Abstract
The genomic segment encoding the putative hemagglutinin of infectious salmon anemia virus (ISAV) is described. Expression of the putative hemagglutinin in a salmon cell line demonstrated hemadsorptive properties of the protein for salmon erythrocytes. The polypeptide was recognized by an ISAV-specific monoclonal antibody. Nucleotide sequencing indicated the occurrence of a variable region in the hemagglutinin gene.
Infectious salmon anemia virus (ISAV), an important pathogen for the Atlantic salmon (Salmo salar) farming industry, shows many properties that closely resemble those of the Orthomyxoviridae (1, 4, 7, 9). As with the influenza A and B viruses, the genome of ISAV consists of eight single-stranded RNA segments of negative polarity with partially complementary 3′- and 5′-end noncoding terminal sequences (9), and the mRNA synthesis requires 8- to 18-nucleotide 5′-cap structures cleaved from cellular heteronuclear RNAs (11, 13). Transfer of convalescent-phase sera from surviving fish has been shown to reduce mortality in fish exposed to ISAV (2). This indicates that humoral immune responses of the salmon may be protective against ISAV infection.
Molecular cloning.
The ISAV isolates Glesvaer and 390/98, isolated from ISA epizootics in western Norway in 1990 and in western Scotland in 1998, respectively, were used in this study. For the Glesvaer isolate, mRNA from ISAV-infected salmon head kidney cells (SHK-1 cells) was extracted and used in reverse transcription, adapters were added, and the cDNA was subsequently ligated into pCRII plasmid vector (Invitrogen, Leek, The Netherlands). Plasmid inserts were excised from gels, 32P labeled, and used as probes in Northern hybridisation reactions. For the 390/98 isolate, a subtractive cDNA library was obtained. Total RNA was extracted from both ISAV-infected and noninfected SHK-1 cells, converted into double-stranded cDNA, and used for the subtractive hybridization (PCR-Select cDNA subtraction kit; Clontech Laboratories, Palo Alto, Calif.). Material resulting from this was PCR amplified and ligated into the vector pGEM-T Easy (Promega, Madison, Wis.). Insert fragments were labeled and used as probes in Northern hybridization reactions. To obtain the 3′ and 5′ ends of viral RNA from both isolates, rapid amplification of cDNA ends (RACE) was used (13).
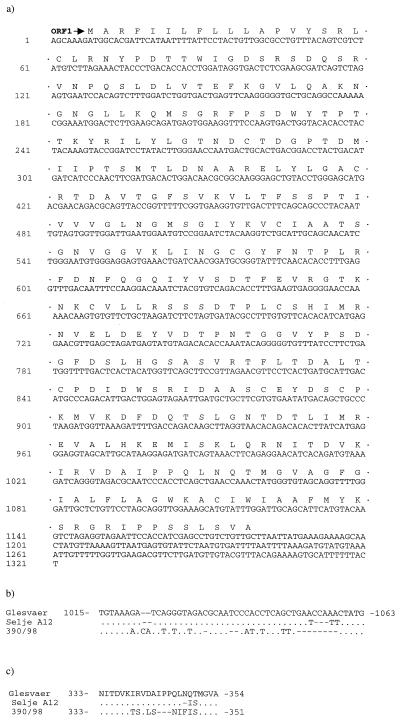
Multiple molecular clones that were tentatively classed as ISAV specific were sequenced, and alignments of nucleotide and amino acid sequences were carried out. One molecular clone from the Glesvaer isolate-infected SHK-1 cells was found to give positive Northern hybridisation with ISAV-infected SHK-1, with a distinct band of approximately 1.3 kb (Fig. 1A). Combining the results of the 5′-end mRNA and 5′-end viral RNA (vRNA) RACE, this genomic segment of the Glesvaer isolate was found to consist of 1,321 nucleotide residues (Fig. 2a) (accession number AF220607). A full-length mRNA, excluding the heterogeneous 5′ primer sequence and poly(A) tail, was deduced to be 1,301 nucleotides long, while the similar full-length mRNA of the Scottish 390/98 isolate was 1,292 nucleotides long (accession number AJ276859). Alignment of the nucleotide sequences of the Glesvaer and 390/98 isolates showed 96.8% identity. However, the discrepancy between the nucleotide sequences of the two isolates was unevenly distributed. Between nucleotide residues 1021 and 1060 (numbers refer to the Glesvaer isolate), there were 22 residue differences, including deletions and gaps, yielding 12 of 13 consecutive residues between residues 339 and 350 in the deduced amino acid sequence nonhomologous (Fig. 2b and c). To determine whether this localized discrepancy indicated genetic diversity or was caused by artifacts introduced through the laboratory procedures, a reverse transcription (RT)-PCR with subsequent cloning and nucleotide sequencing was performed using RNA extracted from the kidney of a salmon in a natural outbreak of ISA in Norway (this clone was named Selje A12) and from Scottish ISAV isolate 1490/98, which is known to differ from 390/98 in the nucleotide sequences of segments 2 and 8 (8). This revealed identity between Glesvaer and Selje A12 isolates up to nucleotide 1054. There is a 6-nucleotide discrepancy (nucleotides 1054 to 1059), including a 3-nucleotide gap, in the Selje A12 sequence between the Glesvaer and Selje A12 isolates (Fig. 2b and c). The sequence from 1490/98 was identical to that from Glesvaer, although these isolates are known to differ in the sequences of segments 2 and 8 (8).
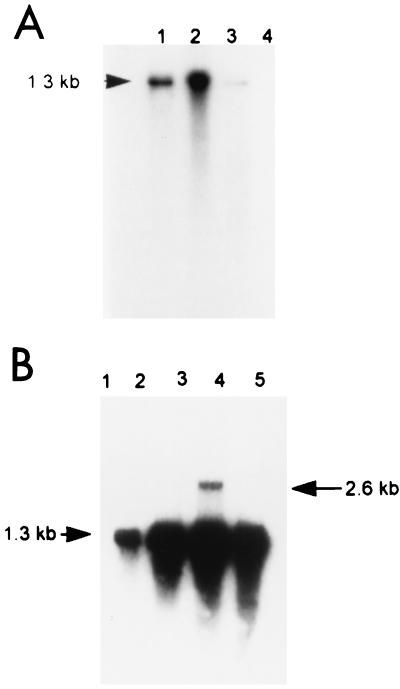
FIG. 1.
Northern blot hybridization using the ISAV-specific clone obtained from the Glesvaer isolate as a probe. Molecular sizes (in kilobases) are indicated. (A) Lanes: 1 RNA extracted from ISAV-infected SHK-1 cells 6 days p.i.; 2, RNA extracted from ISAV-infected SHK-1 cells 7 days p.i.; 3, RNA extracted from pelleted ISAV; 4, noninfected SHK-1 cells. (B) The target was total RNA extracted from ISAV Glesvaer-infected SHK-1 cells. Lanes 1, 2, 3, 4, and 5 correspond to 1, 3, 5, 7, and 9 days p.i., respectively.
FIG. 2.
Complete nucleotide sequence of ISAV Glesvaer HA genomic segment with translated ORFs (a) and alignments of nucleotide (b) and amino acid (c) sequences of segment from ISAV Glesvaer, Selje A12, and 390/98 at the variable region. ·, identical to base in Glesvaer; —, gaps introduced to maximize alignment.
Computer analysis of the nucleotide sequence of this genomic segment found one large open reading frame (ORF) (ORF1) that consisted of 1,176 and 1,167 residues for the Glesvaer and 390/98 isolates, respectively. In both isolates, ORF1 started at the AUG codon at nucleotides 8 to 10. The deduced putative polypeptides from ORF1 were thus 391 and 388 amino acids long and approximately 42.4 to 42.7 kDa, excluding possible posttranslational modifications.
Two hydrophobic domains of the predicted ORF1-encoded polypeptide scored as certain (from amino acids 150 to 170 and 350 to 370) using the TopPred 2 programme (16), and thus represented possible transmembrane regions of the deduced amino acid sequence. Two other regions were also found to be hydrophobic, from amino acids 1 to 21 and 122 to 142, although the latter was only weakly hydrophobic. The N-terminal hydrophobic part, amino acids 1 to 21, was deduced by the SignalP program (10) to be a signal peptide, with the most probable cleavage site between amino acids 16 and 17, and amino acid sequencing of purified ISAV polypeptides supports this (S. Griffiths, personal communication). A cell attachment sequence, RGD (arginine-glycine-apartate), which indicates membrane association, was found from amino acids 119 to 121. Two possible glycosylation sites were predicted at amino acids 333 and 349 (Glesvaer).
No significant homologies between the nucleotide or deduced amino acid sequences of this ISAV genomic segment with other viral sequences was found using the Blast program, which reflects that the evolutionary distance between ISAV and orthomyxoviruses infecting terrestrial animals is vast.
Viral RNA analysis.
A time course study of the occurrence of intracellular RNA specific to this ISAV genomic segment in SHK-1 cells was performed by extracting total RNA from ISAV-infected SHK-1 cells at days 1, 3, 5, 7, and 9 postinfection (p.i.). The RNA was subjected to electrophoresis, blotted, fixed to a nylon membrane, and then used as the target in Northern hybridization as previously described (9). No hybridization signal was found at day 1 p.i., but from day 3 p.i. and onwards, a band representing a genomic segment of approximately 1.3 kb with increasing density was observed. A band of approximately 2.6 kb was also observed at days 5 and 7 p.i. (Fig. 1b). In ISAV-infected cells, there would be three different classes of viral RNA: mRNA, vRNA (genomic RNA), and template RNA for the synthesis of vRNA. The sizes of vRNA, template RNA, and full-length mRNA would correspond to the 1.3-kb fragment that showed a strong hybridization signal. This RNA band appeared much stronger from day 5 p.i., which was probably the result of the production of full-length genomic RNA. The RNA band with a size of approximately 2.6 kb, which mainly appeared at days 5 and 7 p.i., was possibly caused by a binding between vRNA and template RNA.
To investigate the possibility that several different mRNAs could be transcribed from the present ISAV genomic segment, a PCR was carried out using primers corresponding to the 5′ and 3′ ends of mRNAs. This choice of primers is based on the assumption that ISAV mRNA, like influenza virus mRNA, always contains a copy of the 3′ end of the genomic segment in the 5′ end of mRNAs, and similarly the poly(A) addition signal in the 5′ end of the genomic segment is present in the 5′ end of the mRNA. The results from the mRNA RT-PCR did not reveal any mRNA other than the 1.3-kb mRNA after amplification from Glesvaer-infected SHK-1 cells (5 or 7 days p.i.).
Expression.
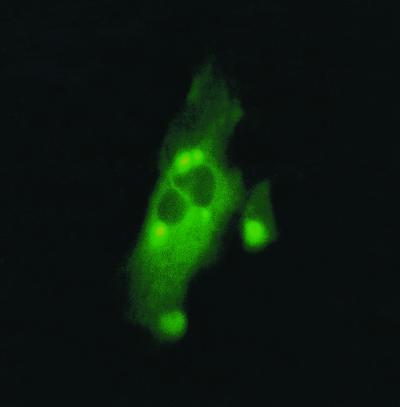
ORF1 was amplified by PCR and cloned into the pEGFP-N1 vector (Clontech Laboratories). The resulting plasmid was named ORF1-pEGFP-N1. The nucleotide sequence of the insert and adjacent sequences in the plasmid were determined to ensure that the insert was in the correct reading frame. The cloned gene was expressed as a fusion to the N terminus of a red-shifted variant of wild-type enhanced green fluorescent protein (EGFP). EGFP was chosen as a fusion partner because it enables gene expression to be observed in living cells as fluorescence without the need for fixation prior to microscopy. No autofluorescence was seen in negative controls. The capacity of the ORF1-pEGFP-N1 plasmid to express proteins in fish cells was tested by transfection of SHK-1 cells. This cell line originates from Atlantic salmon and was chosen in order to obtain correct processing of the expressed ISAV polypeptide. The intracellular location of the EGFP-ORF1 fusion protein was primarily detected as strong fluorescence in the cytoplasm, but later, in some cells, also as small, intense fluorescent points with a perinuclear location (Fig. 3). No fluorescence was observed in the nucleus.
FIG. 3.
Micrograph of fusion proteins encoded by ORF1-pEGFP-N1 plasmid in SHK-1 cells, showing cytoplasmic localization and dense perinuclear granules of the fusion protein.
Immune staining and hemadsorption.
Transfected SHK-1 cells were stained with the ISAV-specific monoclonal antibody (MAb) 3H6F8 (3), and sheep anti-mouse immunoglobulin biotinylated whole antibody (Amersham Pharmacia, Uppsala, Sweden) was used as the secondary antibody (Fig. 4). To test for the specificity of 3H6F8 staining, Alexafluor 594-labeled secondary antibodies were also used (Molecular Probes, Leiden, The Netherlands), and stained cells showed evenly distributed red fluorescence with no staining of the nucleus, and the same cells expressed the green fluorescence of the EGFP (data not shown).
FIG. 4.
Micrograph of ORF1-pEGFP-N1-transfected SHK-1 cells. The cells were incubated with salmon erythrocytes and stained with MAb 3H6F8. A stained cell with adherent salmon erythrocytes is shown.
Salmon erythrocytes were found to bind to single cells in the ORF1-pEGFP-N1-transfected SHK-1 monolayer and were observed as dense aggregations of erythrocytes on the SHK-1 cell surface, demonstrating the hemadsorptive properties of the ORF1 polypeptide (Fig. 4). The same SHK-1 cells that aggregated erythrocytes were also recognized by the 3H6F8 MAb (Fig. 4). It has previously been found that the 3H6M8 MAb inhibits hemagglutination of ISAV and recognizes a surface protein of the virus particle (3). Together, these data indicate that the polypeptide encoded by the ORF1 is the putative ISAV hemagglutinin.
ISAV hemagglutinin.
Hemagglutinin is a high-abundance protein of influenza virus particles and has been calculated to make up approximately 40% of the weight of influenza B virus particles (12). Of the ISAV polypeptides, the most abundant, as observed in gel electrophoresis, had a size of approximately 43 kDa (4), which corresponds well to the deduced polypeptide encoded by ORF1. However, it is less than the hemagglutinin polypeptide of influenza virus, which, prior to modification, has a molecular size of approximately 61 kDa. However, electron microscopic studies have shown that the morphology of the surface spikes of ISAV appear quite different from those of influenza A virus (4), indicating structural differences between the surface proteins. The ISAV surface projections were approximately 10 nm long, while those of influenza A virus were 14 nm long (4).
The receptor-destroying enzyme activity of ISAV has been suggested to be an acetylesterase and not neuraminidase (4). A similar activity is found in influenza C virus, for which receptor binding and cleavage activities are combined in one molecule (6). In order to test for eventual acetylesterase activity of the expressed ORF1 polypeptide 4, methylumbelliferyl acetate was added to both transfected and control SHK-1 cultures, and the turnover of substrate by acetylesterase activity was measured (HTS 7000 Bio Assay reader; Perkin Elmer Applied Biosystems, Foster City, Calif.). No difference in acetylesterase activity was observed between transfected and control cells, indicating that the ORF1 polypeptide did not have acetylesterase activity.
The influenza virus hemagglutinin proteins, in contrast to several other influenza virus proteins, never enter the nucleus during the replication cycle. Similarly, the ORF1 polypeptide was not found in the nucleus. The influence of the ORF1 polypeptide fusion partner, EGFP, on intracellular localization is unknown; however, it follows the localization signal of its fusion partner (5), and without a fusion partner, EGFP is distributed throughout the cell cytoplasm and nucleus. It has been found earlier that ISAV binds to sialic acid residues on the cell surface and that the fusion between virus and cell membrane takes place in the acid environment of endosomes (1), indicating that surface proteins of ISAV have activities similar to those of influenza virus hemagglutinin. The influenza virus hemagglutinin polypeptide has an N-terminal signal peptide that is cleaved by a signal peptidase, and it has two further hydrophobic regions, of which the one at the C terminus crosses the viral envelope. The other hydrophobic region of influenza virus hemagglutinin is an essential component of the hemagglutinin fusion process. The putative ISAV hemagglutinin also has two hydrophobic regions that scored as probable membrane-crossing regions besides the signal peptide, and the locations of these corresponded well with those in influenza virus hemagglutinin. If the putative ISAV hemagglutinin has the same orientation as influenza virus hemagglutinin, it would thus have a cytoplasmatic C-terminal tail of 20 to 22 amino acids (from 391 to approximately 370), and influenza virus A (H3) has a cytoplasmatic tail of 10 amino acids (15). The ISAV hemagglutinin hydrophobic domain from residues 350 to 370 could cross the viral envelope, and the ectodomain would consist of 334 amino acid residues. The two predicted N-linked glycosylation sites on ISAV hemagglutinin would thus be in the ectodomain, compared to between three and nine (depending on the isolate) N-linked oligosaccharide chains per subunit added to the ectodomain of influenza virus A hemagglutinin (14).
The region in the ISAV genomic segment that showed sequence discrepancy between isolates is of particular interest. This region encodes a hydrophilic part of the ORF1 protein just upstream of the hydrophobic domain predicted to cross the viral envelope. Nucleotide sequencing of the Selje A12 sample revealed closer nucleotide sequence homology to the Glesvaer isolate than to the 390/98 isolate in the variable region. This homology may reflect closer genetic and epidemiological relationship, as 390/98 originated from a different geographical area. The Selje A12 sample was amplified from RNA obtained from diseased fish and has thus never been cultivated in SHK-1 cells. The similarity between Glesvaer and Selje A12 indicated that there had been no selection for genetic variation caused by adaptation of ISAV to SHK-1 cells. The variable region of this segment may prove useful in further epidemiological and phylogenetic analyses.
In conclusion, we have described the cloning and nucleotide sequencing of the ISAV genomic segment that encodes the putative hemagglutinin. The putative ISAV hemagglutinin differs remarkably in size from those of influenza viruses, but otherwise conforms to many of the properties of orthomyxoviral hemagglutinins.
Acknowledgments
This work was supported by grant 110562/122 from the Norwegian Research Council.
The skillful technical assistance of Ann Teig and Julie Inglis is appreciated.
REFERENCES
- 1.Eliassen T M, Froystad M K, Dannevig B H, Jankowska M, Brech A, Falk K, Romoren K, Gjoen T. Initial events in infectious salmon anemia virus infection: evidence for the requirement of a low-pH step. J Virol. 2000;74:218–227. doi: 10.1128/jvi.74.1.218-227.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Falk K, Dannevig B H. Demonstration of a protective immune-response in infectious salmon anemia (ISA)-infected atlantic salmon salmo-salar. Dis Aquat Org. 1995;21:1–5. [Google Scholar]
- 3.Falk K, Namork E, Dannevig B H. Characterization and applications of a monoclonal antibody against infectious salmon anaemia virus. Dis Aquat Org. 1998;34:77–85. doi: 10.3354/dao034077. [DOI] [PubMed] [Google Scholar]
- 4.Falk K, Namork E, Rimstad E, Mjaaland S, Dannevig B H. Characterization of infectious salmon anemia virus, an orthomyxo-like virus isolated from Atlantic salmon (Salmo salar L.) J Virol. 1997;71:9016–9023. doi: 10.1128/jvi.71.12.9016-9023.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flach J, Bossie M, Vogel J, Corbett A, Jinks T, Willins D A, Silver P A. A yeast RNA-binding protein shuttles between the nucleus and the cytoplasm. Mol Cell Biol. 1994;14:8399–8407. doi: 10.1128/mcb.14.12.8399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Herrler G, Durkop I, Becht H, Klenk H-D. The glycoprotein of influenza C virus is the haemagglutinin, esterase and fusion factor. J Gen Virol. 1988;69:839–846. doi: 10.1099/0022-1317-69-4-839. [DOI] [PubMed] [Google Scholar]
- 7.Hovland T, Nylund A, Watanabe K, Endresen C. Observation of infectious salmon anemia virus in Atlantic salmon, Salmo-salar L. J Fish Dis. 1994;17:291–296. [Google Scholar]
- 8.Inglis J A, Bruce J, Cunningham C O. Nucleotide sequence variation in isolates of infectious salmon anaemia virus (ISAV) from Atlantic salmon Salmo salar in Scotland and Norway. Dis Aquat Org. 2000;43:71–76. doi: 10.3354/dao043071. [DOI] [PubMed] [Google Scholar]
- 9.Mjaaland S, Rimstad E, Falk K, Dannevig B H. Genomic characterization of the virus causing infectious salmon anemia in Atlantic salmon (Salmo salar L.): an orthomyxo-like virus in a teleost. J Virol. 1997;71:7681–7686. doi: 10.1128/jvi.71.10.7681-7686.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Eng. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- 11.Plotch S J, Bouloy M, Ulmanen I, Krug R M. A unique cap(m7GpppXm)-dependent influenza virion endonuclease cleaves capped RNAs to generate the primers that initiate viral RNA transcription. Cell. 1981;23:847–858. doi: 10.1016/0092-8674(81)90449-9. [DOI] [PubMed] [Google Scholar]
- 12.Ruigrok R W H. Structure of influenza A, B and C viruses. In: Nicholson K G, Webster R G, Hay A J, editors. Textbook of influenza. Oxford, U.K: Blackwell Science Ltd.; 2000. pp. 29–42. [Google Scholar]
- 13.Sandvik T, Rimstad E, Mjaaland S. The viral RNA 3′- and 5′-end structure and mRNA transcription of infectious salmon anaemia virus resemble those of influenza viruses. Arch Virol. 2000;145:1659–1669. doi: 10.1007/s007050070082. [DOI] [PubMed] [Google Scholar]
- 14.Schulze I T. Effects of glycosylation on the properties and functions of influenza virus hemagglutinin. J Infect Dis. 1997;176(Suppl. 1):S24–S28. doi: 10.1086/514170. [DOI] [PubMed] [Google Scholar]
- 15.Verhoeyen M, Fang R, Jou W M, Devos R, Huylebroeck D, Saman E, Fiers W. Antigenic drift between the haemagglutinin of the Hong Kong influenza strains A/Aichi/2/68 and A/Victoria/3/75. Nature. 1980;286:771–776. doi: 10.1038/286771a0. [DOI] [PubMed] [Google Scholar]
- 16.von Heijne G. Membrane protein structure prediction: hydrophobicity analysis and the positive-inside rule. J Mol Biol. 1992;225:487–494. doi: 10.1016/0022-2836(92)90934-c. [DOI] [PubMed] [Google Scholar]