Abstract
Beta-lactamase-producing Enterobacteriaceae present a significant therapeutic challenge. Current developments in phenotypic diagnostics focus primarily on rapid minimum inhibitory concentration (MIC) determination. There is a requirement for rapid phenotypic diagnostics to improve antimicrobial susceptibility tests (AST) and aid prescribing decisions. Phenotypic AST are limited in their ability to characterise beta-lactamase-producing Enterobacteriaceae in detail. Despite advances in rapid AST, gaps and opportunities remain for developing additional diagnostic approaches that facilitate personalised antimicrobial prescribing. In this perspective, we highlight the state-of-the-art in beta-lactamase detection, identify gaps in current practice, and discuss barriers for innovation within this field.
Subject terms: Antimicrobial resistance, Bacteria
In this work, authors aim to address the critical challenges in diagnosing and managing beta-lactamase-producing Enterobacteriaceae, which are pivotal in the context of antimicrobial resistance (AMR).
Introduction
In 2024, WHO updated the bacterial priority pathogens list, highlighting organisms that pose the greatest risk to global health. Priority 1 pathogens (critical) include Acinetobacter baumannii (A.baumannii) and Enterobacteriaceae1. These organisms are often multidrug-resistant and can be difficult to treat, leading to poor outcomes in severe infections such as sepsis2. Beta-lactamase enzyme production is a common mechanism of resistance3 in all priority 1 pathogens4.
Beta-lactamase enzymes inactivate beta-lactam antimicrobials by hydrolysing the beta-lactam ring5. Beta-lactamases are naturally occurring and can be chromosomal or plasmid mediated6. WHO lists third-generation cephalosporin-resistant Enterobacteriaceae (3GCRE) and carbapenem resistant Enterobacteriaceae (CRE) as ‘critical’ healthcare threats1. CRE and 3GCRE have worldwide distribution and an increasing prevalence, at least partially, as a result of the overuse of beta-lactam antimicrobials7.
The announcement of the 2024 winners of the Longitude Prize in antimicrobial resistance (AMR) (Sysmex Astrego)8 has brought significant attention and investment in rapid diagnostics to address AMR. Despite reported advances in rapid AST, there remains a substantial gap and opportunity to develop additional diagnostic tools that facilitate personalised approaches to antimicrobial prescribing.
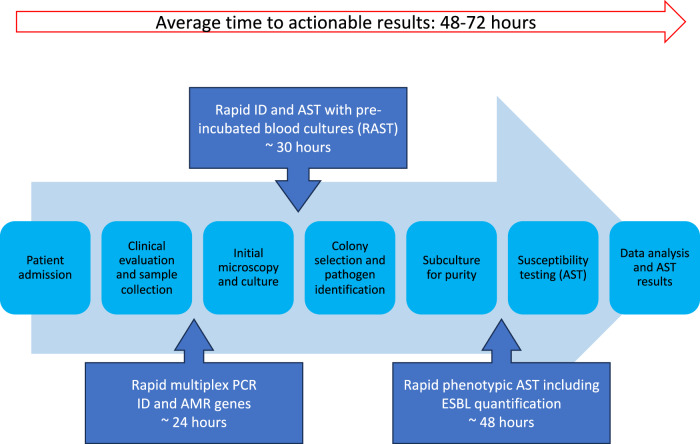
Current efforts in diagnostic development have primarily focused on the development of rapid AST. Implementation of rapid AST technology could significantly reduce turnaround times in clinical microbiology laboratories (Fig. 1). The figure highlights the current workflow and how ongoing advancements in technology are transforming the speed of pathogen identification and antimicrobial susceptibility testing in clinical settings. Yet, there remains a need for innovations that extend beyond these areas, such as beta-lactamase quantification and dynamic diagnostic capabilities, to further combat AMR effectively and move towards individualised prescribing.
Fig. 1. Current workflow in clinical settings, highlighting ongoing advancements.
The figure illustrates the current diagnostic workflow for infection identification and antimicrobial susceptibility testing (AST) in hospitalised patients. Upon patient admission, clinical evaluation and sample collection are initiated. The conventional diagnostic pathway includes initial microscopy and culture, colony selection and pathogen identification, subculture for purity, and AST taking 48–72 h for completion, including data analysis and final AST results. Advancements are being made in rapid diagnostics, including rapid multiplex PCR for pathogen identification and antimicrobial resistance (AMR) gene detection, which takes around 24 h. Rapid identification and AST using pre-incubated blood cultures (RAST) can provide results in about 30 h. In addition, rapid phenotypic AST, including extended-spectrum beta-lactamase (ESBL) quantification, takes approximately 48 h. These integrated approaches aim to reduce diagnostic time and enable more timely and targeted treatment decisions, improving patient outcomes in clinical settings.
Beta-lactamase enzyme classification
Beta-lactamase enzymes have extensively diversified in response to the clinical use of new generations of beta-lactam antibiotics. Beta-lactamases can be characterised by the Ambler molecular and structural classification9 or Bush-Jacoby-Medeiros functional classification10 (Table 1).
Table 1.
The classification of beta-lactamase enzymes using the Ambler and Bush-Jacoby-Medeiros classification systems
| Ambler Molecular Classification | Bush-Jacoby-Medeiros Functional Classification | Type | Characteristics | Examples of Enzymes |
|---|---|---|---|---|
| A | Group 2: serine β - lactamase | Narrow spectrum β – lactamases (penicillinases) | Hydrolyse a limited number of penicillins | PSE, CARB |
| Broad spectrum β – lactamases | Hydrolyse a range of beta-lactam antibiotics, including penicillins, and cephalosporins | TEM-1, TEM-2, SHV-1 | ||
| Extended spectrum β – lactamases (ESBLs) | Hydrolyse extended-spectrum cephalosporins, such as cefotaxime, ceftriaxone, and ceftazidime, in addition to penicillins, monobactams and early-generation cephalosporins. Susceptible to inhibition by clavulanic acid | TEM-3, SHV-5, CTX-M1 | ||
| Class A carbapenemases | Hydrolyse carbapenems but susceptible to inhibition by boronic acid | |||
| B | Group 3: metallo β - lactamase | Metallo β - lactamases | Hydrolyse carbapenems and are resistant to clavulanic acid, sulbactam, and tazobactam. Susceptible to inhibition by EDTA and dipicolinic acid |  |
| C | Group 1: cephalosporinases | AmpC | Hydrolyse cephalosporins and are resistant to clavulanic acid. Susceptible to inhibition by cloxacillin and boronic acid | DHA, CMY-1, ACT-1 |
| D | Group 2: serine β - lactamase | Extended spectrum β – lactamases (ESBLs), Oxacillinases | Hydrolyse cloxacillin and oxacillin and in some cases (OXA-11, OXA-15) hydrolyse extended-spectrum cephalosporins | OXA-1, OXA-10, OXA-11, OXA-15 |
| Carbapenem-hydrolysing class D β – lactamases (CHDLs) | Hydrolyse carbapenems |  |
Carbapenemases shown in red.
The Ambler classification is the most widely accepted classification system as it is considered the simplest. The Ambler classification categorises beta-lactamases into four classes; A, B, C, and D, based on the amino acid sequence in the protein molecules. Classes A, C, and D utilise a serine residue at the active centre of the enzyme, whereas class B uses zinc ions for their enzymatic activity.
The Bush-Jacoby-Medeiros classification groups beta-lactamases into groups 1–3 depending on their ability to degrade beta-lactam substrates and their response to inhibitor effects.
Narrow spectrum beta-lactamases - Penicillinases
Penicillinases are beta-lactamase enzymes that show specificity for penicillin. A key example of a penicillinase enzyme is SHV-1, which is chromosomally found in Klebsiella pneumoniae (K.pneumoniae)11. SHV-1 enzymes confer resistance to penicillins and early-generation cephalosporins (cefalexin, cephaloridine and cephalothin). This Ambler class A penicillinase has spread via plasmids to many Enterobacteriaceae. The majority of SHV enzymes have been detected in K. pneumoniae or Escherichia coli (E. coli)12.
The development of beta-lactamase inhibitors like clavulanate, sulbactam, and tazobactam was a significant breakthrough in combating bacterial resistance to beta-lactam antibiotics, targeting enzymes such as TEM-1, TEM-2, and SHV-1 and their extended-spectrum variants. However, the evolution of bacteria in response led to the emergence of inhibitor-resistant TEM (IRT) enzymes. IRT enzymes confer resistance to penicillins and beta-lactam/beta-lactamase combinations like piperacillin/tazobactam but remain susceptible to third-generation cephalosporins. Standard in vitro AST is often insufficient for reliably identifying IRT enzymes, contributing to a likely underestimation of their prevalence13.
Extended-spectrum beta-lactamases
Extended-spectrum beta-lactamases (ESBLs) are a rapidly evolving group of beta-lactamases. To date, the total number of characterised ESBLs exceeds 40014. Historically, TEM and SHV-type ESBLs were the most prevalent, but since the early 2000s, CTX-M-type enzymes have been the most common ESBL, with the CTX-M-15 variant dominating worldwide7. ESBLs are defined by their ability to hydrolyse third-generation cephalosporins, oxyaminopenicillins, and the monobactam aztreonam, but can be inhibited by clavulanic acid in vitro15. ESBL-producing organisms can also exhibit resistance to many other classes of antibiotics. This often leaves limited treatment options due to co-inheritance of resistance mechanisms on the plasmid, such as plasmid-mediated quinolone resistance and plasmid-mediated dihydrofolate reductase isoforms driving sulphonamide resistance16,17. As a result, carbapenems are considered the drug of choice against ESBL infections18. In 2017, there were an estimated 197,400 cases of ESBL-producing Enterobacteriaceae among hospitalised patients and 9100 estimated deaths in the United States19.
AmpC beta-lactamases
AmpC beta-lactamases are clinically significant cephalosporinases that confer resistance to most beta-lactam antibiotics, with the exception of fourth-generation cephalosporins and carbapenems20. They are resistant to inhibition by beta-lactamase inhibitors, such as clavulanate, sulbactam and tazobactam21 but can be inhibited in vitro by cloxacillin. Mechanisms of AmpC beta-lactamase resistance can be grouped into 3 categories: (1) inducible resistance via chromosomally encoded AmpC genes (e.g., E. cloacae), (2) non-inducible chromosomal resistance due to promoter and/or attenuator mutations (e.g., E.coli, A. baumannii), or (3) plasmid-mediated resistance (e.g., K. pneumoniae, E. coli, Salmonella spp.)20,22. Detecting infections caused by AmpC enzymes can be difficult, leading to possible mismanagement of antibiotic therapy. Boronic acids are well-established inhibitors of AmpC enzymes23, and various boronic acid derivatives have been employed in diagnostic tests to detect these enzymes in bacterial strains. Plasmid-mediated AmpCs are of particular concern due to their ability to spread between bacterial species24,25. A study in Ethiopia reported a 14% detection rate of AmpC beta-lactamases in Enterobacteriaceae species26.
Carbapenemases
Increased carbapenem consumption is a leading driver of carbapenem resistance27. Repeated mutations in ESBL enzymes have led to the emergence of carbapenemases and CRE. Carbapenemases will often hydrolyse all beta-lactam antibiotics, including carbapenems. In 2017, there were an estimated 13,100 cases of CRE in hospitalised patients in the United States, with an estimated 1100 deaths19.
Beta-lactamases with the ability to degrade carbapenems have been identified within Ambler classes A, B and D28. The class A carbapenemases include members of the SME, IMI, NMC, GES and KPC families. Of these, the KPC carbapenemases are the most prevalent, found mostly in K. pneumoniae29. It is estimated that mortality due to KPC-producing K. pneumoniae infections could be as high as 41%30.
The class D carbapenemases, including OXA-type beta-lactamases, are often found in A. baumannii. OXA-48 and ‘OXA-48-like’ enzymes have proliferated to become the most prevalent carbapenemases across much of Europe, Northern Africa and the Middle East produced by Enterobacteriaceae31. In the United Kingdom between 2022–2023, ‘OXA-48-like’ enzymes were the most frequently identified carbapenemase family in E. coli, accounting for 40.7% of cases32. OXA-48 was initially identified in Turkey in 2001, leading to the endemic presence of OXA-producing bacteria in that region33. Among the ‘OXA-48-like’ variants, OXA-181 is the most commonly identified variant so far34. OXA-23 is the most frequently identified carbapenemase in cases of carbapenem-resistant Acinetobacter baumannii (CRAB) and is endemic in India35.
The class B, metallo-beta-lactamase (MBL) carbapenemases, including IMP, VIM and NDM families, have been detected primarily in Pseudomonas aeruginosa (P. aeruginosa), K. pneumoniae and E. coli36. MBL-producing organisms are of particular concern due to their resistance profiles and difficulty in treatment. NDM was first reported in 200837 and is prevalent in India, although it has also been responsible for outbreaks in Eastern Europe, North Africa29 and the United Kingdom38.
Carbapenemases are challenging due to the limited treatment options available, their association with high mortality rates, and their plasmid-mediated nature, which facilitates rapid spread. Consequently, patients with carbapenemase-producing infections require isolation and transmission-based precautions put in place as quickly as possible39.
Beta-lactamase enzyme detection methods
Accurate and timely identification of beta-lactamase-producing bacteria is crucial to support appropriate antimicrobial management, prevent onward transmission, and support public health surveillance27. Various methods are used to detect beta-lactamase enzymes in clinical laboratories and can be broadly grouped into phenotypic or genotypic methods (Table 2). Phenotypic methods include double disc, gradient strip, and broth dilution testing. These methods are relatively simple, cost-effective, and can detect a broad range of resistance mechanisms. However, they may be time-consuming and less specific, sometimes failing to distinguish between different beta-lactamases.
Table 2.
Overview of ESBL detection methods
| Method | Overview | Quantitative (✓ / X) | |
|---|---|---|---|
| Phenotypic | Double Disc Synergy Test (DDST) | Involves placing discs containing beta-lactam antibiotics alone and in combination with a beta-lactamase inhibitor. The presence of an ESBL is indicated by an enhanced inhibition zone around the combination disc | X |
| Combination Disc Test (CDT) | Similar to DDST but utilizes a specific combination of discs to detect ESBL activity | X | |
| Gradient test | Gradient diffusion method that uses strips impregnated with various concentrations of an antibiotic. The intersection of the growth ellipse with the antibiotic strip helps determine ESBL production e.g. ESBL Etest, AB Biodisk | X | |
| Genotypic | Polymerase Chain Reaction (PCR) | Targets specific genes associated with ESBL production, providing a direct molecular confirmation. Common genes include blaCTX-M, blaTEM, and blaSHV e.g. ESBL ELITe MGB® Kit, ELITechGroup MDx | X |
| DNA Microarrays | Enables the simultaneous detection of multiple resistance genes, including ESBLs, offering a high-throughput molecular approach | X | |
| DNA sequencing | Compares the obtained sequence with known ESBL gene sequences in order to identify the specific ESBL gene present in the bacterial isolate | X | |
| Whole Genome Sequencing (WGS) | Offers a comprehensive analysis of the entire bacterial genome, enabling the identification of ESBLs and other resistance mechanisms | X | |
| Proteomics | Matrix-Assisted Laser Desorption/Ionisation Time-of-Flight (MALDI-TOF) | Allows for identification of bacterial species and can detect beta-lactamase activity by co-incubating samples with a beta-lactam antibiotic e.g. MALDI Biotyper® MBT STAR-BL assay, Bruker | X |
| Biochemical | Borate Nitrocefin Test | Utilises the colour change of nitrocefin in the presence of ESBLs, providing a rapid visual confirmation of beta-lactamase activity | X |
| Chromogenic Agar | Incorporates specific substrates that change colour in the presence of ESBLs e.g. chromID® ESBL agar, BioMérieux and Brilliance ESBL agar, ThermoFisher Scientific | X | |
| Immunological | Enzyme-Linked Immunosorbent Assay (ELISA) | Utilizes antibodies to detect ESBLs, providing a rapid and specific immunological confirmation e.g. NG-Test CTX-M MULTI assay, NG Biotech | X |
| Automated | Automated systems | Automated platforms for bacterial identification and susceptibility testing, including ESBL detection e.g. VITEK 2, MicroScan, and Phoenix | X |
Genotypic methods include polymerase chain reaction (PCR) and whole genome sequencing (WGS). Despite their accuracy and speed, genotypic methods are more expensive and may miss novel or uncommon resistance mechanisms not included in the assay design. Genotypic methods do not provide information on the amount of beta-lactamase enzyme being produced and its potential impact on antimicrobial susceptibility.
ESBL Detection methods
In most laboratories, ESBL detection and characterisation is mandatory or strongly recommended as beta-lactamase-producing bacteria tend to harbour broad resistance to many first-line antimicrobials used in clinical practice. Both the Clinical and Laboratory Standards Institute (CLSI) and European Committee on Antimicrobial Susceptibility Testing (EUCAST) guidelines recommend ESBL screening followed by a confirmatory test40,41. Failure to identify and mitigate the transmission of plasmid mediated ESBL can have serious consequences for infection control15. Traditionally, detection of ESBL-producing isolates has relied on phenotypic methods that take advantage of beta-lactamase inhibition in vitro by clavulanate7.
Screening for ESBLs is conducted on bacterial isolates using methods such as disc diffusion and agar or broth dilution, or automated systems. If an isolate is found to be indeterminate or resistant to third-generation cephalosporins (cefotaxime, ceftazidime, or cefpodoxime), ESBL confirmation testing is required40. The confirmation of ESBL production can be performed using several techniques. The most common methods are the combination disc test (CDT), where antibiotic discs containing a beta-lactamase inhibitor show a significant increase in the inhibition zone compared to the antibiotic alone; and the double disc synergy test (DDST), which places cephalosporin discs near a disc with a beta-lactamase inhibitor and looks for an enhanced inhibition zone. Other methods include the ESBL gradient test, which utilises strips with a gradient of a beta-lactam antibiotic and a beta-lactamase inhibitor to reveal differences in minimum inhibitory concentrations (MICs); and broth microdilution methods which compare MICs in the presence and absence of a beta-lactamase inhibitor.
Phenotypic methods, such as disc diffusion and agar or broth dilution, are cost-effective and straightforward but may lack sensitivity and specificity, particularly in distinguishing ESBLs from other beta-lactamases such as AmpC16. In addition, IRT enzymes may not be detected using phenotypic methods as most of the tests rely on inhibition with clavulanate. Automated systems have built-in ESBL screening and confirmation tests but can report false positive ESBL results due to SHV-1 hyperproduction or false negative results due to AmpC production42.
Genotypic methods, including PCR, can provide specific identification of ESBL genes and detect low-level resistance more easily than culture-based methods. Primers to identify genes that code for beta-lactamase enzymes are well-researched and there are several databases available. However, it is important to consider the level of gene detection required (e.g., TEM) and the specific variant type (e.g., IRT). To do this may require multiple primers and differential melting temperature determination to identify specific variants within genes, which may not be feasible on a large scale. For example, the ESBL ELITe MGB® Kit is a multiplex real-time PCR assay specifically designed to detect CTX-M genes, which are the most prevalent group of ESBL genes found in Enterobacteriaceae. Genotypic methods are not typically used in routine AST due to their high cost and the limited availability of CE-certified commercial assays, so are generally reserved for surveillance purposes43.
Recent advances in rapid phenotypic methods have enabled the same-day detection of ESBL producers through colourimetric and immunological lateral flow assays. Rapid colourimetric methods such as Rapid ESBL NP® can provide results within 15 min to 2 h from cultured isolates and can detect any type of ESBL, especially CTX-M producers with good overall sensitivity (93.9%) and specificity (98.5%)44. The Beta Lacta Test is another colourimetric test which broadly detects beta-lactamases (ESBLs, AmpC, carbapenemases) without distinction and demonstrates good sensitivity (95%) and specificity (87%)45. The NG-Test® CTX-M MULTI is a qualitative lateral flow immunoassay for the rapid detection of the five major CTX-M groups. It can detect CTX-M groups 1, 2, 8, 9 and 25 within 15 min from cultured isolates and shows good sensitivity (100%) and specificity (99.6%)46.
AmpC Detection methods
Detecting AmpC enzymes is challenging because they do not always produce a resistant phenotype in conventional disc diffusion or automated susceptibility tests47. AmpC enzymes can be induced or de-repressed, so there is a risk of treatment failure that is not always possible to predict48. The lack of standardised guidelines for their detection in laboratory settings has been a contributing factor to therapeutic failures49.
AmpC enzymes are poorly inhibited by the classical ESBL inhibitors, especially clavulanic acid20. Phenotypic detection generally relies on the inhibition of AmpC by either cloxacillin or boronic acid. Few detection methods exist (Table 3), but two of the most well-recognised are the cefoxitin-cloxacillin disc diffusion test and the AmpC gradient strip test. Resistance to cefoxitin, a cephamycin antibiotic that is selectively hydrolysed by AmpC, is suggestive of an enzyme presence. However, some AmpC variants remain susceptible to cefoxitin (e.g., ACC-like enzymes), and cefoxitin resistance can also result from changes in permeability or species-specific intrinsic resistance mechanisms50.
Table 3.
Overview of AmpC detection methods
| Method | Overview | Quantitative (✓ / X) | |
|---|---|---|---|
| Phenotypic | AmpC Disc Diffusion Test | Involves using antibiotic discs alone and in combination with beta-lactamase inhibitors. Resistance to cefoxitin and enhancement of the inhibition zone with inhibitors like boronic acid or cloxacillin suggest AmpC production | X |
| Disc Potentiation Test (DPT) | Similar to DDST, involves testing with three different enzyme inhibitors. Discs of cephalosporin and cephalosporin plus inhibitor are added to a lawn culture and zone size is measured after incubation | X | |
| Double Disc Synergy Test (DDST) | Involves testing with three different enzyme inhibitors (boronic acid, cloxacillin at two concentrations). The test aims to assess AmpC enzyme activity in the organisms by evaluating the inhibitory effect of cephalosporin antibiotics in the presence of these inhibitors. The principle is based on the inactivation of AmpC enzyme by the inhibitors, leading to an increased zone of inhibition around the cephalosporin | ||
| Gradient test | Gradient of cephamycin and cephamycin combined with a gradient of cloxacillin on respective half of the E-strips are used for the detection of AmpC producers A reduction in the MIC of cephamycin for at least three dilutions or deformation of its zone of inhibition or a “Phantom zone” suggests the presence of AmpC enzyme producers | X | |
| Genotypic | Polymerase Chain Reaction (PCR) | Utilizes polymerase chain reaction to target genes associated with AmpC production, such as blaCMY, blaDHA, and blaACT e.g. Check-MDR CT103XL, Check Points Health | X |
| DNA Microarrays | Enables the simultaneous detection of multiple resistance genes, including AmpC, offering a high-throughput molecular approach | X | |
| DNA sequencing | Involves sequencing the AmpC gene to identify specific mutations or variations linked to increased expression | X | |
| Whole Genome Sequencing (WGS) | Offers a comprehensive analysis of the entire bacterial genome, enabling the identification of ESBLs and other resistance mechanisms | X | |
| Proteomics | Matrix-Assisted Laser Desorption/Ionisation Time-of-Flight (MALDI-TOF) | Allows for identification of bacterial species and can detect the most actively expressed ESBL and AmpC beta-lactamases in multi-drug-resistant (MDR) gram-negative Enterobacteriaceae e.g. MALDI Biotyper® MBT STAR-Cepha assay, Bruker | X |
| Biochemical | Chromogenic Agar | Incorporates specific substrates that change colour in the presence of ESBLs including most of those carrying AmpC type resistance e.g. CHROMagar™ ESBL, CHROMagar | X |
| Immunological | Enzyme-Linked Immunosorbent Assay (ELISA) | Utilizes antibodies to detect AmpC, providing a rapid and specific immunological confirmation | X |
| Automated | Automated systems | Automated platforms for bacterial identification and susceptibility testing, including AmpC detection e.g. VITEK 2, MicroScan, and Phoenix | X |
The cefoxitin-cloxacillin disc diffusion test works by comparing the zone sizes of cefoxitin with and without cloxacillin. A significant reduction in zone size (≤4 mm) around the cefoxitin disc when compared to the cloxacillin disc indicates the presence of AmpC beta-lactamase production. The AmpC gradient strip test consists of a strip containing cefotetan on one side and cefotetan-cloxacillin on the other side. Ratios of the MICs of cefotetan and cefotetan-cloxacillin of ≥ 8 are considered positive for AmpC beta-lactamase production51. It has long been recognised that improved methods for AmpC detection are needed20.
Carbapenemase detection methods
Carbapenemase detection is essential for the appropriate treatment of CRE infections. Screening for carbapenemase production in bacteria often involves susceptibility testing to identify isolates with high MICs for carbapenem antibiotics (meropenem, ertapenem and imipenem). EUCAST guidelines suggest meropenem as the chosen screening antibiotic as it offers the best compromise between sensitivity and specificity40.
Phenotypic methods (Table 4) for carbapenemase confirmatory testing include the carbapenem inactivation method (CIM), Modified Hodge Test (MHT) and combination disc test (CDT). The CDT method is the most common and involves placing discs of meropenem in the presence/absence of various inhibitors (e.g., EDTA, boronic acid). Boronic acid should inhibit Class A carbapenemases, and EDTA should inhibit Class B carbapenemases (MBLs)52. There is currently no available inhibitor for class D carbapenemases with high-level temocillin resistance often used as a screening test for OXA-48 presence. The main disadvantage is that such phenotypic methods take 18–24 hours. This delay in the detection of carbapenemase-producing organisms can impact on delivery of effective therapy and implementation of appropriate infection control procedures. The CIM test is popular for its specificity, sensitivity, and cost-effectiveness, demonstrating a superior ability to detect class D carbapenemases like OXA-48 compared to other phenotypic tests53. Like the CDT, one of the limitations of the CIM method is its incubation period before results are available, typically taking 8 hours on average.
Table 4.
Overview of carbapenemase detection methods
| Method | Overview | Quantitative (✓ / X) | |
|---|---|---|---|
| Phenotypic | Modified Hodge Test (MHT) | Assesses the ability of a carbapenem-resistant strain to enhance the growth of a carbapenem-susceptible indicator strain on agar plates. It provides a visual confirmation of carbapenemase activity | X |
| Combination disc test (CDT) | Involves placing discs of a carbapenem antibiotic in the presence/absence of various inhibitors (e.g., EDTA, boronic acid) and measuring zone sizes | X | |
| Carbapenem Inactivation Method (CIM) | A carbapenem is incubated with the bacterial isolate, the resulting hydrolysis of the carbapenem is assessed. If carbapenemase is present, it will inactivate the carbapenem antibiotic, leading to the growth of the bacterial isolate | X | |
| Genotypic | Polymerase Chain Reaction (PCR) | Targets specific carbapenemase-encoding genes, such as blaKPC, blaNDM, blaVIM, and blaOXA, providing direct molecular confirmation e.g., GeneXpert Carba-R, Cepheid and BIOFIRE® Blood Culture Identification 2 (BCID2) Panel, BioMérieux | X |
| Multiplex PCR | Allows simultaneous detection of multiple carbapenemase genes, aiding in the identification of the specific enzyme responsible for resistance | X | |
| Whole-Genomes Sequencing (WGS) | Offers a comprehensive analysis of the entire bacterial genome, enabling the identification of carpapenemases and other resistance mechanisms | X | |
| Proteomics | Matrix-Assisted Laser Desorption/Ionisation Time-of-Flight (MALDI-TOF) | Allows for identification of bacterial species and detect Class A, B, or D carbapenemase activity in gram-negative Enterobacteriaceae e.g., MALDI Biotyper® MBT STAR-Carba assay, Bruker | X |
| Biochemical | Carba NP colormetric assay | Involves the incubation of the bacterial isolate with a pH indicator and a carbapenem antibiotic. If carbapenemase is present, it hydrolyses the carbapenem, causing a change in pH that is visually observed as a colour change (red to yellow) e.g., RAPIDEC® Carba NP, BioMérieux | X |
| Immunological | Lateral flow immunochromatographic assay | Can detect the five most prevalent carbapenemases (NDM, IMP, VIM, OXA-48 and KPC) in less than 15 minutes from a bacterial colony e.g., NG-test Carba 5, NG Biotech and RESIST-4 O.K.N.V, Coris BioConcept | X |
| Automated | Automated systems | Automated platforms for bacterial identification and susceptibility testing, including carbapenemase detection e.g., VITEK 2, MicroScan, and Phoenix | X |
Genotypic methods for carbapenemase detection include PCR-based techniques and WGS. Due to their high costs, clinical laboratories do not often utilise these techniques and not all carbapenemases and variants can be identified as commercial panels are often made to find only the most common enzymes54. Examples include: the BioFire Filmarray® Blood Culture Identification Panel, capable of identifying 26 bacterial pathogens and 10 resistance mechanisms, including five carbapenemases (KPC, NDM, VIM, IMP, and OXA-48-like), and the Xpert® Carba-R PCR test, which detects carbapenemases (KPC, NDM, VIM, IMP-1, and OXA-48) directly from rectal swabs, blood, urine, or sputum in 50 min.
Advancements in rapid phenotypic techniques now allow for same-day carbapenemase detection using colourimetric and immunochromatographic lateral flow assays. The Carba NP test can detect carbapenemase production in 30 min to 2 h and works by measuring the hydrolysis of imipenem through changes in pH values using the indicator phenol red (red to yellow)55. Studies have favourable sensitivity (97.5%) and specificity (99.0%) for Pseudomonas spp. along with high sensitivity (95.8%) and specificity (93.3%) for Enterobacteriaceae56. The lateral flow immunochromatographic assays, NG-test Carba 5, and RESIST-4 O.K.N.V can detect the five most prevalent carbapenemases families (NDM, IMP, VIM, OXA-48 and KPC) in less than 15 min. NG-test Carba 5 demonstrated 97.7% sensitivity and 96.1% specificity from positive blood culture57 and RESIST-4 O.K.N.V showed good (95.3 %) and specificity (100%) from cultured isolates58.
Clinical implications of beta-lactamase detection methods on clinical practice
Managing infections with beta-lactamase-producing organisms is challenging due to increased bacterial resistance resulting in higher mortality rates59. It is common to find single isolates that express multiple beta-lactamase enzymes, further complicating treatment options60.
Clinicians may benefit from identifying the specific beta-lactamase enzymes produced by an organism to inform treatment decisions. However, genotypic testing is uncommon in most laboratories and typically reserved for difficult cases or epidemiological surveillance61. Instead, beta-lactamase production can be inferred from phenotypic antibiotic susceptibility data and existing detection methods62. The inference of beta-lactamase type is not always reliable as the spectrum of activity of the different enzyme classes often overlaps. For example, MBLs demonstrate broad-spectrum beta-lactamase activity, including carbapenemase activity, but are susceptible to monobactams. Nonetheless, this characteristic is not often clinically useful, as most MBL producers also produce ESBL, which results in monobactam resistance63.
Of all the methods described, no purely quantitative methods for beta-lactamase detection are available. Current beta-lactamase quantification methods are used to measure the level of beta-lactamase (ESBL, AmpC, carbapenemase) gene expression or the abundance of beta-lactamase genes in bacterial isolates. Methods to quantify beta-lactamase gene production in clinical isolates could provide valuable supplementary information about susceptibility to guide treatment decisions and could be useful to monitor antibiotic resistance. Gene quantity does not always correlate directly with gene expression and enzyme activity. For example, in the case of the M43 K. pneumoniae strain, although it harbours a SHV gene that would be detected genotypically, this gene is not phenotypically expressed64. In clinical practice, many factors can influence antimicrobial susceptibility, including porin loss and increased efflux. Although beta-lactamase quantification should not replace established AST methods, it may provide additional information to help inform the clinician. For example, Enterobacter cloacae (E.cloacae) can have strong permeability defects as well as derepressed AmpC expression65. Both mechanisms are responsible for reduced antimicrobial susceptibility, but routine AST cannot always clearly and rapidly differentiate these mechanisms. The quantification of beta-lactamase activity in response to different beta-lactam exposure alongside standard AST may provide greater phenotypic information to support clinical and laboratory decision-making.
Quantifying the absolute amount of beta-lactamase enzyme produced by bacterial isolates and its level of activity on specific antibiotics is more challenging than measuring gene expression or gene abundance. These enzymes are often present in small quantities, and their activity can be influenced by various factors, including type of beta-lactamase, site of infection and bacterial species. There are a few indirect methods to estimate beta-lactamase enzyme activity, including colourimetric assays66 and western blot, but these methods only provide a relative measure of enzyme activity rather than absolute quantification.
The MERINO trial and post-hoc analysis highlighted a need for better data on enzyme activity in relation to minimal inhibitory concentration in ESBL-producing Enterobacteriaceae. The trial failed to demonstrate that piperacillin-tazobactam was non-inferior to meropenem, with the former associated with a higher mortality rate (12.3% vs. 3.7%)67. Concerns about the trial methodology, particularly the determination of MIC and susceptibility reporting, were raised. The post-hoc analysis revealed a higher nonsusceptibility rate to piperacillin-tazobactam using broth microdilution compared to gradient MIC test strips, demonstrating the need for accurate and consistent susceptibility testing approaches68. The trial also found that 30-day mortality was significantly higher for patients with piperacillin-tazobactam MICs > 16 mg/L. Further analysis found that genotypic factors, such as the presence of AmpC and OXA-1 genes with CTX-M, could be associated with elevated MICs and increased mortality69.
Studies demonstrate that poor or inaccurate ESBL detection methods can lead to treatment failure70,71. With the lack of readily available genotypic methods, the focus should be on developing rapid phenotypic detection and quantification methods that could provide additional information to the clinician. For example, in current practice, most ESBL and AmpC infections are treated with carbapenems72. Carbapenem-sparing options are being explored to prevent the emergence of CRE73. In response to the challenge posed by CRE, novel beta-lactam/beta-lactamase inhibitor combinations, such as ceftazidime-avibactam, meropenem-vaborbactam, and imipenem-relebactam, have been developed74. Given the diverse antimicrobial effect exhibited by these different combinations against various CRE strains, the ability to accurately detect carbapenemases is essential for tailoring effective treatment strategies. Beta-lactam/beta-lactamase inhibitors could be a suitable alternative to carbapenems to treat less serious infections such as UTIs. For example, bacteria producing a low level of beta-lactamase enzyme may be able to be treated with beta-lactam/beta-lactamase inhibitors instead of a carbapenem. In addition, alternative agents such as ciprofloxacin or levofloxacin could potentially be used to treat these infections, but there is limited efficacy and safety data available18.
Improving beta-lactamase detection methods may help mitigate the development and impact of AMR on human health. Current AST methods have limitations (Fig. 2), including static results (MIC) that overlook dynamic changes in pathogen susceptibility and challenges in detecting emerging or low-level resistance. Phenotypic detection methods like broth microdilution and disc diffusion assays are widely used for their simplicity and cost-effectiveness but often require extended incubation times and can be affected by the presence of multiple resistance mechanisms, leading to false positives or negatives. In contrast, genotypic methods such as PCR and WGS offer faster results and precise identification of resistance genes but are costly, require specialised equipment and expertise, and may not reflect gene expression and phenotype accurately. Emerging AST technologies may face regulatory challenges during implementation and integration with existing infrastructure. Choosing the appropriate method depends on the clinical context, resource availability, and the need for accurate and timely detection of beta-lactamase-producing bacteria.
Fig. 2. Challenges faced by existing and emerging antimicrobial susceptibility testing (AST) methods.
The figure highlights several key issues in current and developing AST approaches. Traditional AST methods often provide static, one-time-point results, potentially missing dynamic changes in microbial susceptibility over time. These methods may also have difficulty detecting emerging or low-level resistance, leading to incomplete resistance profiles and potentially inadequate treatment decisions. Variability in result interpretation across different laboratories due to a lack of standardisation can impact the consistency and reliability of reporting and clinical decision-making. Emerging AST methods incorporate diverse technologies, which complicates integration and standardisation efforts. Advanced techniques may produce complex data sets that are challenging to interpret. Regulatory approval for novel methods can be time-consuming, and implementing and maintaining new technologies may require substantial resources. Achieving cost-effectiveness and accessibility, along with ensuring reliability and consistency across varied clinical settings, remains a significant challenge.
To bridge this gap, standardised rapid phenotypic methods that offer high sensitivity, specificity, and quick turnaround times while being cost-effective should be explored75. Methods supporting phenotypic beta-lactamase quantification76 linked to enhanced molecular detection methods may offer information to support personalised approaches to antimicrobial prescribing77. Automated susceptibility testing systems could be incorporated with machine learning to identify new resistance patterns78. A focus on point-of-care testing with portable and accessible devices, especially in resource-limited settings, may provide equity in access to diagnostics and support improved global detection and management of beta-lactamase-producing organisms.
Acknowledgements
The research was funded by the National Institute for Health Research Health Protection Research Unit (NIHR HPRU) in Healthcare Associated Infections and Antimicrobial Resistance at Imperial College London in partnership with the UK Health Security Agency (previously PHE), in collaboration with, Imperial Healthcare Partners, University of Cambridge and University of Warwick. This report is independent research funded by the National Institute for Health Research. The views expressed in this publication are those of the author(s) and not necessarily those of the NHS, the National Institute for Health Research, the Department of Health and Social Care or the UK Health Security Agency. This researcher is affiliated with the Wellcome Trust-funded programme CAMO-Net (grant ref: 226691/Z/22/Z).
Author contributions
J. Lawrence contributed to the conception, design, and draughting of the manuscript. D. O’Hare, J. van Batenburg-Sherwood, M. Sutton, A. Holmes, and T. Miles Rawson critically reviewed and provided feedback on the manuscript, contributing to the refinement of the final version. All authors read and approved the final manuscript.
Peer review
Peer review information
Nature Communications thanks the anonymous, reviewers for their contribution to the peer review of this work.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.WHO, WHO bacterial priority pathogens list, 2024: Bacterial pathogens of public health importance to guide research, development and strategies to prevent and control antimicrobial resistance. (2024).
- 2.Póvoa, P. et al. Optimizing antimicrobial drug dosing in critically Ill patients. Microorganisms (2021). [DOI] [PMC free article] [PubMed]
- 3.Worthington, R. J. & Melander, C. Overcoming resistance to β-lactam antibiotics. J. Org. Chem.78, 4207–4213 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Breijyeh, Z., Jubeh, B. & Karaman, R. Resistance of gram-negative bacteria to current antibacterial agents and approaches to resolve It. Molecules25, (2020). [DOI] [PMC free article] [PubMed]
- 5.Bush, K. & Bradford, P. A. β-Lactams and β-Lactamase inhibitors: An overview. Cold Spring Harb. Perspect. Med.6, a025247 (2016). [DOI] [PMC free article] [PubMed]
- 6.Hussain, H. I. et al. Genetic basis of molecular mechanisms in β-lactam resistant gram-negative bacteria. Micro. Pathog.158, 105040 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castanheira, M., Simner, P. J. & Bradford, P. A. Extended-spectrum β-lactamases: an update on their characteristics, epidemiology and detection. JAC Antimicrob. Resist.3, dlab092 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baltekin, Ö. et al. Antibiotic susceptibility testing in less than 30 min using direct single-cell imaging. Proc. Natl. Acad. Sci. USA114, 9170–9175 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ambler, R. P. The structure of beta-lactamases. Philos. Trans. R. Soc. Lond. B Biol. Sci.289, 321–331 (1980). [DOI] [PubMed] [Google Scholar]
- 10.Bush, K. & Jacoby, G. A. Updated functional classification of beta-lactamases. Antimicrob. Agents Chemother.54, 969–976 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tzouvelekis, L. S. & Bonomo, R. A. SHV-type beta-lactamases. Curr. Pharm. Des.5, 847–864 (1999). [PubMed] [Google Scholar]
- 12.Bradford, P. A. Extended-spectrum beta-lactamases in the 21st century: characterization, epidemiology, and detection of this important resistance threat. Clin. Microbiol. Rev.14, 933–951 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantón, R. et al. IRT and CMT beta-lactamases and inhibitor resistance. Clin. Microbiol Infect.14, 53–62 (2008). [DOI] [PubMed] [Google Scholar]
- 14.Mohammed, A. B. & Anwar, K. A. Phenotypic and genotypic detection of extended spectrum beta lactamase enzyme in Klebsiella pneumoniae. PLoS ONE17, e0267221 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Paterson, D. L. & Bonomo, R. A. Extended-spectrum beta-lactamases: a clinical update. Clin. Microbiol. Rev.18, 657–686 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rawat, D. & Nair, D. Extended-spectrum β-lactamases in Gram Negative Bacteria. J. Glob. Infect. Dis.2, 263–274 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mammeri, H. et al. Emergence of plasmid-mediated quinolone resistance in Escherichia coli in Europe. Antimicrob. Agents Chemother.49, 71–76 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tamma, P. D. et al. Infectious Diseases Society of America 2022 guidance on the treatment of extended-spectrum β-lactamase producing enterobacterales (ESBL-E), carbapenem-resistant enterobacterales (CRE), and pseudomonas aeruginosa with difficult-to-treat resistance (DTR-P. aeruginosa). Clin. Infect. Dis.75, 187–212 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.CDC, Antibiotic Resistance Threats in the United States, 2019, U.S. Department of Health and Human Services: Atlanta, GA. (2019).
- 20.Jacoby, G. A. AmpC beta-lactamases. Clin. Microbiol. Rev.22, 161–182 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Meini, S. et al. AmpC β-lactamase-producing Enterobacterales: what a clinician should know. Infection47, 363–375 (2019). [DOI] [PubMed] [Google Scholar]
- 22.Tamma, P. D. et al. A Primer on AmpC β-Lactamases: Necessary Knowledge for an Increasingly Multidrug-resistant World. Clin. Infect. Dis.69, 1446–1455 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Beesley, T. et al. The inhibition of class C beta-lactamases by boronic acids. Biochem. J.209, 229–233 (1983). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Estaleva, C. E. L. et al. High prevalence of multidrug resistant ESBL- and plasmid mediated AmpC-producing clinical isolates of Escherichia coli at Maputo Central Hospital, Mozambique. BMC Infect. Dis.21, 16 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aryal, S. C. et al. Plasmid-mediated AmpC β-Lactamase CITM and DHAM genes among gram-negative clinical isolates. Infect. Drug Resist.13, 4249–4261 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Legese, M. H. et al. Molecular epidemiology of extended-spectrum Beta-Lactamase and AmpC producing Enterobacteriaceae among sepsis patients in Ethiopia: A prospective multicenter study. Antibiotics11, 10.3390/antibiotics11020131 (2022). [DOI] [PMC free article] [PubMed]
- 27.Murray, C. J. L. et al. Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis. Lancet399, 629–655 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Queenan, A. M. & Bush, K. Carbapenemases: the versatile beta-lactamases. Clin. Microbiol. Rev.20, 440–458 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush, K. & Bradford, P. A. Epidemiology of β-Lactamase-producing pathogens. Clin. Microbiol. Rev.33, 10.1128/cmr.00047-19 (2020). [DOI] [PMC free article] [PubMed]
- 30.Ramos-Castañeda, J. A. et al. Mortality due to KPC carbapenemase-producing Klebsiella pneumoniae infections: Systematic review and meta-analysis: Mortality due to KPC Klebsiella pneumoniae infections. J. Infect.76, 438–448 (2018). [DOI] [PubMed] [Google Scholar]
- 31.Boyd, S. E. et al. OXA-48-Like β-Lactamases: Global epidemiology, treatment options, and development pipeline. Antimicrob. Agents Chemother.66, e0021622 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.UKHSA, Quarterly laboratory surveillance of acquired carbapenemase producing Gram-negative bacteria in England:October 2020 to June 2023. (2023).
- 33.Mairi, A. et al. OXA-48-like carbapenemases producing Enterobacteriaceae in different niches. Eur. J. Clin. Microbiol Infect. Dis.37, 587–604 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Carfora, V. et al. The hazard of carbapenemase (OXA-181)-producing Escherichia coli spreading in pig and veal calf holdings in Italy in the genomics era: Risk of spill over and spill back between humans and animals. Front. Microbiol.13, 1016895 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Vijayakumar, S. et al. Insights into the complete genomes of carbapenem-resistant Acinetobacter baumannii harbouring bla (OXA-23,) bla (OXA-420) and bla (NDM-1) genes using a hybrid-assembly approach. Access Microbiol.2, acmi000140 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dogonchi, A. A. et al. Metallo-β-lactamase-mediated resistance among clinical carbapenem-resistant Pseudomonas aeruginosa isolates in northern Iran: A potential threat to clinical therapeutics. Ci Ji Yi Xue Za Zhi.30, 90–96 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yong, D. et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother.53, 5046–5054 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Koo, V. S., O’Neill, P. & Elves, A. Multidrug-resistant NDM-1 Klebsiella outbreak and infection control in endoscopic urology. BJU Int.110, E922–E926 (2012). [DOI] [PubMed] [Google Scholar]
- 39.Magiorakos, A. P. et al. Infection prevention and control measures and tools for the prevention of entry of carbapenem-resistant Enterobacteriaceae into healthcare settings: guidance from the European Centre for Disease Prevention and Control. Antimicrob. Resist. Infect. Control6, 113 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.EUCAST, Guidelines for Detection of Resistance Mechanisms and Specific Resistance of Clinical and/or Epidemiological Importance, Version 2.0. (2017).
- 41.CLSI, Performance Standards for Antimicrobial Susceptibility Testing—Thirtieth Edition: M100. (2020).
- 42.Spanu, T. et al. Evaluation of the new VITEK 2 extended-spectrum beta-lactamase (ESBL) test for rapid detection of ESBL production in Enterobacteriaceae isolates. J. Clin. Microbiol.44, 3257–3262 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.El-Jade, M. R. et al. ESBL Detection: Comparison of a commercially available chromogenic test for third generation cephalosporine resistance and automated susceptibility testing in Enterobactericeae. PLoS ONE11, e0160203 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Demord, A. et al. Rapid ESBL NP test for rapid detection of expanded-spectrum β-Lactamase producers in enterobacterales. Micro. Drug Resist.27, 1131–1135 (2021). [DOI] [PubMed] [Google Scholar]
- 45.Laurent, T. et al. Evaluation of the βLACTA test, a novel commercial chromogenic test for rapid detection of ceftazidime-nonsusceptible Pseudomonas aeruginosa isolates. J. Clin. Microbiol.51, 1951–1954 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fang, H. et al. Evaluation of a lateral flow immunoassay for rapid detection of CTX-M producers from blood cultures. Microorganisms11, 1 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gupta, G., Tak, V. & Mathur, P. Detection of AmpC β Lactamases in Gram-negative Bacteria. J. Lab Physicians6, 1–6 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Macdougall, C. Beyond susceptible and resistant, Part I: Treatment of infections due to gram-negative organisms with inducible β-Lactamases. J. Pediatr. Pharm. Ther.16, 23–30 (2011). [PMC free article] [PubMed] [Google Scholar]
- 49.Maraskolhe, D. L. et al. Comparision of three laboratory tests for detection of AmpC β Lactamases in Klebsiella species and E. Coli. J. Clin. Diagn. Res. 8, Dc05–Dc08 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Derbyshire, H. et al. A simple disc diffusion method for detecting AmpC and extended-spectrum β-lactamases in clinical isolates of Enterobacteriaceae. J. Antimicrob. Chemother.63, 497–501 (2009). [DOI] [PubMed] [Google Scholar]
- 51.Peter-Getzlaff, S. et al. Detection of AmpC beta-lactamase in Escherichia coli: comparison of three phenotypic confirmation assays and genetic analysis. J. Clin. Microbiol.49, 2924–2932 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Giske, C. G. et al. A sensitive and specific phenotypic assay for detection of metallo-β-lactamases and KPC in Klebsiella pneumoniae with the use of meropenem disks supplemented with aminophenylboronic acid, dipicolinic acid and cloxacillin. Clin. Microbiol. Infect.17, 552–556 (2011). [DOI] [PubMed] [Google Scholar]
- 53.Glupczynski, Y. et al. Evaluation of two new commercial immunochromatographic assays for the rapid detection of OXA-48 and KPC carbapenemases from cultured bacteria. J. Antimicrob. Chemother.71, 1217–1222 (2016). [DOI] [PubMed] [Google Scholar]
- 54.Rabaan, A. A. et al. An overview on phenotypic and genotypic characterisation of carbapenem-resistant enterobacterales. Medicina58, 1675 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Dortet, L., Poirel, L. & Nordmann, P. Rapid identification of carbapenemase types in Enterobacteriaceae and Pseudomonas spp. by using a biochemical test. Antimicrob. Agents Chemother.56, 6437–6440 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Literacka, E. et al. Evaluation of the Carba NP test for carbapenemase detection in Enterobacteriaceae, Pseudomonas spp. and Acinetobacter spp., and its practical use in the routine work of a national reference laboratory for susceptibility testing. Eur. J. Clin. Microbiol. Infect. Dis.36, 2281–2287 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Takissian, J. et al. NG-Test carba 5 for rapid detection of carbapenemase-producing enterobacterales from positive blood cultures. Antimicrob. Agents Chemother.63, 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.MacDonald, J. W. & Chibabhai, V. Evaluation of the RESIST-4 O.K.N.V immunochromatographic lateral flow assay for the rapid detection of OXA-48, KPC, NDM and VIM carbapenemases from cultured isolates. Access Microbiol.1, e000031 (2019). [DOI] [PMC free article] [PubMed]
- 59.Husna, A. et al. Extended-spectrum β-Lactamases (ESBL): Challenges and opportunities. Biomedicines11, 2937 (2023). [DOI] [PMC free article] [PubMed]
- 60.Upadhyay, S., Sen, M. R. & Bhattacharjee, A. Presence of different beta-lactamase classes among clinical isolates of Pseudomonas aeruginosa expressing AmpC beta-lactamase enzyme. J. Infect. Dev. Ctries.4, 239–242 (2010). [DOI] [PubMed] [Google Scholar]
- 61.Pereckaite, L., Tatarunas, V. & Giedraitiene, A. Current antimicrobial susceptibility testing for beta-lactamase-producing Enterobacteriaceae in clinical settings. J. Microbiol. Methods152, 154–164 (2018). [DOI] [PubMed] [Google Scholar]
- 62.Livermore, D. M. & Brown, D. F. J. Detection of β-lactamase-mediated resistance. J. Antimicrobia. Chemother.48, 59–64 (2001). [DOI] [PubMed] [Google Scholar]
- 63.Lutgring, J. D. & Limbago, B. M. The problem of carbapenemase-producing-carbapenem-resistant-enterobacteriaceae detection. J. Clin. Microbiol.54, 529–534 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wand, M. E. et al. Characterization of pre-antibiotic era Klebsiella pneumoniae isolates with respect to antibiotic/disinfectant susceptibility and virulence in Galleria mellonella. Antimicrob. Agents Chemother.59, 3966–3972 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Davin-Regli, A. & Pagès, J. M. Enterobacter aerogenes and Enterobacter cloacae; versatile bacterial pathogens confronting antibiotic treatment. Front. Microbiol.6, 392 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cohenford, M. A., Abraham, J. & Medeiros, A. A. A colorimetric procedure for measuring beta-lactamase activity. Anal. Biochem.168, 252–258 (1988). [DOI] [PubMed] [Google Scholar]
- 67.Harris, P. N. A. et al. Effect of piperacillin-tazobactam vs meropenem on 30-day mortality for patients with E coli or klebsiella pneumoniae bloodstream infection and ceftriaxone resistance: A randomized clinical trial. Jama320, 984–994 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Henderson, A. et al. Association between minimum inhibitory concentration, beta-lactamase genes and mortality for patients treated with piperacillin/tazobactam or meropenem from the MERINO study. Clin. Infect. Dis.73, e3842–e3850 (2021). [DOI] [PubMed] [Google Scholar]
- 69.Grant, J. & Afra, K. Point-counterpoint: The MERINO trial and what it should imply for future treatment of ESBL bacteremia. J. Assoc. Med. Microbiol. Infect. Dis. Can.4, 125–130 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wong-Beringer, A. et al. Molecular correlation for the treatment outcomes in bloodstream infections caused by Escherichia coli and Klebsiella pneumoniae with reduced susceptibility to ceftazidime. Clin. Infect. Dis.34, 135–146 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Robberts, F. J., Kohner, P. C. & Patel, R. Unreliable extended-spectrum beta-lactamase detection in the presence of plasmid-mediated AmpC in Escherichia coli clinical isolates. J. Clin. Microbiol.47, 358–361 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rodríguez-Baño, J. et al. Treatment of infections caused by extended-spectrum-beta-lactamase-, AmpC-, and carbapenemase-producing Enterobacteriaceae. Clin. Microbiol. Rev.31, e00079-17 (2018). [DOI] [PMC free article] [PubMed]
- 73.Vasikasin, V. et al. Can precision antibiotic prescribing help prevent the spread of carbapenem-resistant organisms in the hospital setting? JAC Antimicrob. Resist.5, dlad036 (2023). [DOI] [PMC free article] [PubMed]
- 74.Yahav, D. et al. New β-Lactam-β-Lactamase inhibitor combinations. Clin. Microbiol. Rev.34, e00115-20 (2020). [DOI] [PMC free article] [PubMed]
- 75.Tayşi, M. R. et al. Implementation of the EUCAST rapid antimicrobial susceptibility test (RAST) for carbapenemase/ESBL-producing Escherichia coli and Klebsiella pneumoniae isolates, and its effect on mortality. J. Antimicrob. Chemother.79, 1540–1546 (2024). [DOI] [PubMed]
- 76.Lawrence, J. et al. A NOVEL METHOD FOR IN - VITRO BETA-LACTAMASE QUANTIFICATION: PROOF-OF-CONCEPT. Int. J. Infect. Dis.130, S110 (2023). [Google Scholar]
- 77.Spencer, D. C. et al. A fast impedance-based antimicrobial susceptibility test. Nat. Commun.11, 5328 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Batisti Biffignandi, G. et al. Optimising machine learning prediction of minimum inhibitory concentrations in Klebsiella pneumoniae. Microb. Genom.10, 10.1099/mgen.0.001222 (2024). [DOI] [PMC free article] [PubMed]