Abstract
Because the replication of hepatitis B virus (HBV) proceeds via an obligatory reverse transcription step in the viral capsid, cDNA is potentially vulnerable to editing by cytidine deaminases of the APOBEC3 family. To date only two edited HBV genomes, referred to as G → A hypermutants, have been described in vivo. Recent work suggested that HBV replication was indeed restricted by APOBEC3G but by a mechanism other than editing. The issue of restriction has been explored by using a sensitive PCR method allowing differential amplification of AT-rich DNA. G → A hypermutated HBV genomes were recovered from transfection experiments involving APOBEC3B, -3C, -3F, and -3G indicating that all four enzymes were able to extensively deaminate cytidine residues in minus-strand DNA. Unexpectedly, three of the four enzymes (APOBEC3B, -3F, and -3G) deaminated HBV plus-strand DNA as well. From the serum of two of four patients with high viremia, G → A hypermutated genomes were recovered at a frequency of ≈10-4, indicating that they are, albeit relatively rare, part of the natural cycle of HBV infection. These findings suggest that human APOBEC3 enzymes can impact HBV replication via cytidine deamination.
Keywords: hypermutation, DNA editing
G → A hypermutated retroviral genomes result from the editing of nascent DNA by APOBEC3 cytidine deaminases (1-7). This was originally demonstrated for HIV-1 and the APOBEC3G member of a cluster of seven genes (3A-3H) on human chromosome 22 (8, 9). Editing occurs on the background of a Δvif genome (1-5, 10). The HIV Vif protein prevents packaging of either APOBEC3F or -3G resulting in their ubiquitination and degradation by the proteasomal pathway (11-14). Editing of cytidine residues in neo-synthesized minus-strand cDNA results in the formation of multiple uracil residues that are read as T during plus-strand synthesis. Albeit referred to as G → A hypermutants by reference to the viral plus strand, mechanistically the action is occurring on the minus strand and independently of reverse transcriptase (7). Up to 60% of G residues in a lentiviral genome can be substituted resulting in the total loss of information (15-17). As such, APOBEC3F and -3G constitute a powerful restriction mechanism to reverse transcription. Retroviruses have either to avoid replication in cells expressing APOBEC3 molecules or else evolve a mechanism that neutralizes their effect. The vif gene of human and primate lentiviruses, as well as their homologues in most of the other lentiviruses, reflect the latter solution.
Hepatitis B viruses (HBVs) replicate via an obligate reverse transcription step occurring in a capsid structure close to the endoplasmic reticulum. A pair of G → A hypermutated genomes were identified in the blood of a chronically infected patient yet have remained unique despite a burgeoning database (18). Furthermore, gene chip analyses of liver tissue have failed to show significant expression of APOBEC3 molecules, apart from APOBEC3C (ref. 19; http://genecards.bcgsc.bc.ca). Together, these findings suggest that HBV might have adopted the alternative route and sought replication in cells with little or no APOBEC3 expression.
Recent reports demonstrated that HBV replication could be strongly restricted by APOBEC3G and -3F in an experimental setting (20, 21). Restriction was highlighted by a strong reduction in the proportion of pregenomic RNA and DNA in the cytoplasm. Despite this, no G → A hypermutated genomes were identified, suggesting that the two enzymes curtailed replication via a nonediting mechanism. Using the same experimental system, another group confirmed restricted HBV replication but, like the original report, could not find a significant increase in the proportion of G → A hypermutants when the hepatoma Huh7 cell line was used (22). However, using the HepG2 cell line they found a 5-fold increase in the frequency of G → A hypermutants. Yet it is not obvious that there was a cell line effect. When Huh7 was cotransfected by an HIV-1Δvif genome and an APOBEC3G expression plasmid, restricted HIV-1 replication was observed when the supernatant was cultured on a susceptible cell line (20, 21).
These findings suggested that HBV replication may indeed be restricted in an experimental context by APOBEC3G, perhaps in an unconventional manner. However, the discrepancies between the findings and the dearth of such G → A hypermutated genomes in vivo suggests that the picture is incomplete.
Recently, a novel PCR technique was reported that allows differential DNA amplification of G → A hypermutants (23). This technique relies on the fact that the DNA of an AT-rich variant will melt at a slightly lower temperature than parental DNA. Using this technique, we have investigated the effect of various APOBEC3 family members on the integrity of neo-synthesized HBV DNA in Huh7 cells. The results unequivocally demonstrate that HBV DNA is sensitive to extensive cytidine deamination by four APOBEC3 molecules. When the technique was applied to DNA extracted from the serum of four chronically HBV-infected patients, numerous G → A hypermutants were identified in two cases. In short, HBV G → A hypermutation parallels more closely the HIV-1 paradigm than previously thought.
Materials and Methods
Plasmids. APOBEC3B and -3G cDNAs were a gift from Naveenan Navaratnam (Imperial College, London), and APOBEC3C and -3F were obtained as IMAGE clones from the HGMP Resource Centre. The different APOBEC3 cDNAs were subcloned in the expression vector pcDNA3.1D/V5-His-TOPO (Invitrogen). Plasmid pCayw allows expression of pregenomic HBV RNA under control of the powerful cytomegalovirus immediate early promoter (CMV-IE) and was provided by Frank Chisari (The Scripps Research Institute) (24). The plasmid pHBVΔcore harbors the same ayw genome and allows HBsAg expression, but is unable to replicate because of a deletion in the core gene (25).
Cells and Transfections. Huh7 and HepG2 human hepatoma cell lines were maintained in DMEM complemented with 10% FCS, 50 units/ml penicillin, and 50 μg/ml streptomycin (GIBCO/BRL). HepG2.2.15 cells, a derivative of HepG2 stably expressing HBV (26), were maintained in Williams medium complemented with 10% FCS, 50 units/ml penicillin, 50 μg/ml streptomycin, 0.35 μM hydrocortisone (Sigma), and 5 μg/ml bovine pancreatic insulin (Sigma). Transfections were performed by using Fu-GENE 6 (Roche). Briefly, 105 Huh7 cells were cotransfected with equal amounts (2 μg) of the pCayw and individual plasmids encoding human APOBEC3B, -3C, -3F, or -3G cDNAs. Controls were performed in parallel with pHBVΔcore. Total DNA was recovered 3 days after transfection and extracted.
Viral Capsid Purification and Western Blotting. Cells were collected and incubated in PBS with 1% Nonidet P-40 at 4°C overnight (27). After low-speed centrifugation, the lysates were collected and centrifuged at 27,000 × g for 2.5 h. The viral nucleocapsids were resuspended in 60 μl of 10 mM Tris·HCl (pH 7.5), 100 mM NaCl, and 1 mM EDTA. Thirty microliters of purified virus nucleocapsids (≈106 cell equivalents) per lane were separated on SDS-polyacrylamide gels and transferred to nitrocellulose (Schleicher & Schuell). As primary antibodies, a mouse monoclonal antibody specific for the HBcAg (provided by Marie-Louise Michel, Institut Pasteur) or a mouse monoclonal anti-serum specific for the V5 epitope tag (Invitrogen) were applied overnight after blocking of the membrane in 5% dried milk in PBS. After incubation with the respective horseradish peroxidase-coupled secondary antibody, the membrane was subjected to detection by enhanced chemoluminescence (Pierce).
PCR Amplification, Cloning, and Sequencing. A fragment of the HBV genome just 5′ to ORF X was amplified by employing a nested procedure. To increase sensitivity and specificity, hot start PCR was performed. The first-round primers were 5′-CGCAAATATACATCGTATCCAT and 5′-AAGAGTYYTYTTATGTAAGACYTT, where Y is T/C and R is A/G, and primers 5′-ATGGCTGCTARGCTGTGCTGCCAA and 5′-AAGTGCACACGGTYYGGCAGAT were used for the second round. Hypermutated genomes were identified by using differential DNA denaturation PCR (3D-PCR) in a two-round procedure (23). The first reaction involved standard amplification, the reaction parameters were 95°C for 5 min, followed by 35 cycles (95°C for 30 s, 55°C for 30 s, and 72°C for 30 s), and finally for 10 min at 72°C for the first round. Differential amplification occurred in the second round, using the equivalent of 0.5 μl of the first-round reaction as input. Cycling conditions were 90°C or 88°C for 5 min, followed by 35 cycles (90°C or 88°C for 60 s, 45°C for 30 s, and 72°C for 30 s), and finally 10 min at 72°C. The buffer conditions were 2.5 mM MgCl2, 50 mM KCl, 10 mM Tris·HCl (pH 8.3), 200 μM each dNTP, 100 μM each primer, and 2.5 units of Taq DNA polymerase (Applied Biosystems) in a final volume of 50 μl. Second-round PCR yielded a 213-bp fragment.
PCR products were purified from agarose gels (Qiaex II kit, Qiagen) and ligated in the TOPO TA cloning vector (Invitrogen). After transformation of XL1 Blue cells, up to 50 clones were picked. Sequencing was performed by using Thermosequenase (United States Biochemical, Amersham Pharmacia). The 245-bp segment of the ampicillin resistance gene was amplified by using ampi1, 5′-ATRCARTRCTRCCATAACCAT, and ampi2, 5′-GGGAAGYTAGAGTAAGTAGTT. The minimum denaturation temperature was 89°C.
Patient Material. Sera were obtained from four chronic carriers. For patient 130.71 the viral load was 3 × 109 DNA copies per ml; for patient 12763 the viral load was 1 × 109 copies per ml; for patient 129.5 the viral load was 8 × 108 copies per ml; and for patient 12741 the viral load was 6 × 108 copies per ml. Viral load was determined by the Chiron bDNA method. DNA was phenol extracted and taken up in 10 mM Tris·HCl (pH 8)/0.1 mM EDTA. The equivalent of 7 μl of serum was used for each PCR reaction.
Results
APOBEC3 Proteins Are Associated with HBV Capsids. The cytidine deaminase APOBEC3G has recently been shown to be associated with HBV core antigen (HBcAg) in the cytoplasm of virus-producing cells (20). To ascertain whether the four APOBEC3 enzymes can be associated with the HBV capsid, HepG2.2.15 cells were transfected with expression plasmids encoding V5-tagged forms of APOBEC3B, -3C, -3F, and -3G. Western blot analysis performed 3 days posttransfection confirmed that the purified fractions did indeed contain HBcAg (Fig. 1A). Importantly, all APOBEC3 proteins tested were associated with the purified capsids (Fig. 1B). The specificity of immunodetection is shown by the negative HepG2 controls (Figs. 1 A and B).
Fig. 1.
APOBEC3 molecules are associated with HBV capsids. (A) Western blot analysis of the purified HBV capsids from HepG2.2.15 transfected by APOBEC3 expression plasmids and detected by anti-HBcAg antibodies. HepG2 cells transfected by APOBEC3 plasmids were used as negative controls. (B) Western blot analysis of the same lysates using a monoclonal antibody to the APOBEC3-tagged V5 epitope. Molecular weight markers (kDa) are shown to the left.
Four APOBEC3 Deaminases Can Massively Edit HBV DNA. Huh7 cells were transfected by the same pCayw HBV plasmid used by Turelli et al. (20) along with APOBEC3B, -3C, -3F, and -3G expression plasmids. Given the ability of the 3D-PCR to differentially amplify HIV-1 G → A hypermutants, we reasoned that despite residual plasmid DNA from transfection, the method should be able to selectively amplify de novo synthesized and hyperdeaminated HBV DNA from a total DNA extraction. As hyperdeaminated DNA contains multiple uracil residues, Taq polymerase was used, as opposed to Pfu (20), because the latter has a read-ahead function and stalls at uracil residues in template DNA (28, 29).
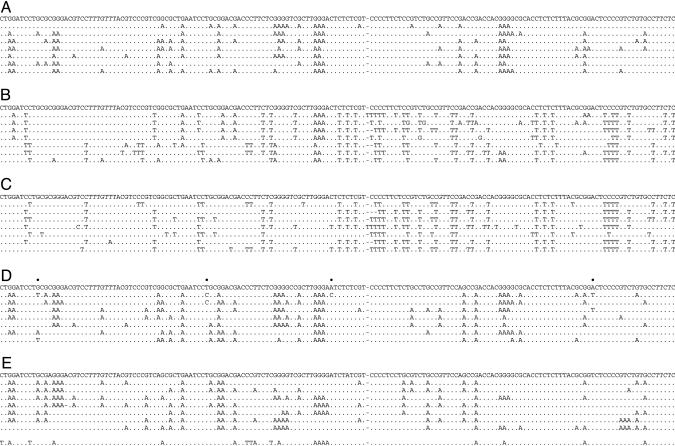
Total DNA was recovered from Huh7 cells 3 days after cotransfection. By slowly decreasing the denaturation temperature (Td), it was shown that 93°C was the minimum temperature allowing amplification of the fragment from the pCayw transfection alone. By performing PCR below the minimal denaturation temperature, for example 90°C, we were able to amplify DNA from all of the APOBEC3 cotransfections, i.e., APOBEC3B, -3C, -3F, and -3G, but not from pCayw alone. These PCR products were cloned, and DNA from up to 50 clones were sequenced per sample, a small selection being shown for the APOBEC3G sample in Fig. 2A. A complete collection of hypermutated sequences is available in Figs. 4-7, which are published as supporting information on the PNAS web site. As expected from the fact that the DNA was amplified at 90°C, all sequences were extensively substituted. All sequences were unique, indicating that they represented independent molecular events. The number of transitions ranged from 14-48 per clone, translating into G → A and/or C → T substitution frequencies of between 13% and 44%.
Fig. 2.
Massive deamination of HBV DNA by APOBEC3 cytidine deaminases. A selection of hypermutated HBV sequences mapping just 5′ to ORF X are shown compared with their reference. The sequence is given with respect to the viral plus strand. Only differences are shown. Hyphens denote gaps introduced to maximize sequence identity. All sequences were unique, indicating that they corresponded to distinct molecular events. (A) Selection of G → A hypermutations resulting from APOBEC3G deamination of minus-strand DNA. (B) A selection of mixed G → A and C → T hypermutants from APOBEC3B, -3F, and -3G cotransfections. (C) A selection of uniquely C → T hypermutants from APOBEC3B, -3F, and -3G cotransfections. (D and E) Hypermutated sequences derived from virions in the sera of patients 130.71 and 12763, respectively. The reference sequence given was the major sequence among 10 derived from PCR products amplified at 95°C. Dots above the reference sequences indicate polymorphic sites among the 10 nonhypermutated sequences. For patient 12763 (Fig. 3E), one of the sequences was a mixed G → A + C → T hypermutant.
Because the APOBEC3 genes were overexpressed by way of the powerful CMV-IE promoter, it might be argued that a very small proportion of APOBEC3 molecules found their way to the nucleus. If so, single-stranded DNA in a transcription bubble might be vulnerable to deamination. Given the sensitivity of 3D-PCR, it was possible that it was picking up such rare events. To this end, Huh7 cells were cotransfected by pHBVΔcore and APOBEC3 plasmids. The pHBVΔcore plasmid allows transcription through the locus amplified by 3D-PCR but, due to a deletion in the core gene, was not replication-competent. Total DNA was harvested at 3 days posttransfection and 3D-PCR performed at 90°C. No DNA was recovered from any of the four cotransfections with pHBVΔcore. These negative findings were confirmed when 3D-PCR was applied to a small locus within the ampicillin resistance gene. Together these observations rule out rare deamination of nuclear HBV DNA in a transcription bubble or deamination of single-stranded DNA resulting from shearing during transfection. Hence, all sequences described here arose from reverse transcription of pregenomic HBV transcripts.
To ascertain the frequency of APOBEC3 editing, limiting dilution of the second-round PCR products (i.e., those at 90°C) showed that the signal titrated out at a frequency of ≈10-8 for the HBV/APOBEC3 cotransfection experiments (data not shown). By contrast limiting dilution of the first-round products (95°C) titrated out to ≈10-11. Given that there must be a little residual plasmid DNA in the samples as well as de novo replication of HBV from a few singly transfected cells, the proportion of edited HBV genomes is probably >10-3.
C → T Hypermutants. Among the APOBEC3 edited HBV sequences were some harboring multiple G → A and C → T substitutions, a selection being shown in Fig. 2B. Such sequences were observed for APOBEC3B, -3F, and -3G but not APOBEC3C and represented 7-19% of the total (Figs. 4-7). The simplest interpretation is that the HBV plus strand is also vulnerable to deamination. Because the plus strand of this segment harbors more C than G residues (36.6% vs. 27.2%), it was possible that lowering further the Td might allow amplification of C → T hypermutants alone. Accordingly, the same first-round PCR products, amplified at 95°C, were subjected to PCR at 88°C, as opposed to the 90°C used previously. Again, 3D-PCR products were recovered for the APOBEC3B, -3F, and -3G cotransfections but not for APOBEC3C. Upon cloning and sequencing, uniquely C → T hypermutants were identified, a selection being shown in Fig. 2C.
To rule out editing of HBV RNA that would also produce C → T transitions in the reference strand, RNA was purified from a fraction of the same cell pellets and purified by using an RNase-free DNase protocol. cDNA was made, and the same locus was amplified by PCR at both 95°C and 90°C. Ample DNA was recovered from all four APOBEC3 cotransfections and the HBV control, whereas none could be recovered at the lower temperature, ruling out RNA editing. The C → T hypermutants indicate that HBV plus-strand DNA can also be edited without prior editing of the minus strand, which would give rise to the mixed sequences seen in Fig. 2B.
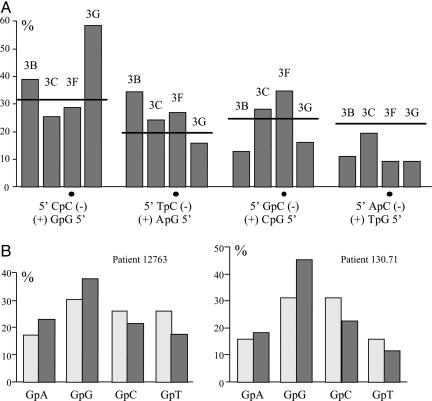
Dinucleotide Target Context of APOBEC3 Family Members. With a large number of hypermutated sequences, it was possible to compare the relative sequence context associated with APOBEC3B, -3C, -3F, and -3G cytidine deamination. As can be seen from Fig. 3A, all four molecules showed subtle differences. The most pronounced effect was for APOBEC3G, where there was a strong preference for CpC (target is italic). APOBEC3B preferred RpC (R is A,G) over YpC (Y is C,T), APOBEC3F slightly preferred TpC and GpC, whereas APOBEC3C showed no significant bias. Except for APOBEC3F these findings are in keeping with what has been described in the literature (7, 30-34). The same target site specificities were also observed among the G → A and C → T hypermutants, indicating that there was no asymmetry during editing of (+)- and (-)-strand DNA (data not shown).
Fig. 3.
Dinucleotide context associated with editing. (A) The ordinate represent the substitution frequency as a function of the 5′ nucleotide. Dots indicate the deaminated C residue. The expected value is represented by a horizontal bar and corresponds to the base composition of the locus. A χ2 analysis showed that the observed frequencies APOBEC3B, -3F, and -3G deviated significantly from the expected values (P < 0.001). (B) Dinucleotide context for G → A hypermutants derived from the serum of patients 12763 and 130.71, respectively. The observed frequencies (dark gray) encompass 481 and 505 transitions for the two patients, respectively. Expected frequencies are shown in light gray.
G → A Hypermutations in HBV-Infected Patients. Given the success of 3D-PCR at identifying G → A hypermutants, the technique was applied to DNA extracted from the serum of four chronically HBV-infected patients with high viremia (0.6-3 × 109 DNA copies per ml). Two of the four samples (130.71 and 12763) proved positive when 3D-PCR was performed at a Td = 90°C. The products were cloned and ≈20 clones sequenced. As can be seen from a small sample from each patient the sequences were extensively G → A hypermutated (Fig. 2 D and E; and see Figs. 8 and 9, which are published as supporting information on the PNAS web site). Interestingly, the extent of hypermutation was comparable to those obtained experimentally (Fig. 2 A).
The dinucleotide sequence context for both patients was CpC > TpC > GpC ≈ ApC (Fig. 3B), which is reminiscent of APOBEC3G (Fig. 3A), although this does not preclude a melange of APOBEC3 molecules. Close inspection of the sequences from patient 12763 showed a single G → A and C → T hypermutant (Fig. 2E, bottom sequence) similar to those shown in Fig. 2B. Although the substitutions were mainly G → A, there were five C → T transitions, four of which occurred in CpC and TpC dinucleotides, typical of APOBEC3B, -3F, and -3G target motifs. Because these sites were not polymorphic in 10 nonhypermutated sequences from this patient, it is probable that these C → T transitions resulted from cytidine deamination of plus-strand HBV DNA.
An outstanding question was at what frequency these highly hypermutated genomes were occurring in vivo. PCR material from the first-round amplification at 95°C were serially diluted up to 106-fold and subjected to 3D-PCR at Td = 90°C as described above. For patients 12763 and 130.71 the last positive dilutions were 10-4 and 5 × 10-4, respectively. Given the viral loads for these patients, these numbers translate to ≈105 and 1.5 × 106 heavily hypermutated HBV genomes per ml of serum.
Discussion
The APOBEC3 family of cytidine deaminases has recently been shown to constitute a powerful intracellular barrier to the replication of classical retroviruses, particularly the lentiviruses of which HIV-1 is the most infamous (1-5, 10). The present experimental findings show that gene products from four of the seven APOBEC3 genes can extensively edit both plus and minus HBV DNA strands. Along with the in vivo findings it is clear that pregenomic viral RNA can indeed be packaged and ultimately secreted into blood in the presence of APOBEC3 expression. The absence or scarcity of G → A hypermutated sequences that lead to the suggestion of a nonediting role for APOBEC3G restriction of HBV replication in the earlier studies (20-22) may reflect remaining plasmid DNA or productive growth from a small fraction of cells transfected by pCayw alone. Other variables could include negative selection due to the use of Pfu polymerase and amplification primers with one or several G residues at the 3′ end.
Restriction of HBV replication via deamination of nascent cDNA can be envisaged without evoking a packaging defect as was suggested (20-22). An essential step in the HBV life cycle is the return of capsids harboring neo-synthesized DNA genomes to the nucleus (35), resulting in a 10- to 50-fold amplification of nuclear HBV DNA. Any genetic editing by APOBEC3 molecules would probably result in defective genomes, particularly given its compact organization. On an experimental background of APOBEC3 expression, transcription will proceed from the original pCayw plasmid, but progeny genomes returning to the nucleus will be overwhelmingly defective. If normalized to the pCayw control alone it will appear that there is less HBV RNA and DNA in the cytosol leading to the notion of a packaging defect. What is actually happening is that intracellular amplification of HBV is strongly curtailed.
HBV departs from the HIV paradigm in that four APOBEC3 molecules, 3B, 3C, 3F and 3G, can massively edit HBV DNA. Furthermore, this occurs in the context of a complete infectious molecular clone, suggesting that HBV does not encode a gene product that can neutralize APOBEC3 function. By contrast, HIV cDNA is edited by only two APOBEC3 molecules, 3F and 3G, and then on a Δvif background (1-5, 7, 30-35). Because numerous papers suggest that there are active HIV packaging requirements involving RNA and capsid elements (36-39), the relative promiscuity of HBV capsid assembly to the inclusion of four distinct APOBEC3 molecules suggests, but does not prove, that there may be no specific requirements beyond, perhaps, RNA. Although the HBcAg capsid is fenestrated, the pore sizes are too small to allow entry of APOBEC3 molecules (40), it follows that APOBEC3 molecules must enter during capsid assembly.
Large numbers of C → T hypermutants and mixed G → A + C → T hypermutants represent another departure from the HIV paradigm. To date, only one HIV-1 C → T hypermutant has been reported (41). In the experimental context, APOBEC3F generated as many C → T hypermutated genomes as “conventional” G → A hypermutated genomes. Albeit a single example, the mixed G → A + C → T hypermutant derived from patient 12763 serum (Fig. 2E) indicates that these experimental observations are not irrelevant. The locus analyzed maps to the 3′ end of the HBV plus strand. It is possible that the two strands may breathe somewhat, allowing deamination of plus-strand DNA.
With the identification of extensively hypermutated genomes from two of four chronically HBV-infected patients, it is clear that the dearth of hypermutated HBV genomes to date is probably related to their relatively low frequency in serum, here ≈10-4. As HBV virions in serum are turning over rapidly with a half-life of ≈24 h (42), the probability of scoring hundreds of thousands of hypermutants in two of four randomly chosen samples, albeit with high viremia (0.6-3 × 109 DNA copies per ml) must be exceedingly small unless a fraction of cells actively supporting HBV replication are also expressing APOBEC3 molecules on a daily basis. Because the proportion of G → A hypermutants in vivo is very low, this indicates that HBV essentially replicates in hepatocytes where APOBEC3 molecules are not expressed.
Assuming that the proportion of productively infected cells supporting transcription of one or more APOBEC3 genes is proportional to the fraction of hypermutated genomes in serum (≈10-4), then it is a small proportion indeed and unlikely to have much impact on viral transmission and pathology. Because the degree of G → A hypermutation was comparable for naturally occurring genomes as well as those generated in vitro, it is likely that the levels of APOBEC3 gene expression must be rather elevated in this small proportion of cells in vivo because the CMV-IE promoter is considered to be among the most powerful when used in transfection studies. It is not possible to comment on the nature of this small subset of cells expressing APOBEC3G and/or -3B based on the dinucleotide context of the natural hypermutants (Fig. 3B). It could reflect clonal variation among hepatocytes, or another cell type bearing the receptor for the virus.
Might APOBEC3 editing have any long-term impact on the HBV genome? The major argument against is the small fraction of infected cells in which APOBEC3 molecules are expressed, ≈10-4 for the moment. In addition, HBV does not have an A-rich genome like the lentiviruses (HBV 23% A and lentiviruses 32-37% A on the plus strand) while third base codon usage in the nonoverlapping regions is not A-rich, as is the case for HIV (43). It was noted that the in-phase stop codon barring expression of the PreC form of HBcAg is in a sequence vulnerable to APOBEC3G editing, notably 5′-GCCCCA on minus-strand DNA (20). A stop codon in the plus strand arises if the underlined cytidine residue is deaminated. Given the hundreds of cytidine residues in the HBV genome vulnerable to APOBEC3 editing in vivo (see Fig. 2 D and E and ref. 18), it is unlikely that a single C will be deaminated. A simpler scenario would be selection from a mutant spectrum generated by error-prone DNA polymerization where the genomic mutation rate is much less than one per genome per cycle.
Albeit theoretical, the exquisite sensitivity of HBV replication to cytidine deamination, coupled with the very compact HBV genome organization and unusually low mutation fixation rate, suggests that any means to up-regulate any of the APOBEC3B, -3C, -3F, or -3G genes might result in enhanced control of viral replication.
Note Added in Proof. Using a novel PCR method, Noguchi et al. (44) have described G → A hypermutations in numerous serum samples.
Supplementary Material
Acknowledgments
We thank Drs. Nicole Chenciner, Marie-Louise Michel, and Frank Chisari for making their plasmids readily available and Valérie Thiers for the four sera. This work was supported by grants from the Pasteur Institute. R.S. is a recipient of a Boehringer Ingelheim Fonds Fellowship. P.S. is supported by a SIDACTION fellowship.
Author contributions: P.S., S.W.-H., and J.-P.V. designed research; R.S., D.G., and M.H. performed research; P.S., S.W.-H., and J.-P.V. analyzed data; and P.S., S.W.-H., and J.-P.V. wrote the paper.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: HBV, hepatitis B virus.
References
- 1.Lecossier, D., Bouchonnet, F., Clavel, F. & Hance, A. J. (2003) Science 300, 1112. [DOI] [PubMed] [Google Scholar]
- 2.Zhang, H., Yang, B., Pomerantz, R. J., Zhang, C., Arunachalam, S. C. & Gao, L. (2003) Nature 424, 94-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mangeat, B., Turelli, P., Caron, G., Friedli, M., Perrin, L. & Trono, D. (2003) Nature 424, 99-103. [DOI] [PubMed] [Google Scholar]
- 4.Harris, R. S., Bishop, K. N., Sheehy, A. M., Craig, H. M., Petersen-Mahrt, S. K., Watt, I. N., Neuberger, M. S. & Malim, M. H. (2003) Cell 113, 803-809. [DOI] [PubMed] [Google Scholar]
- 5.Mariani, R., Chen, D., Schrofelbauer, B., Navarro, F., Konig, R., Bollman, B., Munk, C., Nymark-McMahon, H. & Landau, N. R. (2003) Cell 114, 21-31. [DOI] [PubMed] [Google Scholar]
- 6.Yu, Q., Konig, R., Pillai, S., Chiles, K., Kearney, M., Palmer, S., Richman, D., Coffin, J. M. & Landau, N. R. (2004) Nat. Struct. Mol. Biol. 11, 435-442. [DOI] [PubMed] [Google Scholar]
- 7.Suspène, R., Sommer, P., Henry, M., Ferris, S., Guetard, D., Pochet, S., Chester, A., Navaratnam, N., Wain-Hobson, S. & Vartanian, J. P. (2004) Nucleic Acids Res. 32, 2421-2429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jarmuz, A., Chester, A., Bayliss, J., Gisbourne, J., Dunham, I., Scott, J. & Navaratnam, N. (2002) Genomics 79, 285-296. [DOI] [PubMed] [Google Scholar]
- 9.Conticello, S. G., Thomas, C. J., Petersen-Mahrt, S. K. & Neuberger, M. S. (2005) Mol. Biol. Evol. 22, 367-377. [DOI] [PubMed] [Google Scholar]
- 10.Sheehy, A. M., Gaddis, N. C., Choi, J. D. & Malim, M. H. (2002) Nature 418, 646-650. [DOI] [PubMed] [Google Scholar]
- 11.Sheehy, A. M., Gaddis, N. C. & Malim, M. H. (2003) Nat. Med. 9, 1404-1407. [DOI] [PubMed] [Google Scholar]
- 12.Marin, M., Rose, K. M., Kozak, S. L. & Kabat, D. (2003) Nat. Med. 9, 1398-1403. [DOI] [PubMed] [Google Scholar]
- 13.Yu, X., Yu, Y., Liu, B., Luo, K., Kong, W., Mao, P. & Yu, X. F. (2003) Science 302, 1056-1060. [DOI] [PubMed] [Google Scholar]
- 14.Conticello, S. G., Harris, R. S. & Neuberger, M. S. (2003) Curr. Biol. 13, 2009-2013. [DOI] [PubMed] [Google Scholar]
- 15.Wain-Hobson, S., Sonigo, P., Guyader, M., Gazit, A. & Henry, M. (1995) Virology 209, 297-303. [DOI] [PubMed] [Google Scholar]
- 16.Vartanian, J. P., Henry, M. & Wain-Hobson, S. (2002) J. Gen. Virol. 83, 801-805. [DOI] [PubMed] [Google Scholar]
- 17.Janini, M., Rogers, M., Birx, D. R. & McCutchan, F. E. (2001) J. Virol. 75, 7973-7986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Günther, S., Sommer, G., Plikat, U., Iwanska, A., Wain-Hobson, S., Will, H. & Meyerhans, A. (1997) Virology 235, 104-108. [DOI] [PubMed] [Google Scholar]
- 19.Wieland, S., Thimme, R., Purcell, R. H. & Chisari, F. V. (2004) Proc. Natl. Acad. Sci. USA 101, 6669-6674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Turelli, P., Mangeat, B., Jost, S., Vianin, S. & Trono, D. (2004) Science 303, 1829. [DOI] [PubMed] [Google Scholar]
- 21.Turelli, P., Jost, S., Mangeat, B. & Trono, D. (2004) Science 305, 1403. [DOI] [PubMed] [Google Scholar]
- 22.Rösler, C., Köck, J., Malim, M. H., Blum, H. E. & von Weizsäcker, F. (2004) Science 305, 1403. [DOI] [PubMed] [Google Scholar]
- 23.Suspène, R., Henry, M., Guillot, S., Wain-Hobson, S. & Vartanian, J. P. (2005) J. Gen. Virol. 86, 125-129. [DOI] [PubMed] [Google Scholar]
- 24.Scaglioni, P. P., Melegari, M. & Wands, J. R. (1997) J. Virol. 71, 345-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chenciner, N., Delpeyroux, F., Israel, N., Lambert, M., Lim, A., Streeck, R. E. & Houssais, J. F. (1990) Biotechnology 8, 858-862. [DOI] [PubMed] [Google Scholar]
- 26.Sells, M. A., Chen, M. L. & Acs, G. (1987) Proc. Natl. Acad. Sci. USA 84, 1005-1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beames, B. & Lanford, R. E. (1995) J. Virol. 69, 6833-6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Greagg, M. A., Fogg, M. J., Panayotou, G., Evans, S. J., Connolly, B. A. & Pearl, L. H. (1999) Proc. Natl. Acad. Sci. USA 96, 9045-9050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shuttleworth, G., Fogg, M. J., Kurpiewski, M. R., Jen-Jacobson, L. & Connolly, B. A. (2004) J. Mol. Biol. 337, 621-634. [DOI] [PubMed] [Google Scholar]
- 30.Beale, R. C., Petersen-Mahrt, S. K., Watt, I. N., Harris, R. S., Rada, C. & Neuberger, M. S. (2004) J. Mol. Biol. 337, 585-596. [DOI] [PubMed] [Google Scholar]
- 31.Zheng, Y. H., Irwin, D., Kurosu, T., Tokunaga, K., Sata, T. & Peterlin, B. M. (2004) J. Virol. 78, 6073-6076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wiegand, H. L., Doehle, B. P., Bogerd, H. P. & Cullen, B. R. (2004) EMBO J. 23, 2451-2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liddament, M. T., Brown, W. L., Schumacher, A. J. & Harris, R. S. (2004) Curr. Biol. 14, 1385-1391. [DOI] [PubMed] [Google Scholar]
- 34.Bishop, K. N., Holmes, R. K., Sheehy, A. M., Davidson, N. O., Cho, S. J. & Malim, M. H. (2004) Curr. Biol. 14, 1392-1396. [DOI] [PubMed] [Google Scholar]
- 35.Tuttleman, J. S., Pourcel, C. & Summers, J. (1986) Cell 47, 451-460. [DOI] [PubMed] [Google Scholar]
- 36.Alce, T. M. & Popik, W. (2004) J. Biol. Chem. 279, 34083-34086. [DOI] [PubMed] [Google Scholar]
- 37.Svarovskaia, E. S., Xu, H., Mbisa, J. L., Barr, R., Gorelick, R. J., Ono, A., Freed, E. O., Hu, W. S. & Pathak, V. K. (2004) J. Biol. Chem. 279, 35822-35828. [DOI] [PubMed] [Google Scholar]
- 38.Luo, K., Liu, B., Xiao, Z., Yu, Y., Yu, X., Gorelick, R. & Yu, X. F. (2004) J. Virol. 78, 11841-11852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schafer, A., Bogerd, H. P. & Cullen, B. R. (2004) Virology 328, 163-168. [DOI] [PubMed] [Google Scholar]
- 40.Wynne, S. A., Crowther, R. A. & Leslie, A. G. (1999) Mol. Cell 3, 771-780. [DOI] [PubMed] [Google Scholar]
- 41.Vartanian, J. P., Meyerhans, A., Asjo, B. & Wain-Hobson, S. (1991) J. Virol. 65, 1779-1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nowak, M. A., Bonhoeffer, S., Hill, A. M., Boehme, R., Thomas, H. C. & McDade, H. (1996) Proc. Natl. Acad. Sci. USA 93, 4398-4402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bronson, E. C. & Anderson, J. N. (1994) J. Mol. Evol. 38, 506-532. [DOI] [PubMed] [Google Scholar]
- 44.Noguchi, C., Ishino, H., Tsuge, M., Fujimoto, Y., Imamura, M., Takahashi, S. & Chayama, K. (2005) Hepatology 41, 626-633. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.