Abstract
Polyphasic approach has become a generally accepted method for the classification of cyanobacteria. In this study, we present a detailed characterisation of two strains, KUCC C1 and KUCC C2, isolated from the Curonian Lagoon and classified to the Aphanizomenon genus. Despite phylogenetic similarity, the strains differ with respect to morphology, ultrastructure characteristics, and the metabolite profile. In the KUCC C1 extract, three unknown peptides and eight anabaenopeptins were detected, while KUCC C2 produced one unknown peptide and one aeruginosin. In both strains, a total of eleven pigments were detected. The production of myxoxantophyll, chlorophyll-a, chlorophylide-a, and zeaxanthin was higher in KUCC C2 than in KUCC C1. Extracts from both strains of Aphanizomenon also had different effects in antibacterial, anticancer and enzyme inhibition assays. Comprehensive analyses of Aphanizomenon strains performed in this study showed significant diversity between the isolates from the same bloom sample. These differences should be considered when exploring the ecological significance and biotechnological potential of a given population.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-76064-y.
Subject terms: Microbiology, Molecular biology
Introduction
The genus Aphanizomenon (Cyanoprokaryota) was first identified by A. Morren ex Bornet & Flahault in 1886 (1888) (type species: Aphanizomenon flos-aquae)1. This filamentous cyanobacterium was classified under the International Code of Nomenclature for algae, fungi, and plants (ICN) and Komárek et al.2 to the order Nostocales and family Aphanizomenonaceae3. Application of technological advancements, better understanding of genetic diversity, and a comprehensive evaluation of morphological and physiological characteristics led to a taxonomic revision of Aphanizomenon species. Given modern phylogenomic analyses, there is no justification for separating this genus. Aphanizomenon strains group with Anabaena and Dolichospermum isolates into one intermixed, monophyletic cluster that by some researchers is suggesting to be one genus4. This is however a scientific proposal that requires further research. To date, 17 species have been classified to this distinguished genus1. They usually form free-floating planktonic or metaphytic single trichome bundles. Aphanizomenon is a nitrogen-fixing cyanobacterium, that forms heterocysts under nitrogen deficiency. Cyanobacteria of this genus are widely distributed and can be found in fresh and brackish waters worldwide, mainly in temperate regions2. Aphanizomenon blooms can be formed by species producing saxitoxins, cylindrospermopsins or anatoxins1. A. flos-aquae can be used as a dietary supplement5, or as a source of natural products (phycocyanin, mycosporine-like amino acids) used in the cosmeceutical or nutraceutical sector6–8.
Historically, Aphanizomenon sp. was described in the Baltic Sea for the first time in 1887 (known as Limnochlide)9. The prevalence of blooms in the Baltic Sea during the late 19th century prompted two opposite hypotheses. According to the first one, A. flos-aquae had spread into the Baltic Sea from river estuaries, while the other hypothesis postulated the brackish character of Aphanizomenon species9. Analysis of the ultrastructure composition of Aphanizomenon sp., showed that the isolates from the Baltic Sea are significantly different from the freshwater species10. These results were also supported by Laamanen et al.11 and Barker et al.12, showing differences in the sequences of the 16–23 S rRNA internal transcribed spacer region (ITS), and the 16 S rRNA gene between the Baltic Sea and freshwater isolates.
In the Curonian Lagoon, which is connected to the Baltic Sea by a narrow strait, blooms of Aphanizomenon spp. have formed almost every summer since the 1930s13–19. The lagoon receives freshwater from the River Nemunas and, under certain meteorological and hydrodynamic conditions, the river water can be transferred to the Baltic coastal area through the strait. Under the dominance of north or north-west wind, the intrusion of the Baltic Sea waters into the Curonian Lagoon can also be observed17. As a consequence, the exchange of organisms between the two ecosystem can occur20,21. It is very likely that the Aphanizomenon population observed in the Curonian Lagoon might be the same or closely related to the one from the Baltic Sea. The taxonomic classification of Aphanizomenon from the Curonian Lagoon was previously based on morphological characteristics, 16 S rRNA gene sequencing, and multi-locus sequencing18,22,23. Whole genome sequencing of A. flos-aquae 2012/KM1/D3 from the Curonian Lagoon was performed by Šulčius et al.24 and Dreher et al.4. However, it is still not clear whether the analysed strain represents the entire population of the Curonian Lagoon and whether it is ecologically and genetically separate from the Baltic Sea population.
Today, the taxonomic classification of cyanobacteria based solely on the morphological characteristics is considered insufficient. Also, the standard methods used for species identification following the bacteriological code do not reflect the real diversity within population. This was improved with the application of more advanced molecular methods. However, the new insight into the cyanobacterial diversity provided a serious challenge for their correct classification25. Therefore, today, a more integrated and comprehensive approach, that combines genetic and phenotypic characteristics is applied to discover the differences within organisms belonging to the same population26.
The aim of this study was to characterize two Aphanizomenon strains from the Curonian Lagoon, southeastern Baltic Sea. Phenotypic characteristics included morphology, ultrastructure, pigments, and metabolite profiles. Phylogenetic analyses were based on the 16 S rRNA/cpcBA/rbcLX genomic regions, commonly used in molecular cyanobacterial taxonomy. The presence of selected genes (sxtA, mcyE) involved in biosynthesis of cyanotoxins in other Aphanizomenon strains was checked. Furthermore, in vitro bioassays were applied to assess the biotechnological potential of the two strains. The observed differences in morphology, phylogeny, and secondary metabolite profile indicate an existing diversity in the Baltic Aphanizomenon population.
Results
Morphology and ultrastructure of Aphanizomenon strains
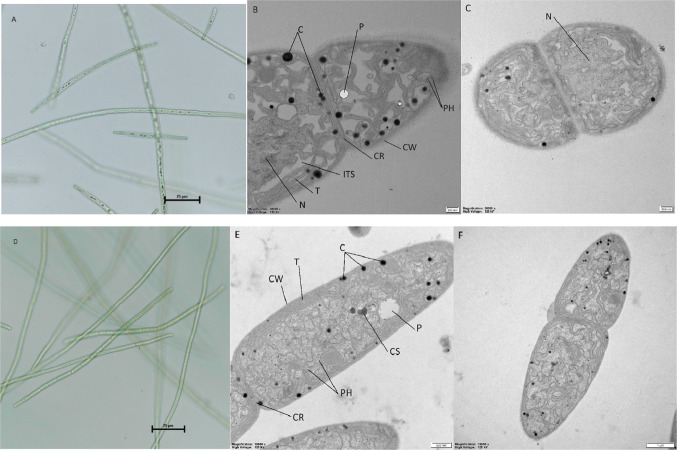
The cells of KUCC C1 and KUCC C2 maintained in culture were light green. KUCC C2 cells always formed a thin layer on the surface of the medium, while KUCC C1 cells were homogenously suspended in the medium or concentrated at the bottom of the flasks. The trichomes of both strains were straight and arranged separately (Fig. 1A,D). Vegetative cells of the KUCC C2 strain were cylindrical or barrel-shaped, sometimes slightly shorter, or longer than wide – from 2.56 to 4.92 μm in width and from 4.16 to 9.24 μm in length (mean value 3.67 ± 0.47 μm in width and 7.00 ± 0.98 μm in length) (Fig. 1D,E). Heterocysts intercalary, solitary, usually one in a trichome, cylindrical, from 2.89 to 5.12 μm in width and 4.52 to 8.59 μm in length (mean value 3.72 ± 0.51 in width and 6.60 ± 1.50 in length) (Supplementary Fig. S3). Vegetative cells of KUCC C1 were cylindrical, intensely elongated and narrow (Fig. 1A). In the culture the terminal cells were sometimes solitary and distinct from the trichome. The width ranged from 2.01 to 3.93 μm and 5.50 to 13.62 μm in length (mean value 2.75 ± 0.41 μm in width and 8.83 ± 1.64 μm in length) (Fig. 1B,C). Heterocysts of KUCC C1 were very similar to KUCC C2 – also intercalary, solitary, usually one in a trichome and cylindrical, from 2.76 to 3.99 in width and 6.75 to 9.38 in length (mean value 3.48 ± 0.36 in width and 8.45 ± 0.57 in length). The apical cells of KUCC C1 were conical, slightly attenuated, from 1.91 to 3.08 μm in width and 7.26 to 10.91 μm in length (mean value 2.53 ± 0.33 μm in width, 9.23 ± 0.90 μm in length) (Fig. 1B), while KUCC C2 – cylindrical and slightly elongated, from 1.64 to 2.75 μm in width and 9.07 to 11.76 μm in length (mean value 2.19 ± 0.30 μm in width, 10.33 ± 0.75 μm in length) (Fig. 1F). Akinetes of both strains were larger than vegetative cells, elongated, with rounded ends, from 12.34 μm to 13.71 μm in length and from 2.46 to 2.84 μm in width for KUCC C1; from 11.45 to 13.45 μm in length and from 2.46 to 3.12 μm in width for KUCC C2 (Supplementary Fig. S3).
Fig. 1.
Morphology and ultrastructure of Aphanizomenon sp. KUCC C1 and KUCC C2 cells: light microscopy of KUCC C1 (A) and KUCC C2 (D), ultrathin TEM sections: general view of the longitudinal section of KUCC C1 (B) and KUCC C2 (E) cells, and dividing cells of KUCC C1 (C) and KUCC C2 (F). Cyanophycin granules (C), polyphosphate granules (P), phycobilisomes (PH), cell wall (CW), cross wall (CR), nucleoplasm (N), intrathylakoid space (ITS), thylakoids (T), carboxysome (CS). Scale bars: 25 μm (A, D), 200 nm (B, C), 500 nm (E), 1 μm (F).
Ultrastructural studies showed that the thylakoids of KUCC C1 were curved, irregular and predominantly distributed within the cell, occasionally concentrated along the cellular peripheries (Fig. 1B). KUCC C2 thylakoids were mainly distributed at the cellular peripheries, usually rarely present in the center of the cell (Fig. 1E). Phycobilisomes, cyanophycin granules, and polyphosphate granules were observed in both strains, while carboxysomes were only present in KUCC C2 (Fig. 1E). The cyanophycin granules in both strains were located primarily in the cellular peripheries. In KUCC C1 the more aggregated granules were located near the cross wall, while in KUCC C2 – they were far from the cross walls (Fig. 1B,E). Intrathylakoid spaces and nucleoplasm were very pronounced and prominent in KUCC C1 (Fig. 1B). The content of the apical cells of KUCC C2 was very similar to vegetative cells, however, a lower number of thylakoids was observed in the developing stage than in the fully developed apical cells (Supplementary Figs. S1 and S2). In KUCC C1, the apical cells were similar to that of vegetative cells, but had more pronounced intrathylakoid spaces compared to vegetative cells.
Genetic analysis
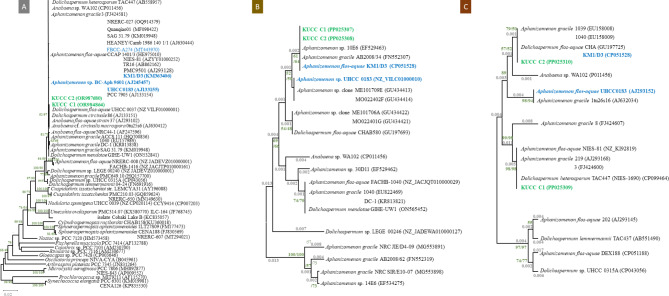
The phylogenetic relationships of the studied strains, their closest relatives, and other representative from the Nostocales order and the cyanobacterial phylum were analysed (Fig. 2). 16 S rDNA sequences from organisms classified to Aphanizomenon, Dolichospermum, and Anabaena genera formed a well-supported cluster, demonstrating a mean similarity of 99.6% (Fig. 2A). The phylogenetic tree supported their separation from other Nostocales (bootstrap ≥ 90). Four Baltic Aphanizomenon strains, two of which were analysed in this study, were incorporated into the cpcBA and rbcLX trees, and only some of the others that were included in the 16 S rDNA tree. This discrepancy was caused by a lack of relevant sequences in the available databases. Analyses revealed a 100% similarity of the cpcBA fragment between KUCC strains, another isolate from the Curonian Lagoon (A. flos-aquae KM1/D3), and two freshwater isolates (Aphanizomenon sp. 10E6, A. gracile AB2008/34) from German lakes (Fig. 2B). Another Baltic isolate (Aphanizomenon sp. UHCC 0183) clustered together with the freshwater sequences from the USA (clone ME101709E, and MO022402F); these strains differ from the Curonian isolates at a single position in the cpcA gene. The divergence among the rbcLX sequences was greater, reaching a maximum of 5%. The KUCC C2 sequences formed a cluster with the another Curonion strain (KM1/D3), and freshwater Chinese isolates (A. gracile 1039, 1040, Dolichospermum flos-aquae CHA). On the other hand, KUCC C1 formed a highly supported group with the other isolates from Dolichospermum and Aphanizomenon genera (Fig. 2C).
Fig. 2.
Phylogenetic relationships of studied KUCC strains (marked in bold green), other Baltic (marked in bold blue), brackish (marked in blue) Aphanizomenon isolates, and their closest relatives, based on the alignment of 16 S rDNA (1177 bp) (A), cpcBA (553 bp) (B), and rbcLX (746 bp) (C) sequences. Subsequent numbers above branches indicate the bootstrap values for NJ/ ML.
The primers used for the amplification of mcyE, sxtA, and geoA genes failed to produce detectable products in the PCR reactions using DNA extracted from the investigated strains (Supplementary Fig. S4).
Metabolic diversity of Aphanizomenon strains
Pigments
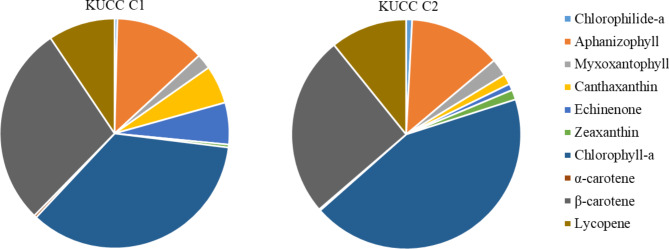
The pigment composition (chlorophyll-a, carotenoids, and phycobilins) of KUCC C1 and KUCC C2 was examined. In both strains, a total of eleven pigments were detected. In addition to chlorophyll-a, chlorophilide-a, aphanizophyll, myxoxantophyll, canthaxanthin, echinenone, zeaxanthin, α-carotene, β-carotene, lycopene and phycocyanin were present in both extracts (Table 1; Fig. 3). Both strains synthesised similar quantities of aphanizophyll, α-carotene, β-carotene, lycopene and phycocyanin (Table 1). Canthaxanthin and echinenone concentrations were 60 and 80% higher, respectively, in KUCC C1 than in KUCC C2. On the other hand, KUCC C2 was characterised by a higher production of myxoxantophyll, chlorophyll-a, chlorophylide-a, and zeaxanthin than KUCC C1 (35, 40, 65, and 80%, respectively).
Table 1.
Concentrations of pigments (µg g− 1 d.w.) and pigment to chlorophyll-a ratios determined for two Aphanizomenon strains isolated from the Curonian Lagoon.
| Pigment | KUCC C1 | KUCC C2 | ||
|---|---|---|---|---|
| Concentration [µg g–1 d.w.] | Pigment to chlorophyll-a ratio | Concentration [µg g–1 d.w.] | Pigment to chlorophyll-a ratio | |
| Chlorophyll and carotenoids | ||||
| Chlorophilide-a | 0.28 | 0.01 | 0.80 | 0.02 |
| Aphanizophyll | 8.96 | 0.36 | 12.44 | 0.30 |
| Myxoxantophyll | 1.50 | 0.06 | 2.33 | 0.06 |
| Canthaxanthin | 3.78 | 0.15 | 1.40 | 0.03 |
| Echinenone | 4.13 | 0.17 | 0.87 | 0.02 |
| Zeaxanthin | 0.29 | 0.01 | 1.32 | 0.03 |
| Chlorophyll-a | 24.61 | 1.00 | 41.40 | 1.00 |
| α-carotene | 0.25 | 0.01 | 0.20 | 0.01 |
| β-carotene | 19.86 | 0.81 | 24.32 | 0.59 |
| Lycopene | 6.62 | 0.27 | 10.31 | 0.25 |
| Phycobilins | ||||
| Phycocyanin | 142.42 | 5.79 | 157.01 | 3.79 |
Fig. 3.
The content (%) of pigments in two Aphanizomenon strains, KUCC C1 and KUCC C2, isolated from the Curonian Lagoon.
Cyanopeptides
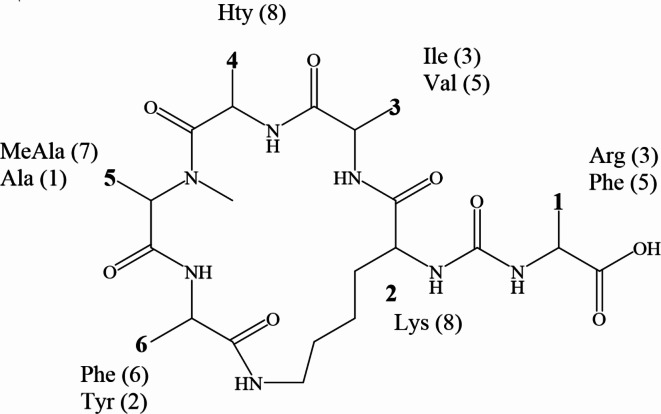
Under the MS conditions used in the study, thirteen peptides were detected in the extracts of the KUCC C1 and KUCC C2 strains (Supplementary Table S2). The cyanobacteria clearly differ with respect to their peptide profile; neither of the peptides was present in both strains. In the KUCC C2 extract, only the unknown peptide with m/z at 873 and the aeruginosin with m/z of [M + H]+ at 526 were detected. The aeruginosin spectrum has fragment ions at m/z 122, 140, 221 and 266 and was partially identified as ?-Leu-Choi-Argal (where ? stands for unknown residue). KUCC1 produces three unknown peptides with m/z of [M + H]+ at m/z 1018, 969 and 951, and eight anabaenopeptins. The suggested structures of the anabaenopeptins and the list of the most intense ions in their spectra are presented in the supplementary materials (Supplementary Table S.2). Structure elucidation of the peptides was based on the collected fragmentation spectra with characteristic ions, including immonium ions (e.g. 120 as Phe immonium ion) and ions that correspond to a specific fragment of their structure (e.g. the cyclic part or the [M + H – (AA1 + AA2 + AA3)] fragment) (Fig. 4). When possible, the elucidation process was supported by a comparison with the published spectra of the peptides27–30. None of the four tested cyanobacterial toxins (microcystin, cylindrospermopsin, saxitoxin, anatoxin-a) was detected in studied strains.
Fig. 4.
General structure of anabaenopeptins produced by Aphanizomenon KUCC C1. The number of variants with specific amino acids (AA) is given in brackets.
Activity of extracts
In the cytotoxicity assay only the KUCC C2 extract showed activity against breast cancer cells T45D and only at the highest concentration used in the assay (cell viability reduced by 97% at 200 µg mL–1) (Table 2). Both Aphanizomenon extracts were active against trypsin and chymotrypsin (60 – 80% at 45 µg mL–1), but no activity against thrombin and elastase was observed. Neither of the extracts affected the growth of the eleven bacterial strains tested. However, KUCC C1 inhibited violacein production by Ch. violaceum ATCC12472. Quorum quenching activity was observed at the extract concentrations of 250 µg mL–1. Diminished production of violacein, (by approximately 50%) was observed at concentrations up to 63 µg mL–1.
Table 2.
Antibacterial, cytotoxic, and enzymatic inhibition activity of Aphanizomenon KUCC C1 and KUCC C2 extracts.
| Bioassays | Cell lines | KUCC C1 | KUCC C2 |
|---|---|---|---|
| 1. Cytotoxic | T45D | - | + |
| HeLa | - | - | |
| Enzymes | |||
| 2. Enzymatic | Trypsin | + | + |
| Chymotrypsin | + | + | |
| Thrombin | - | - | |
| Elastase | - | - | |
| Bacterial strains | |||
| 3. Antibacterial | Chromobacterium violaceum ATCC12472 | + | - |
| Vibrio cholerae 2329 | - | - | |
| Vibrio vulnificus ATCC29306 | - | - | |
| Vibrio diazotrophicus Cd1 | |||
| Aeromonas salmonicida 2013 | - | - | |
| Enterococcus faecium 45 | - | - | |
| Staphylococcus aureus CCNPB/1505 | - | - | |
| Acinetobacter baumannii CCNPB/O | - | - | |
| Pseudomonas aeruginosa CCNPB/MBL | - | - | |
| Escherichia coli ATCC25922 | - | - | |
| Escherichia coli CCNPB/ESBL | - | - |
“-“ no inhibition, “+” indicates bacterial growth/enzymatic activity/cell viability inhibition by more than 50%.
Discussion
Due to fragmentary descriptions of isolates, and type material, the taxonomy of cyanobacteria poses a serious challenge31. One of the issues is that the application of ICN did not uncover genotypic traits, while the International Code of Nomenclature of Prokaryotes (ICNP) indicates a taxonomic system that usually is not consistent with morphology26. Therefore, both ICN and ICNP need to be used for correct identification of cyanobacteria. Today, the polyphasic approach in cyanobacterial taxonomy includes application of molecular markers, characteristics of ultrastructures, morphology, biochemical and ecophysiological traits2,32. In this work, this approach was used to characterise two Aphanizomenon strains isolated from the same bloom sample from the Curonian Lagoon and, deposited in the KUCC collection.
Based on morphological features (shape and size of vegetative cell, size and position of heterocysts, size and content of apical cells, shape of the trichomes), Aphanizomenon KUCC C1 has been found to be more similar to A. gracile and KUCC C2 to A. flos-aquae. The mean values of width and length of both strains corresponded to cell size according to Komárek2 and the Baltic Sea HELCOM Phytoplankton Expert Group (PEG)33 size classes. However, some of the terminal cells of Aphanizomenon KUCC C1 were slightly longer compared to previously described A. gracile cells. Morphologically distinct terminal cells of A. gracile can be considered an additional feature to separate this species from other species of this genus34.
Ultrastructure studies, specifically arrangement of thylakoids, polyphosphate, and cyanophycin granules, revealed a high similarity of the Aphanizomenon KUCC C2 to previously studied Baltic Sea Aphanizomenon cells10,35. Aphanizomenon KUCC C1 has a completely different cell arrangement in comparison to KUCC C2; characteristic pattern of the organelles and terminal cells is very similar to the A. gracile previously described by Wejnerowski et al.34,36. Large intrathylakoid spaces present in the KUCC C1, as well as the arrangement of thylakoids with a well-defined nucleoplasm are the main ultrastructure characteristics that showed similarities to A. gracile.
Between the strains significant differences in pigments contents were also observed, providing valuable information on their ecological and physiological characteristics. Due to unique ecological preferences, individual species or strains significant differences in pigment profiles can be observed37,38. Also, in the case of the two studied Aphanizomenon strains, their specific requirements were expressed by the way they grow in culture medium and produced pigments. For example, two-fold higher chlorophyll-a production was determined in KUCC C2 compared to KUCC C1. A higher concentration of chlorophyll-a in microalgae is usually correlated with their increased biological productivity and greater contribution to primary production in the ecosystem38. A. flos-aquae, which appears to be similar to KUCC C2, forms annual blooms in the Curonian Lagoon19 and the Baltic Sea39. This seasonal dominance of A. flos-aquae can be attributed to competitive mechanisms40, such as higher photosynthetic efficiency and biological productivity. Definitely, more detailed research and experimental work with a larger number of Aphanizomenon isolates from the Curonian Lagoon must be performed to reach more definite conclusions about the environmental importance of the specific traits of KUCC C1 and C2.
Research on the Baltic Sea Aphanizomenon has been hampered by the difficulties in establishing the cultures of this taxon and fragmented data on most of the available isolates. To the best of our knowledge, there are only two strains maintained in culture collections, which are well described in the scientific literature, their genomes have been sequenced: UHCC 0183 (also published as Tr183)41, and A. flos-aquae 2012/KM/D324. Other studies on the genetic diversity of this genus in the Baltic Sea have relied only on DNA isolated from environmental samples (e.g11,35,42). In a few papers, the authors included other Baltic Aphanizomenon isolates. However, these strains lack genetic characterisation e.g., Aphanizomenon sp. BA6943,44, A. flos-aquae 2012/KM1/C423, or Aphanizomenon 2015/KM/E845.
Traditionally, it is considered that according to botanical taxonomy, only one species of the genus Aphanizomenon occurs in the Baltic Sea – A. flos-aquae11. However, the accurate taxonomic assignment of isolates within the Aphanizomenon genus, as determined by traditional taxonomic methodologies, remains as an unresolved challenge up to the present time. The need for taxonomic validation of the morphologically distinct, however, phylogenetically closely related genera: Anabaena, Dolichospermum, and Aphanizomenon, was suggested for the first time more than 20 years ago41. In recent phylogenomic studies, a distinct group of some of the isolates belonging to these genera was separated and named the ADA clade46–48. Similarly, in our work, the KUCC isolates were mixed with other ADA sequences in the 16 S rDNA tree. They clustered with other isolates assigned by Österholm et al.48 to the β supergroup in the ADA clade, which contains the most diverse strains in terms of the diversity of the encoded secondary metabolites, as well as the geographical range of occurrence and water salinity preference. Additional analyses of more variable cpcBA and rbcLX sequences also did not support the separation of Aphanizomenon KUCC C1 and KUCC C2 as A. gracile and A. flos-aquae, respectively. Studies on the Baltic Aphanizomenon suggest that the populations are genetically homogenous (e.g11,42). KUCC strains exhibited similar genotypes, but the significant differences between the two strains are reflected, among others, in their metabolic profiles. Although the phylogenomic approach is receiving more acceptance in recent years, as the most robust method to resolve cyanobacterial taxonomy, in our opinion it cannot be overlooked that genetically similar strains vary so significantly in the production of bioactive compounds.
Cyanobacteria are characterised by production of many secondary metabolites49. Incorporation of these compounds in the polyphasic approach, provides valuable insights and complements traditional classification methods26. The presence, absence, or variation of the cyanometabolite profiles can serve as molecular markers for the accurate characterisation and classification of cyanobacterial strains50. LC-MS/MS analysis did not show the presence of any of the known cyanotoxins in Aphanizomenon KUCC C1 and KUCC C2. Also, the mcyE and sxtA genes involved in the synthesis of microcystins and saxitoxins, respectively, were not detected. Saxitoxins have been frequently detected in freshwater Aphanizomenon strains, such as A. gracile51,52. Instead, in KUCC C1 and KUCC C2 thirteen peptides were detected. However, the metabolite profiles of the strains differed greatly. KUCC C1 produced mainly anabaenopeptins, while none of them was detected in KUCC C2. There are only two published papers showing the ability of Aphanizomenon to produce anabaenopeptins. In one of them, the detection of anabaenopeptin-like clusters in A. gracile was revealed53. The same report showed that phylogenetically similar A. gracile strains were very different with respect of the produced metabolites53. The second report showed the production of anabaenopeptins by Aphanizomenon from Japanese waters54 and the peptide structure were very similar to the anabaenopeptins identified in KUCC C1. Among the detected anabaenopeptins, rare cases of non-methylated amino acids in position 5 (Ala) and non-homo amino acids in position 4 were observed, which gives an additional characteristic feature for Aphanizomenon KUCC C1 from the Curonian Lagoon. Anabaenopeptins are cyclic peptides produced by cyanobacteria through non-ribosomal peptide synthetases (NRPS) enzymes, and the adenylation domains are responsible for selecting and activating specific amino acids during the biosynthesis process. In the case of anabaenopeptins, the adenylation domains of the NRPS select and activate specific amino acids, contributing to the diversity of the peptide structures55. The Aphanizomenon genus is not a frequent producer of this class of peptides, and to our knowledge this is the first report of anabaenopeptins producing Aphanizomenon in European waters. In the other strain, KUCC C2, the number of metabolites detected in the extract was much lower compared to KUCC C1. Among two metabolites, only one was partially identified and assigned to the class of aeruginosins. This class of non-ribosomal peptides (NRPs) is usually produced by the Microcystis and Planktothrix genera49. Previously, similar, partly identified aeruginosins were detected by Šulčius et al.56, Overlinge et al.22, Pilkaitytė et al.18 in the Curonian Lagoon during the blooms of A. flos-aquae. The Baltic Sea Aphanizomenon is a non-toxins producing population57, moreover, there are no published data on NRPs produced by the Baltic Sea Aphanizomenon. There is one work describing two new metabolites, aphapeptin F1 (m/z 562) and F2 (m/z 592), isolated from the Aphanizomenon X008a and X0023 strains, respectively58.
Even though, in the bioassay performed in our work the activity of the extracts was tested, it is highly likely that the inhibition of trypsin and chymotrypsin by KUCC C2 can be attributed to the presence of aeruginosin with m/z at 526. To our knowledge, no data has been published on the production of aeruginosins by Aphanizomenon. This class of peptides are known as potent serine proteases inhibitors59, i.e. compounds that were widely explored as potential drugs used for the treatment of many metabolic disorders59. This fact indicates that Aphanizomenon KUCC C2 may have a some biotechnological potential due to its nontoxic properties and its ability to produce aeruginosins. Aphanizomenon has been known as a microorganism of high biotechnological potential with positive effect on human health60. For example, Klamath Algae Products offers nutraceutical products containing A. flos-aquae powders. The extract of the same strain was also shown to be a rich source of vitamin B12 for human consumption61 and a positive mechanical activity on human distal colon62. Furthermore, Aphanizomenon extracts have the potential to be used as antioxidants63 and probiotic substrates64. Pigments (phycocyanin, phycocyanobilin) extracted from Aphanizomenon showed anticancer activity65. The unique metabolic features of the two Aphanizomenon strains from the Curonian Lagoon and their antibacterial, anticancer, and enzyme inhibition activities indicate the need for further studies on their biotechnological potential.
Materials and methods
Culture conditions
Aphanizomenon KUCC C1 and Aphanizomenon KUCC C2 were isolated from the Curonian Lagoon (Sautheastern Baltic Sea) by dr. Donata Overlingė in 2018 and deposited in the Culture Collection of Klaipeda University (KUCC). The strains were maintained in Z8 medium (with nitrogen, no salts were added)66, at 21 °C (± 0.5 °C), with constant illumination (10 µM photons m− 2 s− 1). In order to stimulate the formation of akinetes for morphological measurements, the cultures were grown in Z8 medium supplemented with NaCl (the final salinity 10 psu). The biomass of non-axenic monospecies cultures was harvested between 3 and 4 weeks after inoculation, in the mid-exponential/early stationary growth phase. For genetic analyses, 80 ml subsamples were centrifuged at 2,000 × g, 10 min, 4 °C; the pellet was frozen at -20 °C overnight before the DNA extraction. For all other analyses, the cultures were centrifuged (3,041 × g; 5 min; 4 °C) and lyophilised.
Genetic analysis
Genomic DNA was extracted from wet biomass using the DNeasy PowerSoil Pro Kit (QIAGEN, Hilden, Germany) according to the manufacturer’s protocol, altering only the last step, i.e. DNA was eluted with sterile water instead of the buffer.
All polymerase chain reactions were performed in a Mastercycler® nexus GSX1 (Eppendorf, Hamburg, Germany). For each reaction: 30 ng of genomic DNA, 5 pmol of the respective forward and reverse oligonucleotide primers, MyTaqTM Red Mix (BiolineReagents Ltd, London, UK) were used.
For taxonomic identification PCR amplifications of 16 S rRNA/cpcBA/rbcLX were performed according to Jungblut et al.67, Neilan68, Rudi et al.69, respectively. The PCR products were purified and commercially sequenced (Genomes S.A., Warszawa, Poland). Both forward and reverse electropherograms of each PCR product were trimmed and assembled using Finch TV, and Mega 7.070. The sequences are available in the DDBJ/EMBL/GenBank databases under accession numbers incorporated in Fig. 2. The obtained nucleotide sequences were edited with Finch TV and compared to the sequences in the NCBI GenBank (http://www.ncbi.nlm.nih.gov) using the blastn algorithm (http://blast.ncbi.nlm.nih.gov). A total of 58 operational taxonomic units (OTUs) were included in the phylogenetic analyses of the 16 S rRNA gene. Mega 7.0 was used to align sequences and reconstruct trees using the maximum likelihood estimation. Bootstrap support was obtained from 1,000 replicates. For each set of aligned sequences, distance matrices were calculated by applying a K80 substitution model.
Both strains were also checked for the presence of genes involved in the biosynthesis potential of geosmin (geoA) and cyanotoxins (sxtA, mcyE) previously identified in some of the Aphanizomenon strains48. For the geoA gene, primers and PCR conditions described by John et al.71 were applied; and genomic DNA from Nostoc edaphicum CCNP1411 was the positive control72. The amplifications of the sxtA and mcyE genes were performed according to Ballot et al.73 and Rantala et al.74, respectively. Microcystin-producing Microcystis sp. CCNP1101 and saxitoxin-producing Aphanizomenon gracile NRC_SIR/C31-09 were included in the PCRs as positive controls22,51. All PCR products were separated on 1% TAE agarose gels stained with Midori Green Advance (NIPPON Genetics, Düren, Germany), and visualised using the InGenius 3 documentation system (Syngen, Cambridge, UK).
Morphological and ultrastructural analysis
Morphological parameters were described using an inverted Nikon Eclipse Rs2R microscope (Tokyo, Japan) equipped with a Nikon DS-Ri2 camera (Tokyo, Japan) and operated with NIS-Elements Documentation software, version number 5.01.00. Taxonomic characterisation was based on a key by Komárek et al.2. The width and length of a hundred vegetative and apical cells were measured. Altered salinity in the cultures (see “Culture conditions”) resulted in the production of several akinetes, therefore the data set of morphological dimensions were smaller than vegetative and apical cells.
To visualise the cellular ultrastructures, transmission electron microscopy (TEM) was used. The samples were fixed in ice-cold 2.5% electron microscopy grade glutaraldehyde (Polysciences) in 0.1 M PBS (pH 7.4). Then, post-fixed in 1% osmium tetroxide, dehydrated through a graded series of ethanol (30–100%), and embedded in Epon (Fluka). Ultrathin (65 nm) sections were cut using a Leica UC7 ultramicrotome and stained with Uranyless (Delta Microscopies) and Reynold’s lead citrate. Cells were examined with a Tecnai G2 Spirit BioTWIN transmission electron microscope.
Analyses of pigments
Chlorophyll-a and carotenoids were extracted from lyophilised Aphanizomenon cells (20 mg) with 90% acetone in MiliQ water (4 mL) by grinding75. Qualitative and quantitative analyses of the pigments were performed using a Shimadzu HPLC system (Shimadzu Corporation, Kyoto, Japan) equipped with a photodiode array and fluorescence detectors. Pigments were separated in a C18 LichroCART™LiChrospher™ 100 column (250 × 4 mm; 5 μm; 100 Å) (Merck, Germany) using a mixture of a methanol: 1 M ammonium acetate (80 : 20) (phase A) and methanol: acetone (60 : 40) (phase B). The gradient started at 0% B and went to 100% B in 10 min, the flow rate was 1 mL− 1 76,77.
Phycobilins were extracted from lyophilised Aphanizomenon cells (20 mg) using a buffer solution (pH = 5.5; 4 mL) of Trizma Base (0.25 M), hydrated disodium EDTA (10 mM) and lysozyme (2 mg mL− 1)78. The qualitative and quantitative analyses were performed using a spectrofluorimeter (Cary Eclipse, Agilent Technologies)79.
Extraction and analyses of cyanometabolites by LC-MS/MS
Freeze-dried biomass of KUCC C1 and KUCC C2 was extracted with 75% methanol in water by vortexing for 15 min and centrifuged (19,837 × g; 4 °C; 20 min). After centrifugation the supernatant was collected and analysed by an Agilent HPLC system (Agilent Technologies, Waldboronn, Germany) coupled to a hybrid triple quadrupole/linear ion trap mass spectrometer QTRAP LC-MS/MS (QTRAP5500, Applied Biosystems, Sciex, Canada) using the non-target method previously optimised for cyanopeptide detection80. In short, the chromatographic separation was performed in a Zorbax Eclipse XDB-C18 column (4.6 mm × 150 mm, 5 μm; Agilent Technologies, Santa Clara, CA, USA). For the elution, a mixture of Milli-Q water (solvent A) and acetonitrile (solvent B), both containing 0.1% formic acid were used. The Turbo Ion Source operated in positive ion mode. Mass spectra were acquired using Information Dependent Acquisition (IDA, 500–1200 Da), at capillarity of 5.5 kV, ion source temperature 550 °C. The curtain gas was set at 20 psi, the cone gas pressure and desolvation gas pressure at 60 psi. The declustering potential and collision energy were 80 V and 60 V, respectively. Additionally, to check the presence of microcystin, anatoxin-a, saxitoxins, and cylindrospermopsin in the samples, enhanced ion product mode at collision energy 40 eV was used for precursor ions with m/z 135, 166, 300, and 416, respectively. Data were processed with Analyst QS (Version 1.5.1, Applied Biosystems/MDS Analytical Technologies, Concord, ON, Canada, 2008).
Bioactivity assays
The biological activity was assessed using three different bioassays. For antibacterial activity, the broth microdilution assay was performed according to the European Committee on Antimicrobial Susceptibility Testing (EUCAST) recommendations (http://www.eucast.org). Eleven different bacteria strains were tested (6 clinical and 5 environmental) (Supplementary Table S1). Four clinical isolates (no. 1–4) were kindly provided by dr. Kamila Korzekwa, Medical Laboratories Center Dialab (Wrocław, Poland). The environmental isolates were obtained from dr. Ewa Kotlarska, Institute of Oceanology Polish Academy of Sciences (isolates no. 5–7 from IO PAN MB Strain Collection Institute of Oceanology, Polish Academy of Sciences, Molecular Biology Laboratory) and prof. Aneta Łuczkiewicz, Gdańsk University of Technology (isolate no. 8 from the Department of Water and Wastewater Technology Strain Collection). Other isolates were purchased from the commercial collection. All bacterial strains mentioned above are deposited in the Culture Collection of Northern Poland at the Laboratory of Marine Biotechnology (CCNP), University of Gdańsk81–83.
Cyanobacteria biomass was extracted in the same way as for the LC-MS/MS analyses (see Sect. “Extraction and analyses of cyanometabolites by LC-MS/MS”). After extraction, the supernatants were dried in miVac QUATTRO centrifugal vacuum concentrator (SP Scientific, Ipswich, UK). For the antibacterial bioassay, dry extracts were dissolved in 2% v/v DMSO and tested in two-fold serial microdilutions; their final concentration in the experimental wells ranged from 1.95 mL–1 to 1,000 µg mL–1.
Enzymatic inhibition (trypsin, chymotrypsin, thrombin, elastase) assays were performed according to Pluotno and Carmeli84, Ocampo Bennet85, and Kwan et al.86. For full details on trypsin, chymotrypsin, thrombin inhibition assay see Overlingė et al.22. In the case of elastase, elastatinal (Sigma Aldrich; St. Louis, MO USA) was used as an inhibitor and N-succinyl-Ala-Ala-Ala-p-nitroanilide (Sigma Aldrich) served as a substrate87. For cytotoxicity assay human breast adenocarcinoma T47D and HeLa cell lines (Merck KGaA, Darmstadt, Germany) were used. The assay was performed according to the colorimetric method described by Felczykowska et al.88. For full details, see Overlingė et al.22.
Conclusions
This study has provided new data on the genetic and phenotypic features of Aphanizomenon KUCC C1 and KUCC C2 isolated from the Curonian Lagoon. The morphological and ultrastructural analyses indicate that the strains can be classified as A. gracile (KUCCC1) and Aphanizomenon flos-aquae (KUCC C2). Phylogenetic analyses however did not support this classification. On the other hand, the isolates differ with respect of the produced metabolites, and thus, they can be classified to separate chemotypes. This discrepancy in metabolic profiles can have a significant effect on the ecotype and biotechnological potential of the two cyanobacterial strains. The comprehensive resolution of the taxonomic classification of the Aphanizomenon genus is beyond the scope of this study, however detailed characteristics of the two new brackish isolates might be useful in further studies on the Baltic cyanobacteria classified to this genus.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Acknowledgements
The research was funded by Lithuanian National Science Foundation, grant number SMIP‐20‐31, by Doctorate Study program in Ecology and Environmental Sciences, Marine Research Institute, Klaipėda University (for D.O.), the statutory program of the University of Gdańsk (grant No. 531-O303-D879-23) and the statutory program of the Institute of Oceanology, PAN (grant no. II.3). We would like to express our gratitude to hab. dr. Magdalena Narajczyk for her assistance in capturing the Transmission Electron Microscopy images.
Author contributions
D.O. Conceptualization, Investigation, Resources, Writing—Original Draft; A.T.-S. Formal analysis, Investigation, Writing—Review & Editing; M.C. Formal analysis, Investigation, Writing—Review & Editing; K. S. Investigation, Writing—Review & Editing, Resources; H.M.-M. Conceptualization, Resources, Writing—Review & Editing.
Data availability
Sequence data that support the findings of this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers PP025307, PP025308, PP025309, PP025310, OR987680, OR984864.
Declarations
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Guiry, M. D. & AlgaeBase World-wide Electronic Publication (National University of Ireland, 2023). https://www.algaebase.org
- 2.Komárek, J. Cyanoprokaryota: 3rd part: Heterocystous Genera. in Süßwasserflora Von Mitteleuropa (eds Büdel, B., Gärtner, G., Krienitz, L. & Schagerl, M.) (Springer Spektrum, 2013).
- 3.Turland, J. N. et al. International Code of Nomenclature for Algae, fungi, and Plants (Shenzhen Code) Adopted by the Nineteenth International Botanical Congress Shenzhen, China, July 2017 (Koeltz Botanical Books, 2018).
- 4.Dreher, T. W., Davis, E. W. & Mueller, R. S. Complete genomes derived by directly sequencing freshwater bloom populations emphasize the significance of the genus level ADA clade within the Nostocales. Harmful Algae. 103, 102005 (2021). [DOI] [PubMed] [Google Scholar]
- 5.Gilroy, D. J., Kauffman, K. W., Hall, R. A., Huang, X. & Chu, F. S. Assessing potential health risks from microcystin toxins in blue-green algae dietary supplements. Environ. Health Perspect.108, 435–439 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, H. et al. Occurrence of mycosporine-like amino acids (MAAs) from the bloom-forming cyanobacteria Aphanizomenon strains. Molecules27, 1–14 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Amato, A., Terzo, S. & Mulè, F. Natural compounds as beneficial antioxidant agents in neurodegenerative disorders: A focus on Alzheimer’s disease. Antioxidants8, (2019). [DOI] [PMC free article] [PubMed]
- 8.Nuzzo, D. et al. Effects of the Aphanizomenon flos-aquae extract (Klamin®) on a neurodegeneration cellular model. Oxid. Med. Cell. Longev. (2018). [DOI] [PMC free article] [PubMed]
- 9.Finni, T., Kononen, K., Olsonen, R. & Wallström, K. The history of cyanobacterial blooms in the Baltic Sea. Ambio30, 172–178 (2001). [PubMed] [Google Scholar]
- 10.Janson, S., Carpenter, E. J. & Bergman, B. Fine structure and immunolocalisation of proteins in Aphanizomenon sp. from the Baltic Sea. Eur. J. Phycol.29, 203–211 (1994). [Google Scholar]
- 11.Laamanen, M. J., Forsström, L. & Sivonen, K. Diversity of Aphanizomenon flos-aquae (cyanobacterium) populations along a Baltic Sea salinity gradient. Appl. Environ. Microbiol.68, 5296–5303 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Del Campo, F. F. & Ouahid, Y. Identification of microcystins from three collection strains of Microcystis aeruginosa. Environ. Pollut.158, 2906–2914 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Schmidt-Ries, H. Untersuchungen Zur Kenntnis Des Pelagials eines Strangewassers (Kurisches Haff). Zeitchriften fur Fischerei Und Deren Hilfwisstnschrift6, 183 (1940). [Google Scholar]
- 14.Olenina, I. Long-term changes in the Kuršių Marios lagoon: Eutrophication and phytoplankton response. Ekologija1, 56–65 (1998). [Google Scholar]
- 15.Lesutiene, J., Bukaveckas, P. A., Gasiunaite, Z. R., Pilkaityte, R. & Razinkovas-Baziukas, A. Tracing the isotopic signal of a cyanobacteria bloom through the food web of a Baltic Sea Coastal Lagoon. Estuar. Coast Shelf Sci.138, 47–56 (2014). [Google Scholar]
- 16.Zilius, M. et al. The influence of cyanobacteria blooms on the attenuation of nitrogen throughputs in a Baltic Coastal Lagoon. Biogeochemistry141, 143–165 (2018). [Google Scholar]
- 17.Overlingė, D. et al. Are there concerns regarding cHAB in coastal bathing waters affected by freshwater-brackish continuum? Mar. Pollut Bull.159, 111500 (2020). [DOI] [PubMed] [Google Scholar]
- 18.Pilkaitytė, R., Overlingė, D., Gasiūnaitė, Z. R. & Mazur-Marzec, H. Spatial and temporal diversity of cyanometabolites in the Eutrophic Curonian lagoon (SE Baltic Sea). Water13, 1760 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vaičiūtė, D. et al. Hot moments and hotspots of cyanobacteria hyperblooms in the Curonian lagoon (SE Baltic Sea) revealed via remote sensing-based retrospective analysis. Sci. Total Environ.769, 145053 (2021). [DOI] [PubMed] [Google Scholar]
- 20.Zakaria, M. A. & Abdulrahman, A. S. M. Biodiversity and toxin production of cyanobacteria in mangrove swamps in the Red Sea off the southern coast of Saudi Arabia. Bot. Mar.58, 23–34 (2015). [Google Scholar]
- 21.Bormans, M. et al. Demonstrated transfer of cyanobacteria and cyanotoxins along a freshwater-marine continuum in France. Harmful Algae87, (2019). [DOI] [PubMed]
- 22.Overlingė, D. et al. Phytoplankton of the Curonian lagoon as a new interesting source for bioactive natural products. Special impact on Cyanobacterial metabolites. Biomolecules11, 1139 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Šulčius, S. et al. Increased risk of exposure to microcystins in the scum of the filamentous cyanobacterium Aphanizomenon flos-aquae accumulated on the western shoreline of the Curonian lagoon. Mar. Pollut Bull.99, 264–270 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Šulčius, S. et al. Draft genome sequence of the cyanobacterium Aphanizomenon flos-aquae strain 2012/KM1/D3, isolated from the Curonian lagoon (Baltic Sea). Genome Announc.3, 1–2 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kumari, N. & Rai, L. C. Cyanobacterial diversity: Molecular insights under multifarious environmental conditions. Adv. Cyanobact. Biol.INC10.1016/B978-0-12-819311-2.00002-4 (2020).
- 26.Komárek, J. A polyphasic approach for the taxonomy of cyanobacteria: Principles and applications. Eur. J. Phycol.51, 346–353 (2016). [Google Scholar]
- 27.Erhard, M., von Döhren, H. & Jungblut, P. Rapid typing and elucidation of new secondary metabolites of intact cyanobacteria using MALDI-TOF mass spectrometry. Nat. Biotechnol.15, 906–909 (1999). [DOI] [PubMed] [Google Scholar]
- 28.Spoof, L., Błaszczyk, A., Meriluoto, J., Cegłowska, M. & Mazur-Marzec, H. Structures and activity of new anabaenopeptins produced by Baltic Sea cyanobacteria. Mar. Drugs14, (2016). [DOI] [PMC free article] [PubMed]
- 29.Nowruzi, B. et al. Identification and toxigenic potential of a Nostoc Sp. Algae. 27, 303–313 (2012). [Google Scholar]
- 30.Zervou, S. K., Kaloudis, T., Gkelis, S., Hiskia, A. & Mazur-Marzec, H. Anabaenopeptins from cyanobacteria in freshwater bodies of Greece. Toxins (Basel)14, (2022). [DOI] [PMC free article] [PubMed]
- 31.Schmelling, N. M. & Bross, M. Too many big promises: what is holding back cyanobacterial research and applications? bioRxiv (2023). 10.1101/2023.06.05.543618
- 32.Johansen, J. R. & Casamatta, D. A. Recognizing cyanobacterial diversity through adoption of a new species paradigm. Algol. Stud. für Hydrobiol. Suppl. Vol.117, 71–93 (2005). [Google Scholar]
- 33.HELCOM. Quality assurance of phytoplankyon monitoring in the Baltic Sea (PEG QA) (2023). https://helcom.fi/helcom-at-work/projects/peg/
- 34.Wejnerowski, L. et al. Solitary terminal cells of Aphanizomenon gracile (cyanobacteria, Nostocales) can divide and renew trichomes. Phycol. Res.65, 248–255 (2017). [Google Scholar]
- 35.Palińska, K. A. & Surosz, W. Population of Aphanizomenon from the Gulf of Gdańsk (Southern Baltic Sea): Differences in phenotypic and genotypic characteristics. Hydrobiologia610, 355 (2008). [Google Scholar]
- 36.Wejnerowski, L., Cerbin, S., Wojciechowicz, M. K. & Dziuba, M. K. Differences in cell wall of thin and thick filaments of cyanobacterium Aphanizomenon Gracile SAG 31.79 and their implications for different resistance to Daphnia grazing. J. Limnol.75, 634–643 (2016). [Google Scholar]
- 37.Stoń, J., Kosakowska, A., Łotocka, M. & Łysiak-Pastuszak, E. Pigment composition in relation to phytoplankton community structure and nutrient content in the Baltic Sea. Oceanologia. 44, 419–437 (2002). [Google Scholar]
- 38.Wright, S. W. & Jeffrey, S. W. Pigment markers for phytoplankton production. Handb. Environ. Chem. Vol. 2 React. Process.2N, 71–104 (2006).
- 39.Kahru, M., Elmgren, R., Kaiser, J., Wasmund, N. & Savchuk, O. Cyanobacterial blooms in the Baltic Sea: Correlations with environmental factors. Harmful Algae92, 101739 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Bartoli, M. et al. Drivers of cyanobacterial blooms in a hypertrophic lagoon. Front. Mar. Sci.5, (2018).
- 41.Gugger, M. et al. Phylogenetic comparison of the cyanobacterial genera Anabaena and Aphanizomenon. Int. J. Syst. Evol. Microbiol.52, 1867–1880 (2002). [DOI] [PubMed] [Google Scholar]
- 42.Barker, G. L. A., Konopka, A., Handley, B. A. & Hayes, P. K. Genetic variation in Aphanizomenon (Cyanobacteria) colonies from the Baltic Sea and North America. J. Phycol.36, 947–950 (2000). [Google Scholar]
- 43.Wejnerowski, Ł. et al. In vitro toxicological screening of stable and senescing cultures of Aphanizomenon, Planktothrix, and Raphidiopsis. Toxins (Basel)12, (2020). [DOI] [PMC free article] [PubMed]
- 44.Budzałek, G., Sliwinska-Wilczewska, S., Klin, M., Wisniewska, K. & Latała, A. Changes in growth, photosynthesis performance, pigments, and toxin contents of bloom-forming cyanobacteria after exposure to macroalgal allelochemicals. Toxins (Basel)13, 1–20 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kosiba, J., Krztoń, W., Koreiviené, J., Tarcz, S. & Wilk-Woźniak, E. Interactions between Ciliate species and Aphanizomenon flos-aquae vary depending on the morphological form and biomass of the diazotrophic cyanobacterium. Int. J. Environ. Res. Public. Health19, (2022). [DOI] [PMC free article] [PubMed]
- 46.Driscoll, C. B. et al. A closely-related clade of globally distributed bloom-forming cyanobacteria within the Nostocales. Harmful Algae77, 93–107 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Teikari, J. E. et al. Insight into the genome and brackish water adaptation strategies of toxic and bloom-forming Baltic Sea Dolichospermum sp. UHCC 0315. Sci. Rep.9, 1–13 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Österholm, J., Popin, R. V., Fewer, D. P. & Sivonen, K. Phylogenomic analysis of secondary metabolism in the toxic cyanobacterial genera Anabaena, Dolichospermum and Aphanizomenon. Toxins (Basel)12, 248 (2020). [DOI] [PMC free article] [PubMed]
- 49.Jones, M. et al. Comprehensive database of secondary metabolites from cyanobacteria. BioRxiv - Prepr Serv. Biol.10.1101/2020.04.16.038703 (2020). [Google Scholar]
- 50.Suurnäkki, S. et al. Identification of geosmin and 2-methylisoborneol in cyanobacteria and molecular detection methods for the producers of these compounds. Water Res.68, 56–66 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Karosienė, J. et al. First report of saxitoxins and anatoxin-a production by cyanobacteria from Lithuanian lakes. Eur. J. Phycol.55, 327–338 (2020). [Google Scholar]
- 52.Ballot, A., Fastner, J. & Wiedner, C. Paralytic shellfish poisoning toxin-producing cyanobacterium Aphanizomenon gracile in Northeast Germany. Appl. Environ. Microbiol.76, 1173–1180 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Halary, S. et al. Intra-population genomic diversity of the bloom-forming cyanobacterium, Aphanizomenon Gracile, at low spatial scale. ISME Commun.3, 1–4 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murakami, M., Suzuki, S., Itou, Y., Kodani, S. & Ishida, K. New anabaenopeptins, carboxypeptidaze-A inhibitors from the cyanobacterium Aphanizomenon Flos-Aquae. J. Nat. Prod.63, 1280–1282 (2000). [DOI] [PubMed] [Google Scholar]
- 55.Welker, M. & Von Döhren, H. Cyanobacterial peptides - Nature’s own combinatorial biosynthesis. FEMS Microbiol. Rev.30, 530–563 (2006). [DOI] [PubMed] [Google Scholar]
- 56.Šulčius, S. et al. The profound effect of harmful cyanobacterial blooms: From food-web and management perspectives. Sci. Total Environ.609, 1443–1450 (2017). [DOI] [PubMed] [Google Scholar]
- 57.Repka, S., Meyerhöfer, M., Von Bröckel, K. & Sivonen, K. Associations of cyanobacterial toxin, nodularin, with environmental factors and zooplankton in the Baltic Sea. Microb. Ecol.47, 350–358 (2004). [DOI] [PubMed] [Google Scholar]
- 58.Ferreira Ferreria, A. H. Peptides in cyanobacteria under different environmental conditions. 1–120 (2006).
- 59.Ersmark, K., Valle, D., Hanessian, S. & J. R. & Chemistry and biology of the aeruginosin family of serine protease inhibitors. Angew Chemie - Int. Ed.47, 1202–1223 (2008). [DOI] [PubMed] [Google Scholar]
- 60.Bishop, M. & Zubeck, M. W. H. Evaluation of microalgae for use as nutraceuticals and nutritional supplements. J. Nutr. Food Sci.02, (2012).
- 61.Baroni, L. et al. Effect of a Klamath algae product (‘AFA-B12’) on blood levels of vitamin B12 and homocysteine in vegan subjects: a pilot study. Int. J. Vitam. Nutr. Res.79, 117–123 (2009). [DOI] [PubMed] [Google Scholar]
- 62.Amato, A. et al. Spasmolytic effects of Aphanizomenon Flos-Aquae (Afa) extract on the human colon contractility. Nutrients13, (2021). [DOI] [PMC free article] [PubMed]
- 63.Caro, V. et al. Enhanced in situ availability of Aphanizomenon flos-aquae constituents entrapped in buccal films for the treatment of oxidative stress-related oral diseases: Biomechanical characterization and in vitro/ex vivo evaluation. Pharmaceutics11, (2019). [DOI] [PMC free article] [PubMed]
- 64.Campana, R. et al. Influence of Aphanizomenon flos-aquae and two of its extracts on growth ability and antimicrobial properties of Lactobacillus acidophilus DDS-1. Lwt81, 291–298 (2017). [Google Scholar]
- 65.Scoglio, S. et al. Inhibitory effects of Aphanizomenon flos-aquae constituents on human UDP-glucose dehydrogenase activity. J. Enzyme Inhib. Med. Chem.31, 1492–1497 (2016). [DOI] [PubMed] [Google Scholar]
- 66.Kótai, J. Instructions for preparation of modified nutrient solution Z8 for algae. NIVA B-11/69. NIVA 1976. Estimation of algal growth potential. Nor. Inst. Water Res.D2, (1972).
- 67.Jungblut, A. D. et al. Diversity within cyanobacterial mat communities in variable salinity meltwater ponds of McMurdo Ice Shelf, Antarctica. Environ. Microbiol.7, 519–529 (2005). [DOI] [PubMed] [Google Scholar]
- 68.Neilan, B. A. Detection and identification of cyanobacteria associated with toxic blooms: DNA amplification protocols. Phycologia35, 147–155 (1996). [Google Scholar]
- 69.Rudi, K., Skulberg, O. M. & Jakobsen, K. S. Evolution of cyanobacteria by exchange of genetic material among phyletically related strains. J. Bacteriol.180, 3453–3461 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Kumar, S., Stecher, G. & Tamura, K. MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol.33, 1870–1874 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.John, N. et al. An improved method for PCR-based detection and routine monitoring of geosmin-producing cyanobacterial blooms. Water Res.136, 34–40 (2018). [DOI] [PubMed] [Google Scholar]
- 72.Fidor, A., Konkel, R. & Mazur-Marzec, H. Bioactive peptides produced by cyanobacteria of the genus Nostoc: A review. Marine Drugs17, (2020). 10.3390/md17100561 [DOI] [PMC free article] [PubMed]
- 73.Ballot, A. et al. Diversity of cyanobacteria and cyanotoxins in Hartbeespoort Dam, South Africa. Mar. Freshw. Res.65, 175–189 (2013). [Google Scholar]
- 74.Rantala, A. et al. Phylogenetic evidence for the early evolution of microcystin synthesis. Proc. Natl. Acad. Sci.101, 568–573 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Parsons, R., Timothy, M. & Yoshiaki, L. M. C. A manual of chemical and biological methods for sea water analysis. Deep Sea Res. Part. Oceanogr. Res. Pap.31 (1984).
- 76.Stoń-Egiert, J. & Kosakowska, A. RP-HPLC determination of phytoplankton pigments-comparison of calibration results for two columns. Mar. Biol.147, 251–260 (2005). [Google Scholar]
- 77.Jean-Pierre, D. Estimation of cyanobacteria biomass by marker pigment analysis. in Handbook of Cyanobacterial Monitoring and Cyanotoxin Analysis (eds Jussi, M., Lisa, S. & Geoffrey, C. A.) 343–349 (Wiley, Ltd, (2017). [Google Scholar]
- 78.Stewart, D. E. & Farmer, F. H. Extraction, identification, and quantitation of phycobiliprotein pigments from phototrophic plankton. Limnol. Oceanogr.29, 392–397 (1984). [Google Scholar]
- 79.Sobiechowska-Sasim, M., Stoń-Egiert, J. & Kosakowska, A. Quantitative analysis of extracted phycobilin pigments in cyanobacteria—an assessment of spectrophotometric and spectrofluorometric methods. J. Appl. Phycol.26, 2065–2074 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Mazur-Marzec, H. et al. Baltic cyanobacteria—a source of biologically active compounds. Eur. J. Phycol.50, 343–360 (2015). [Google Scholar]
- 81.Sadowy, E. & Luczkiewicz, A. Drug-resistant and hospital-associated Enterococcus faecium from wastewater, riverine estuary and anthropogenically impacted marine catchment basin. BMC Microbiol.14, (2014). [DOI] [PMC free article] [PubMed]
- 82.Kotlarska, E., Łuczkiewicz, A., Baraniak, A., Jankowska, K. & Całkiewicz, J. ESBL-producing Aeromonas spp. isolated from wastewater and marine environment. Acta Biochim. Pol. vol. 65 at (2018).
- 83.Kotlarska, E., Łuczkiewicz, A. & Burzyński, A. Vibrio cholerae in the Polish coastal waters of the Baltic Sea—single case or a sign of climate warming? Acta Biochim. Pol.65, (2018).
- 84.Pluotno, A., Carmeli, S. & Banyasin A and banyasides a and B, three novel modified peptides from a water bloom of the cyanobacterium Nostoc Sp. Tetrahedron61, 575–583 (2005). [Google Scholar]
- 85.Ocampo Bennet, X. Peptide au seiner Cyanobakterien Wasserblütte (1998) aus dem Wannsee/Berli: Strukturen and biologische Wirksamkeit. (2007).
- 86.Kwan, J. C., Taori, K., Paul, V. J. & Luesch, H. Lyngbyastatins 8–10, elastase inhibitors with cyclic depsipeptide scaffolds isolated from the marine cyanobacterium Lyngbya semiplena. Mar. Drugs7, 528–538 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Konkel, R. et al. Anabaenopeptins from Nostoc Edaphicum CCNP1411. Int. J. Environ. Res. Public. Health19, (2022). [DOI] [PMC free article] [PubMed]
- 88.Felczykowska, A. et al. Selective inhibition of cancer cells’ proliferation by compounds included in extracts from Baltic Sea cyanobacteria. Toxicon108, 1–10 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequence data that support the findings of this study have been deposited in the DDBJ/EMBL/GenBank databases under accession numbers PP025307, PP025308, PP025309, PP025310, OR987680, OR984864.