Abstract
The tempo and mode of evolution of the extinct giant moas of New Zealand remain obscure because the number of lineages and their divergence times cannot be estimated reliably by using fossil bone characters only. We therefore extracted ancient DNA from 125 specimens and genetically typed them for a 658-bp mtDNA control region sequence. The sequences detected 14 monophyletic lineages, 9 of which correspond to currently recognized species. One of the newly detected lineages was a genetically divergent form of Megalapteryx originally described as a separate species, two more were lineages of Pachyornis in southern and northeastern New Zealand, and two were basal lineages of South Island Dinornis. When results from genetic typing and previous molecular sexing were combined, at least 33.6% of the specimens were incorrectly classified. We used longer sequences of the control region and nine other mtDNA genes totaling 2,814 base pairs to derive a strongly supported phylogeny of the 14 moa lineages. Molecular dating estimated the most recent common ancestor of moas existed after the Oligocene drowning of New Zealand. However, a cycle of lineage-splitting occurred ≈4–10 million years ago, when the landmass was fragmented by tectonic and mountain-building events and general cooling of the climate. These events resulted in the geographic isolation of lineages and ecological specialization. The spectacular radiation of moa lineages involved significant changes in body size, shape, and mass and provides another example of the general influence of large-scale paleoenvironmental changes on vertebrate evolutionary history.
Keywords: adaptive radiation, extinct moas, environmental changes
Geologically young island archipelagos such as the Galápagos Islands and Hawaii are renowned for their relatively recent passerine adaptive radiations, from which the classical allopatric mode and tempo of evolution have been inferred. In contrast, the radiation of the New Zealand moa could potentially be much more ancient, because the landmass broke from Antarctica/Australia ≈80 million years ago (mya). An analysis of lineage diversification in the flightless moa may reveal a contrasting picture to that provided by the classic examples, with the possibility of deep cladogenesis as well as more recent lineage-splitting in different geographic regions.
From both a morphological and ecological perspective, moa represent an ideal group to investigate the tempo and mode of evolution. First, ratites are thought to be basal in birds, and moa represent an ancient ratite group. Second, moa are remarkable because, in addition to the changes in bill morphology, they also underwent substantial changes in body size and proportions (1–3). Third, it has been argued that these changes were adaptive responses to the utilization of different habitats within the New Zealand landscape (4). Fourth, in previous studies we showed that single-copy nuclear genes (3), in addition to mtDNA sequences of moa (3, 5, 6), can be regularly amplified from moa subfossil bones. Finally, the large number of moa subfossil remains provides the necessary material for a large-scale study of this now-extinct group.
Although of general interest in evolutionary biology, the moa radiation is less well understood than some more renowned passerine examples, perhaps because these birds were extinct ≈100 years after human colonization of New Zealand in about A.D. 1300 (7). Consequently, they are known largely from the remains recovered from caves, swamps, and middens (sites of discarded human food). Additionally, the complex nature of morphological variation has increased the difficulty of distinguishing within-species variation from species-level differences, and thus moa taxonomy has been notoriously unstable. At least 64 species and 20 genera of moa have been named. The number of species was reduced in successive taxonomic treatments to 20 (8), 29 (9), and 13 species (1). Recent systematic revisions (cited in ref. 10) list 11 species in six genera (Megalapteryx, Dinornis, Pachyornis, Emeus, Euryapteryx, and Anomalopteryx), but at least one of these previously described species is invalid because it harbors a mixture of large females and small males from two species (3, 11). Whatever the precise number of species was, moa were clearly a speciose group and outnumbered the other ratite groups in species diversity.
To investigate the tempo and mode of evolution in this extinct clade of birds, we used ancient mtDNA control region sequences to genetically type a large number of subfossil bones from museum collections in New Zealand. Using exemplars to represent the resulting control region lineages, we subsequently obtained partial sequences from nine additional mtDNA genes. These concatenated sequences were used to construct a detailed phylogeny of moa, together with appropriate outgroups. Dates of divergence were inferred from the well supported molecular phylogeny, thus providing the first estimates of the timing of the moa radiation. Finally, we examined the tempo and mode of moa evolution in relation to large-scale paleoenvironmental events, the changing dynamics of the New Zealand landmass, and the distribution of moa lineages through time.
Materials and Methods
DNA Extraction, PCR, and Sequencing. Because previous studies have relied on specimens, including many known only from isolated bones, incomplete specimens, or possibly mosaics of one or more species, we confined our DNA extractions to bone samples catalogued as being from different specimens in New Zealand museum collections (see Table 2, which is published as supporting information on the PNAS web site). From over 230 samples kindly made available to us, we were successful in obtaining amplifiable DNA from 125 individuals. These samples represent all currently known moa taxa. DNA was amplified and sequenced in laboratories in both Canada (Royal Ontario Museum) and New Zealand (Allan Wilson Centre for Molecular Ecology and Evolution, Massey University). For a listing of the moa samples used, the regions amplified, and the laboratory in which each sample was analyzed, see Table 2. Details of the methods and precautions for DNA extraction, amplification, and sequencing used at the Allan Wilson Centre are essentially as outlined in the supplemental material in refs. 3 and 12. Briefly, DNA was extracted from ≈0.2 g of moa bone shavings in 0.5 M EDTA with proteinase K. Samples were purified by phenol/chloroform extraction and then concentrated by using Vivaspin retention columns (Vivascience, U.K.). Extracted DNA was amplified for 45 cycles with Platinum Taq (Invitrogen) by using the following cycling parameters: 94°C for 2 min, then 10 cycles of 94°C for 20 s, 54°C for 20 s, and 72°C for 20 s, followed by 35 cycles of 94° for 20 s, 50°C for 20 s, and 72°C for 20 s. The primers used, their annealing temperatures, and subsequent length of the PCR products are outlined in Table 3, which is published as supporting information on the PNAS web site. Sequences were obtained from both strands by using independent amplifications. DNA extraction, amplification, and sequence analysis were repeated for 16 samples (identified in Table 2) at a dedicated ancient DNA laboratory at the University of Auckland. Each of these replicates resulted in identical sequences.
For the samples analyzed at the Royal Ontario Museum, core samples of bone between 0.1 and 0.5 g were taken from lower limb bones by using sterilized 7-mm diamond glass drills. The resulting bone cores had their outer surfaces removed (to a depth of 1–2 mm) by using sterilized aluminium oxide powder propelled from an Airbrasive system (MicroBlaster; Comco, Burbank, CA). All moa DNA extractions were performed in a dedicated ancient biomolecules facility. The bones were then pulverized in a sterile grinder, and the DNA was extracted by using the protocol described in ref. 13. The extracted DNA samples were amplified by using the following PCR amplification profile: 94°C for 45 s, 50°C for 45 s, and 72°C for 90 s. A total of 48 cycles were routinely used, and for samples that had low product yield, the amplifications were repeated from the original DNA isolation. The amplified products were separated by gel electrophoresis and purified by using filter pipet-tip centrifugation. The products were then sequenced by using Thermo-Sequenase (GE Healthcare). DNA sequences were entered manually into a computer by using the program xesee (14). To ensure that the sequences were of mitochondrial origin, all of the autoradiographs produced were examined for evidence of multiple bands indicating more than one sequence. The sequences for the protein-coding genes were translated to confirm that the reading-frames were consistent with amino acid sequences. More specifically, variation across the three codon positions was examined to ensure it followed the expected pattern of decreasing variation from third to first to second positions, which is a by-product of the degeneracy of the genetic code. For RNA-coding genes, the sequences were mapped onto secondary structure models and checked for any substitutions that would be incompatible with the predicted structures. Identical sequences were obtained from specimens (see Table 2) sequenced in both laboratories.
Phylogenetic Analyses. Bayesian inference of phylogeny with Markov chain Monte Carlo sampling was conducted separately for the 125 control region sequences (658 base pairs) and for concatenated sequences of the control region and nine mtDNA genes (ATPase 6 and 8, cyt b, ND3, ND4, ND5, COIII, 12S rRNA, and tRNALys totalling 2,814 base pairs) from 40 exemplars of the major moa lineages. To check the location of the root of the tree, we constructed another phylogeny by using only the latter nine genes, because the control region of moa could not be aligned to that of outgroup ratite and tinamou taxa (Struthio camelus, Casuarius casuarius, Dromaius novaehollandiae, Apteryx haasti, and Eudromia elegans). Analyses were conducted with mrbayes 3.0b4 (15), and in the exemplar data set, each gene was partitioned and assigned its own best-fitting model of evolution, as determined with modeltest 3.06 (16). For both data sets, one cold chain and three heated chains were run simultaneously for 2 million generations, and 1 tree per 1,000 was sampled. The first 100 trees were discarded as burn-in, and Bayesian posterior probabilities were estimated on the 50% majority rule consensus of the remaining 1,900 trees. Analyses were repeated four times and recovered the same topology. Trees were also constructed by using maximum likelihood in phyml (17) and maximum parsimony in paup (18), and bootstrap support at the nodes was computed for 100 replicates of the data. A general time-reversible model of DNA substitution, proportion of invariable sites, and shape parameter of the gamma distribution (GTR + I + G) model was used in maximum likelihood, and the parameters were estimated in phyml. Maximum parsimony was performed with a heuristic search and 10 random taxon additions, with the tree-bisection–reconnection branch-swapping algorithm.
Divergence Time Estimates. Estimates of divergence times were obtained by using penalized likelihood rate-smoothing with the truncated Newton algorithm in the program r8s 1.60 (19). Unfortunately, fossils that might serve as phylogenetically constrained anchor points (20) for dating nodes in the moa tree have not yet been discovered, mainly because of the lack of tertiary exposures in New Zealand (10). To obtain an estimate of the age of the moa radiation, an external geological calibration of 82 million years (myr) was therefore applied to the split between moa and outgroup ratite and tinamou taxa, corresponding to the time when New Zealand separated from Gondwana (6). The appropriate smoothing value of 1.0 was calculated by using cross-validation (19). Because the control region could not be aligned reliably to that of the outgroup taxa, this first analysis was restricted to the tree obtained from sequences of the mitochondrial protein-coding, 12S rRNA, and tRNALys genes. The resultant estimate of 18.5 myr was then fixed at the base of the moa radiation (node a in Fig. 1) in the larger control region tree of 125 specimens. Divergence times and their 95% confidence intervals (CI) at key nodes were estimated with a smoothing value of 0.1 calculated with cross-validation (19). The root of the control region tree was located on the branch leading to Megalapteryx as determined by outgroups in the multiple gene tree (see Fig 3, which is published as supporting information on the PNAS web site).
Fig. 1.
Bayesian tree constructed by using 658-bp control region sequences from 125 moa specimens under a GTR + I + G model of evolution. The tree is shown as a chronogram used in molecular dating by the program r8s. Numbers at the branch tips identify the 14 major lineages as follows: 1, P. mappini, eastern North Island; 2, P. mappini, western North Island; 3, P. elephantopus, Canterbury and Otago; 4, P. australis;5, P. elephantopus, Southland; 6, Euryapteryx geranoides; 7, Emeus crassus; 8, Anomalopteryx didiformis; 9, Dinornis robustus; 10, Dinornis robustus, northwest Nelson; 11, Dinornis robustus, Otago; 12, Dinornis novaezealandiae; 13, Megalapteryx didinus; and 14, M. benhami?. Specimens are color-coded according to geographic locations plotted together with place names. Major mountain ranges are represented by peaks. Unfilled bars at the branch tips indicate specimens without locality data. Asterisks at the nodes indicate posterior probabilities (above the nodes) or maximum likelihood bootstrap values (below the nodes) of 1.0 and 100%, respectively. Letters at nodes refer to divergence times among lineages listed in Table 1. (Lower Insets) Extent of the New Zealand landmass and movement of tectonic plates from 25 mya to present (44). Faults, subduction zones, and seafloor spreading centers are shown in red.
Results and Discussion
Genetic Typing with Control Region Sequences. Building on earlier ancient DNA studies (3, 5, 6), we combined data sets from our laboratories to create a 658-bp alignment of the central conserved domain and a short segment of the flanking 5′ terminus of the control region. This region was chosen because previous studies (3, 5, 6) suggested that this would provide an appropriate level of phylogenetic resolution, given its faster rate of molecular evolution than that found in other mtDNA genes (12). The Bayesian partitioned likelihood tree for the control region sequences of moa (Fig. 1) revealed 14 major phylogenetic lineages. All but Euryapteryx and one of the two new basal lineages in the South Island Dinornis clade had posterior clade probabilities of >0.9. Two genetically divergent lineages of Megalapteryx were identified at the base of the tree. These lineages correspond to Megalapteryx didinus and Megalapteryx benhami of earlier classifications, thus questioning their recent synonomy (21). In addition to the clades representing the remaining accepted genera (Pachyornis, Anomalopteryx, Emeus, Euryapteryx, and Dinornis), we detected two divergent lineages within Pachyornis mappini (clades 1 and 2), a unique lineage (clade 5) that suggests Pachyornis elephantopus is a paraphyletic taxon, and three distinctive lineages (clades 9, 10, and 11) within South Island Dinornis.
There also is evidence from the control region sequences for geographic structuring within existing morphologically defined species. The most obvious splits are between populations in the North and South Islands, as in Anomalopteryx didiformis, implying recent isolation. In Euryapteryx geranoides, the birds from Otago and South Canterbury are grouped together (posterior probability of 1.0), birds from the Pyramid Valley area in North Canterbury are separated in another clade (posterior probability of 0.61), and most of the birds from the far north of the North Island are separated into yet another clade (posterior probability of 0.58). The latter were previously assigned to Euryapteryx curtus, but one specimen from this clade exhibited a haplotype typical of birds in the South Island. The distinctiveness of the two clades at opposite ends of the two main islands suggests that isolation-by-distance promoted differentiation, and that lineage sorting is incomplete. Two separate clades are present within the smallest species, P. mappini, which is restricted to the North Island. Birds from the East Coast region, including Hawkes Bay, are in one clade, whereas birds from the northwest of the North Island are restricted to another clade, and the only geographic overlap was found in the west at Whanganui, where both haplotypes occurred (Fig. 1).
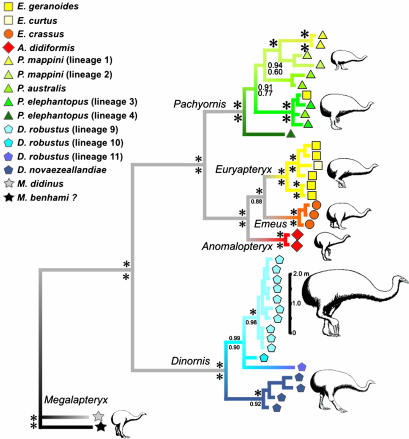
Multiple Gene Support for Lineages. To check support for the phylogenetic relationships of the 14 major lineages of moa detected with the control region sequences, we combined them with sequences from nine mtDNA protein-coding genes and the 12S rRNA and tRNALys genes for exemplars of each lineage, because longer DNA sequences have been shown to have a higher probability of recovering the topology of known trees (22–25). The concatenated data set of 2,814 base pairs recovered a Bayesian partitioned likelihood tree identical in topology to the control region tree. Relationships among lineages were supported with higher posterior probabilities, consistent with the increased signal in these longer sequences (Fig. 2). The same tree was recovered in maximum likelihood and maximum parsimony, and major nodes had strong bootstrap support. Genetic distances ranged from 18.45% between P. mappini and the most divergent new Dinornis lineage to 1.70% between Dinornis robustus and the other new Dinornis lineage (see Table 4, which is published as supporting information on the PNAS web site).
Fig. 2.
Bayesian tree constructed under a GTR + I + G model of evolution with 2,814 base pairs of mtDNA from 40 moa specimens, including exemplars of all 14 lineages detected in Fig. 1. Symbols at the branch tips indicate that even in this selected data set, bones from three specimens were misidentified, and one was placed in the incorrect genus. Asterisks at the nodes indicate posterior probabilities (above the nodes) or maximum likelihood bootstrap values (below the nodes) of 1.0 and 100%, respectively.
In contrast, a previous cladistic analysis of 82 osteological characters of the 11 then-recognized species supported the monophyly of moas but failed to fully resolve the branching order of the genera and relationships among species of Pachyornis (10). Additionally, the lineages of Megalapteryx (4.47% sequence divergence) were not recognized because they had been synonymized based on similarity in bone shape, despite marked differences in size (21). The difficulty in assigning bones unequivocally to taxonomic groups is shown by the phylogenetic tree of control region sequences. Bones from six specimens were assigned to the incorrect genus. Additionally, 62 more have been incorrectly synonymized in the past or are from unrecognized lineages or were shown to be sexual morphs that necessitated classification changes for a total morphological classification error of 33.6%. If the four new lineages are shown eventually to warrant taxonomic recognition, the error would rise to 54.4%. Ancient DNA shows that unless sequences are recovered from almost all specimens, the identity and number of lineages cannot be established within even a relatively small clade of subfossil birds like moa.
Molecular Dating. Estimates of divergence times allow reconstruction of the historical biogeography of these lineages and inferences about vicariance, dispersal, and geographic isolation events that might have promoted divergence. One of the principal problems in dating divergence times in species generally is whether to use a previously calibrated phylogenetic rate based on mtDNA genomes of ratites (5–6), a faster rate of mutation calibrated on pedigrees (26), or an even faster evolutionary rate calculated on serial samples from recent time depths (3). However, there is likely to be a large discrepancy between the rate of fixation (ka) in the ancestral lineage of moas and the rate of mutation (μ) of mtDNA (26). We decided to use phylogenetic rates that we calibrated with fossils and earth history events to date splits in the ratite tree, because these are likely to represent older divergence events (5, 6, 27). A recent attempt (11) to date the split between Dinornis robustus and Dinornis novaezelandiae used a phylogenetic rate (28) calibrated for the 5′ hypervariable part of the control region of the snow goose (Anser caerulescens). This is the fastest rate known for flying birds and thus may not be appropriate for the more slowly evolving ratites.
The current genera of moa are represented in the fossil record of New Zealand 1.8–2.4 mya (6), and phylogenetic rates based on mtDNA genome sequences dated the Emeus–Dinornis split at 13.2 mya (95% CI, 11.9–14.6) (6) and the Emeus–Anomalopteryx split at 5.3 mya (95% CI, 3.7–6.9) (5). We used the concatenated protein-coding, 12S rRNA, and tRNA Lys sequences and the geological split between New Zealand and Australia/Antarctica, estimated at 82 mya (29, 30), to calibrate the divergence of moas from outgroup ratite taxa. Penalized likelihood rate-smoothing dated the moa radiation at ≈18.5 mya (95% CI; 15.1–23.2), possibly a result of a population bottleneck in the Oligocene period (31). The two divergent lineages of Megalapteryx diverged about 12.3 mya (95% CI, 7.2–17.9) when the continuing uplift of the Southern Alps would have isolated them on opposite sides of the mountains (32).
Because the slow rate of evolution in these genes prevented reasonably accurate estimation of divergence times nearer the tips of clades, we also dated the splits in the gene tree constructed from the large collection of faster-evolving control region sequences (Table 1). Based on corrected GTR + I + G-corrected distances selected with Akaike's information criterion (16), the conserved domain of the control region had an estimated rate of evolution that ranged from 0.49%/myr between Emeus crassus and Euryapteryx geranoides to 0.54%/myr between Anomalopteryx didiformis and Dinornis novaezealandiae. These estimates are in general accordance with the slow phylogenetic rate of evolution in mtDNA genes in kiwi (33).
Table 1. Divergence times and 95% CIs of moa lineages estimated with partial sequences of nine mtDNA protein-coding and 12S rRNA and tRNALys genes aligned to outgroup ratite sequences and separately with control region sequences.
| rRNA, tRNA, and protein-coding genes
|
Control region
|
||||
|---|---|---|---|---|---|
| Taxa | Node | Date, mya | 95% CI | Date, mya | 95% CI |
| Moa root | a | 18.5 | 15.1, 23.2 | — | — |
| Megalapteryx species | b | 12.3 | 7.2, 17.9 | — | — |
| Dinornis vs. others | c | 15.0 | 14.5, 15.6 | ||
| Pachyornis vs. Anomalopteryx | d | — | — | 9.7 | 8.9, 10.5 |
| NI vs. SI Dinornis | e | 8.0 | 7.1, 8.9 | ||
| Anomalopteryx vs. Emeus/Euryapteryx | f | — | — | 5.8 | 5.1, 6.5 |
| Pachyornis species | g | — | — | 6.5 | 5.7, 7.3 |
| D. robustus lineages | h | 5.1 | 4.2, 6.0 | ||
| Emeus vs. Euryapteryx | i | — | — | 4.3 | 3.8, 5.0 |
| P. mappini lineages | j | — | — | 4.3 | 3.7, 5.0 |
| P. australis vs. P. elephantopus | k | — | — | 4.4 | 3.8, 5.2 |
| NI. vs. SI Anomalopteryx | l | — | — | 2.8 | 2.2, 3.5 |
| East/west NI D. novaezealandiae | m | — | — | 1.9 | 1.6, 2.3 |
| Euryapteryx populations | n | — | — | 1.6 | 1.3, 1.9 |
| D. robustus populations | o | — | — | 1.7 | 1.3, 2.3 |
| P. elephantopus populations | p | — | — | 1.4 | 1.2, 1.7 |
| P. mappini populations | q | — | — | 1.7 | 1.4, 2.1 |
| P. lineage 2 populations | r | — | — | 0.9 | 0.7, 1.1 |
Moa Diversification and Paleoenvironmental Events. The additional striking perspective evident from the control region tree (Fig. 1) is that much of the radiation of moa evolved within the last 10 myr during the marked global cooling at the end of the Miocene (Table 1). All of the recognized genera other than the upland Megalapteryx diverged in this period, as did the two species of Dinornis and three recognized species of Pachyornis. Around this time, the expanding New Zealand landmass was being reshaped by mountain-building events and was sundered into North and South Islands by the opening of Cook Strait, ≈5 mya, as the plates rotated and separated (Fig. 1). Geographic isolation in the two islands appears sufficient to explain speciation in Dinornis robustus and Dinornis novaezealandiae (8 mya), as well as the divergence of North Island P. mappini from South Island Pachyornis australis and P. elephantopus (6.5 mya). Geographic isolation and ecological specialization within each island also seems to have promoted speciation or phylogeographic structuring within species. The disjunct distribution of the small-statured P. australis isolated by mountains in northwest Nelson from the large-bodied P. elephantopus in the eastern grasslands of Canterbury and Otago (Fig. 1) implies allopatric speciation (4.4 mya). Much of the recent diversification (Table 1) also appears to have occurred by geographic fragmentation within each major island. For example, we detected two lineages each of P. mappini and Dinornis novaezealandiae in the west and east of the North Island (divergence times of 1.7 and 1.9 mya, respectively). There also is a geographic split estimated to have occurred 1.7 mya between the west coast Dinornis lineage on the opposite side of the Southern Alps from the remaining South Island lineages. Additionally, the split between a new P. elephantopus lineage in Southland and central Otago from the lineage in Canterbury and north Otago was estimated to be 1.4 mya. All of these more recent divergences are probably attributable to the fragmentation of the New Zealand landmass by Pleistocene cycles of glaciation. Finally, the estimated time of divergence between North and South Island populations of Euryapteryx geranoides (1.6 mya) and of Anomalopteryx in both islands (2.8 mya) indicates dispersal between the islands just before or in the Pleistocene when sea levels declined and the islands were connected.
Subfossil remains, together with our analysis, indicate that no more than four species existed in a number of the same biogeographic regions of New Zealand (10, 32), and that these species exploited ecologically different habitats and foods. Previous research has shown that moa had diverse diets, based on differences in the shape of the beak and size of the jaw muscles (4), and analysis of gizzard contents (34–35). The large Dinornis browsed primarily on coarse twigs, Euryapteryx and Emeus ate soft leaves and berries, Pachyornis and Anomalopteryx had a diet of tough leaves, and Megalapteryx foraged in forest edges and adjacent high-altitude grasslands (4). In the absence of foregut fermentation, it has been speculated that moa evolved long intestines to ferment their plant diets and correlated large body sizes (4).
The morphological and genetic diversity in moa is much greater than that found in the sister ratite taxa, where a maximum of five species is found in kiwi (33). Hence, moa have clearly undergone an extensive radiation and phylogeographic structuring within species. This diversification parallels cycles of island dispersal, isolation, and allopatric speciation in textbook avian adaptive radiations in the Galápagos and Hawaii. As in most North American birds (36, 37), moa speciation events occurred during the phase of rapid cooling before the Pleistocene, whereas phylogeographic subdivisions within species developed 1–2 mya after the onset of the Pleistocene ≈2.5 mya (38). Paleoenvironmental changes have therefore had a profound influence on the tempo and mode of evolution in moa, adding to the growing body of evidence that the evolutionary history of vertebrates has been shaped by many of the same large-scale environmental events (39–43).
We conclude that ancient DNA methods (45) provide powerful tools for inferring the number of lineages, as well as the tempo and mode of evolution of entire extinct groups of animals. We have shown that at least 14 lineages comprise the radiation of the New Zealand moa. Nine of these lineages correspond to currently recognized species, and the remaining five new lineages represent either deep phylogeographic splits within existing species or separate phylogenetic species. A synthesis of bone characters, DNA sequence variation, and possibly carbon dating will help resolve this issue. We note that the lineage diversity of moa approximates the species diversity in Darwin's finches (13 species) and Hawaiian honeycreepers (14 surviving and 8 extinct species). However, the moa radiation was accompanied by an order-of-magnitude change in body mass (20–250 kg) (4), consistent with the longer period of isolation and evolution of moa relative to Darwin's finches and Hawaiian honeycreepers. This reconstruction of the tempo and mode of evolution in an extinct family of birds by using ancient DNA helps to clarify the confusing morphological variation that has plagued the study of moa over the past 150 years.
Supplementary Material
Acknowledgments
We thank Chris McGowan, Paul Scofield (Canterbury Museum), Brian Gill (Auckland Museum), Michelle Horwood and Trish Nugent (both of the Whanganui Museum), the Otago Museum, and the Waitomo Caves Museum for permission and assistance in sampling bones; Sergio Pereira for molecular dating analyses; and Bob Zink, David Irwin, Chris McGowan, and Sergio Pereira for critical reading of the manuscript. This work was supported by grants from the Natural Sciences and Engineering Research Council (Canada) (to A.J.B.), the Royal Ontario Museum Foundation (to A.J.B.), and the Marsden Fund of New Zealand (to D.M.L.).
Author contributions: A.J.B. and D.M.L. designed research; L.J.H., O.H., and C.D.M. performed research; L.J.H., O.H., and C.D.M. contributed new reagents/analytic tools; A.J.B., L.J.H., O.H., C.D.M., and D.M.L. analyzed data; and A.J.B. wrote the paper, with contributions from D.M.L. and C.D.M.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CI, confidence interval; mya, million years ago; myr, million years.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AY299860, AY299862–AY299864, AY299866, AY299867, AY299869, AY299870, AY299872, AY299873–AY299877, AY299881–AY299885, AY299888, AY299891, AY299895, AY299899–AY299901, AY299903, AY299904, AY299906–AY299908, AY199912, AY299914, AY299916, AY299920, AY299922–AY299930, AY299934, AY299935, AY299938–AY299941, AY299943, AY299944, AY299948, AY299951, AY299954–AY299957, AY299969, AY299971, DQ023671–DQ023695, DQ029126–DQ029193, and DQ055458–DQ055728).
References
- 1.Cracraft, J. (1976) Smithson. Contrib. Paleobiol. 27, 189-205. [Google Scholar]
- 2.Baker, A. J. (1991) Curr. Ornithol. 8, 1-67. [Google Scholar]
- 3.Huynen, L., Millar, C. D., Scofield, R. P. & Lambert, D. M. (2003) Nature 425, 175-178. [DOI] [PubMed] [Google Scholar]
- 4.Worthy, T. H. & Holdaway, R. N. (1999) N.Z. Geogr. 12, 51-68. [Google Scholar]
- 5.Haddrath, O. & Baker, A. J. (2001) Proc. R. Soc. London Ser. B 268, 939-945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cooper, A., Lalueza-Fox, C., Anderson, S., Rambaut, A., Austin, J. & Ward, R. (2001) Nature 409, 704-707. [DOI] [PubMed] [Google Scholar]
- 7.Holdaway, R. N. & Jacomb, C. (2000) Science 287, 2250-2254. [DOI] [PubMed] [Google Scholar]
- 8.Archey, G. (1941) Auck. Inst. Mus. Bull. 1, 1-145. [Google Scholar]
- 9.Oliver, W. R. B. (1949) Dominion Mus. Bull. 15, 1-206. [Google Scholar]
- 10.Worthy, T. H. & Holdaway, R. N. (2002) The Lost World of the Moa: Prehistoric Life of New Zealand (Canterbury Univ. Press, Christchurch, New Zealand).
- 11.Bunce, M., Worthy, T. H., Ford, T., Hoppitt, W., Willislev, E., Drummond, A. & Cooper, A. (2003) Nature 425, 172-175. [DOI] [PubMed] [Google Scholar]
- 12.Lambert, D. M., Ritchie, P. A., Millar, C. D., Holland, B., Drummond, A. J. & Baroni, C. (2002) Science 295, 2270-2273. [DOI] [PubMed] [Google Scholar]
- 13.Hagelberg, E. (1994) in Ancient DNA, eds. Herrmann, B. & Hummel, S. (Springer, New York), pp. 195-204.
- 14.Cabot, E. & Beckenbach, A. T. (1989) Comput. Appl. Biosci. 5, 233-234. [DOI] [PubMed] [Google Scholar]
- 15.Ronquist, F. & Huelsenbeck, J. P. (2003) Bioinformatics 19, 1572-1574. [DOI] [PubMed] [Google Scholar]
- 16.Posada, D. & Crandall, K. A. (1998) Bioinformatics 14, 817-818. [DOI] [PubMed] [Google Scholar]
- 17.Guindon, S. & Gascuel, O. (2003) Syst. Biol. 52, 696-704. [DOI] [PubMed] [Google Scholar]
- 18.Swofford, D. L. (2001) paup*: Phylogenetic Analysis Using Parsimony (*and Other Methods) (Sinauer, Sunderland, MA), Version 4.0b10.
- 19.Sanderson, M. J. (2003) Bioinformatics 19, 301-302. [DOI] [PubMed] [Google Scholar]
- 20.Dyke, G. J. & van Tuinen, M. (2004) Zool. J. Linn. Soc. 141, 153-177. [Google Scholar]
- 21.Worthy, T. H. (1988) Notornis 35, 99-108. [Google Scholar]
- 22.Cao, Y., Adachi, J., Janke, A., Paabo, S. & Hasegawa, M. (1994) J. Mol. Evol. 39, 519-527. [DOI] [PubMed] [Google Scholar]
- 23.Cummings, M. P., Otto, S. P. & Wakeley, J. (1995) Mol. Biol. Evol. 12, 814-822. [DOI] [PubMed] [Google Scholar]
- 24.Russo, C. A., Takezaki, N. & Nei, M. (1996) Mol. Biol. Evol. 13, 525-536. [DOI] [PubMed] [Google Scholar]
- 25.Zardoya, R. & Meyer, A. (1996) Mol. Biol. Evol. 13, 933-942. [DOI] [PubMed] [Google Scholar]
- 26.Siguroardottir, S., Helgason, A., Gulcher, J. R., Stefansson, K. & Donnelly, P. (2000) Am. J. Hum. Genet. 66, 1509-1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Paton, T., Haddrath, O. & Baker, A. J. (2002) Proc. R. Soc. London Ser. B 269, 839-846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Quinn, T. W. (1992) Mol. Ecol. 1, 105-117. [DOI] [PubMed] [Google Scholar]
- 29.Lawver, L. A., Royer, J.-Y., Sandwell, D. T. & Scortese, C. R. (1991) in Geological Evolution of Antarctica, eds. Thomson, M. R. A., Crame, J. A. & Thomson, J. W. (Cambridge Univ. Press, Cambridge, U.K.), pp. 533-539.
- 30.Cooper, R. A. & Millener, P. R. (1993) Trends Ecol. Evol. 8, 429-433. [DOI] [PubMed] [Google Scholar]
- 31.Cooper, A. & Cooper, R. A. (1995) Proc. R. Soc. London Ser. B 261, 293-302. [Google Scholar]
- 32.Cooper, A., Atkinson, I. A. E., Lee, W. G. & Worthy, T. H. (1993) Trends Ecol. Evol. 8, 433-437. [DOI] [PubMed] [Google Scholar]
- 33.Burbidge, M. L., Colbourne, R. M., Robertson, H. A. & Baker, A. J. (2003) Cons. Genet. 4, 167-177. [Google Scholar]
- 34.Burrows, C. J. (1980) N. Z. J. Ecol. 3, 125-130. [Google Scholar]
- 35.Burrows, C. J., McCulloch, B. & Trotter, M. M. (1981) Rec. Canterbury Mus. 9, 309-336. [Google Scholar]
- 36.Klicka, J. & Zink, R. M. (1997) Science 277, 1666-1669. [Google Scholar]
- 37.Zink, R. M., Klicka, J. & Barber, B. R. (2004) Proc. R. Soc. London Ser. B 359, 215-220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Naish, T. R., Abott, S. T., Alloway, B. V., Beu, A. G., Carter, R. M., Edwards, A. R., Journeaux, T. D., Kamp, P. J. J., Pillans, B. J., Saul, G. & Woolfe, K. J. (1998) Quart. Sci. Reviews 17, 695-710. [Google Scholar]
- 39.Delsuc, F., Vizcaíno, S. F. & Douzery, E. J. P. (2004) BMC Evol. Biol. 4, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bossuyt, F. & Milinkovitch, M. C. (2001) Science 292, 93-95. [DOI] [PubMed] [Google Scholar]
- 41.Mercer, M. J. & Roth, V. L. (2003) Science 299, 1568-1572. [DOI] [PubMed] [Google Scholar]
- 42.Douady, C. J. & Douzery, E. J. P. (2003) Mol. Phylogenet. Evol. 28, 285-296. [DOI] [PubMed] [Google Scholar]
- 43.Douady, C. J., Catzeflis, F., Raman, J., Springer, M. S. & Stanhope, M. J. (2003) Proc. Natl. Acad. Sci. USA 100, 8325-8330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.King, P. R. (2000) New Zealand Oil Exploration Conference Proceedings (Ministry of Economic Development, Wellington, New Zealand), pp. 132-146.
- 45.Lambert, D. M., Baker, A., Huynen, L., Haddrath, O., Hebert, P. D. & Millar, C. D. (2005) J. Hered. 96, 279-284. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.