlesinger K, Giavedoni LS, Carrion LD Jr, Kaushal RD. Responses to acute infection with SARS-CoV-2 in the lungs of rhesus macaques, baboons and marmosets. Nat Microbiol. 2020;6(1):73–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.A. Garcia-Vilanova, et al. "The aging human lung mucosa: A proteomics study." J Gerontol: Series A, 2022;091. 10.1093/gerona/glac091 [DOI] [PMC free article] [PubMed]
- 74.Yu J, et al. Complement dysregulation is associated with severe COVID-19 illness. Haematologica. 2022;107(5):1095–105. 10.3324/haematol.2021.279155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Sayit AT, Elmali M, Deveci A, Gedikli O. Relationship between acute phase reactants and prognosis in patients with or without COVID-19 pneumonia. Rev Inst Med Trop Sao Paulo. 2021;63:e51. 10.1590/S1678-9946202163051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Demichev V, et al. A time-resolved proteomic and prognostic map of COVID-19. Cell Syst. 2021;12(8):780-794.e7. 10.1016/j.cels.2021.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Yuan X, Tong X, Wang Y, Wang H, Wang L, Xu X. Coagulopathy in elderly patients with coronavirus disease 2019. Aging Med (Milton). 2020;3(4):260–5. 10.1002/agm2.12133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Skendros P, et al. Complement C3 inhibition in severe COVID-19 using compstatin AMY-101. Sci Adv. 2022;8(33):eabo2341. 10.1126/sciadv.abo2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Overmyer KA, et al. Large-Scale Multi-omic Analysis of COVID-19 Severity. Cell Syst. 2021;12(1):23-40.e7. 10.1016/j.cels.2020.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Messner CB, et al. Ultra-High-Throughput Clinical Proteomics Reveals Classifiers of COVID-19 Infection. Cell Syst. 2020;11(1):11-24.e4. 10.1016/j.cels.2020.05.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Yin S, et al. Integrated Immunopeptidomic and Proteomic Analysis of COVID-19 lung biopsies. Front Immunol. 2023;14:1269335. 10.3389/fimmu.2023.1269335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Wu M, et al. Transcriptional and proteomic insights into the host response in fatal COVID-19 cases. Proc Natl Acad Sci U S A. 2020;117(45):28336–43. 10.1073/pnas.2018030117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Feng L, et al. Proteome-wide data analysis reveals tissue-specific network associated with SARS-CoV-2 infection. J Mol Cell Biol. 2020;12(12):946–57. 10.1093/jmcb/mjaa033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Schweizer L, et al. Quantitative multiorgan proteomics of fatal COVID-19 uncovers tissue-specific effects beyond inflammation. EMBO Mol Med. 2023;15(9):e17459. 10.15252/emmm.202317459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang T, et al. Proteomic and metabolomic characterization of SARS-CoV-2-Infected cynomolgus macaque at early stage. Front Immunol. 2022;13:954121. 10.3389/fimmu.2022.954121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Arthur L, et al. Cellular and plasma proteomic determinants of COVID-19 and non-COVID-19 pulmonary diseases relative to healthy aging. Nat Aging. 2021;1(6):535–49. 10.1038/s43587-021-00067-x. [DOI] [PubMed] [Google Scholar]
- 87.Urata R, et al. Senescent endothelial cells are predisposed to SARS-CoV-2 infection and subsequent endothelial dysfunction. Sci Rep. 2022;12(1):11855. 10.1038/s41598-022-15976-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Tsumita T, et al. Viral uptake and pathophysiology of the lung endothelial cells in age-associated severe SARS-CoV-2 infection models. Aging Cell. 2024;23(2):e14050. 10.1111/acel.14050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mone P, et al. L-Arginine enhances the effects of cardiac rehabilitation on physical performance: New insights for managing cardiovascular patients during the COVID-19 pandemic. J Pharmacol Exp Ther. 2022;381(3):197–203. 10.1124/jpet.122.001149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Zuin M, Rigatelli G, Battisti V, Costola G, Roncon L, Bilato C. Increased risk of acute myocardial infarction after COVID-19 recovery: A systematic review and meta-analysis. Int J Cardiol. 2023;372:138–43. 10.1016/j.ijcard.2022.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.M. Drame, et al. "Relation between Vitamin D and COVID-19 in Aged People: A systematic review." Nutrients, 2021;13(4). 10.3390/nu13041339. [DOI] [PMC free article] [PubMed]
- 92.Gori M, et al. Association between inhibitors of the renin-angiotensin system and lung function in elderly patients recovered from severe COVID-19. Eur J Prev Cardiol. 2022;29(5):e196–9. 10.1093/eurjpc/zwab143. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Data supporting findings are available in Supplementary Materials. Raw Mass Spectrometry files and the Data-Independent Acquisition file are publicly available at MassIVE (ProteomeXchange consortium), http://massive.ucsd.edu, under MassIVE identifier: MSV000094333; ProteomeXchange identifier: PXD050683 (for both rhesus macaque and olive baboon species).