Abstract
Tuberculosis (TB) remains one of the leading infectious disease killers in the world. The ongoing development of novel anti-TB medications has yielded potent compounds that often target single sites with well-defined mechanisms of action. However, despite the identification of resistance-associated mutations through target deconvolution studies, comparing these findings with the diverse Mycobacterium tuberculosis populations observed in clinical settings is often challenging. To address this gap, we constructed an open-access database encompassing genetic variations from > 50,000 clinical isolates, spanning the entirety of the M. tuberculosis protein-encoding genome. This resource offers a valuable tool for investigating the prevalence of target-based resistance mutations in any drug target within clinical contexts. To demonstrate the practical application of this dataset in drug discovery, we focused on drug targets currently undergoing phase II clinical trials. By juxtaposing genetic variations of these targets with resistance mutations derived from laboratory-adapted strains, we identified multiple positions across three targets harbouring resistance-associated mutations already present in clinical isolates. Furthermore, our analysis revealed a discernible correlation between genetic diversity within each protein and their predicted essentiality. This meta-analysis, openly accessible via a dedicated dashboard, enables comprehensive exploration of genetic diversity pertaining to any drug target or resistance determinant in M. tuberculosis.
Keywords: Mycobacterium tuberculosis, Clinical isolate, Genetic diversity, Non-synonymous changes, Conservation
Subject terms: Genome informatics, Genetic databases, Genetic variation, Predictive markers, Antimicrobial resistance
Introduction
Tuberculosis (TB) is caused by infection with the bacterial pathogen Mycobacterium tuberculosis and, after COVID-19, stands as the leading infectious disease killer globally, with an estimated 1.3 million deaths in 20221. The success of existing treatment regimens is challenged by their long duration and associated drug toxicity, leading to reduced compliance and the emergence of drug resistance, with approximately 0.4 million cases of drug-resistant TB infection reported in 20221. To combat the threat of the increasing TB disease burden, drug discovery efforts are focused on developing shorter treatment regimens, improving the treatment of drug-resistant TB infections, and minimising drug resistance2.
Many new antitubercular drugs are being developed by target-based structure-enabled drug discovery2. However, the single-target nature of these compounds renders them more susceptible to the emergence of drug resistance through, for instance, target-based mutations such as single nucleotide polymorphisms. Identification of resistance-conferring genetic changes, such as mutations, is often the first step in target identification studies and subsequent early-stage drug development. However, these mutations and/or genetic changes are rarely compared against genetic data from clinical isolates to investigate whether resistance is already present or bacteria with these mutations are viable in the clinic. This comparison would require determining a baseline of genetic variance of every M. tuberculosis protein using sequencing data from clinical TB strains. This would allow investigation of whether populations with these known resistance-conferring mutations are present and thus could survive and proliferate under future drug pressure.
Previous literature has demonstrated that essential genes tend to exhibit higher levels of genetic conservation compared to non-essential genes, suggesting an evolutionary advantage of maintaining sequence integrity. Evolutionary studies across various organisms, including bacteria like Escherichia coli and Bacillus subtilis, as well as eukaryotes, have consistently observed this trend3,4. The conservation of essential genes across divergent species underscores their fundamental role in cellular processes and highlights their significance in maintaining fitness5. Additional studies have further supported these findings through evolutionary conservation analysis between essential and non-essential genes in bacterial genomes, providing additional evidence for the selective pressure acting on essential genes to maintain their sequence conservation6,7. Understanding the evolutionary dynamics of essential genes provides valuable insights into the genetic determinants of cellular viability and may inform strategies for drug target prioritisation and antimicrobial drug development.
Here, we established an open-access database comprising genetic variations extracted from > 50,000 clinical isolates of M. tuberculosis, encompassing the entire protein-encoding genome. Through comprehensive analysis, we juxtaposed the genetic diversity of each protein against sequence conservation within the Mycobacterium genus and gene vulnerability scores8. Notably, this revealed a robust correlation between essentiality and genetic conservation across the proteome. To illustrate the utility of this dataset in terms of drug discovery, we focused on four targets of compounds currently in phase II clinical development. By comparing the genetic variability of these targets with known resistance-associated mutations derived from lab-adapted strains, we pinpointed 14 amino acid positions across three drug targets containing resistance-conferring mutations observed in clinical isolates. Accessible through a dedicated dashboard, this freely available meta-analysis presents an invaluable resource for interrogating the genetic diversity of drug targets and resistance determinants within M. tuberculosis.
Results
Construction of a genetic diversity dataset from M. tuberculosis clinical isolates
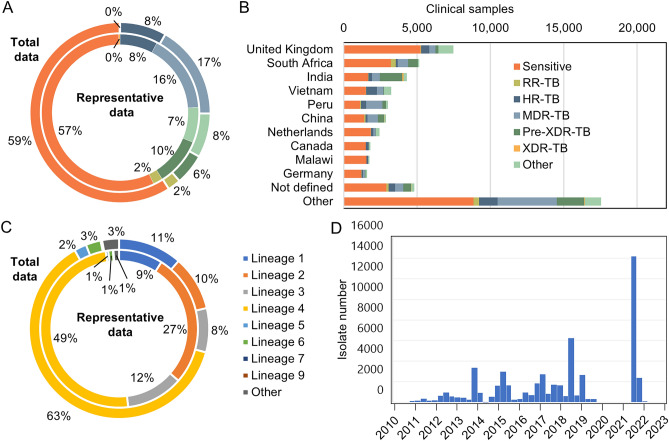
To compile a robust dataset for comprehensive exploration of genetic diversity, we aggregated previously deposited whole-genome sequences from clinical isolates of M. tuberculosis and consolidated them into an accessible dashboard. This dataset encompasses 51,183 genomic sequences obtained from TB clinical isolates derived from infected patients. Our subsequent analysis prioritised non-synonymous mutations, indels, and genomic deletions, facilitating an in-depth meta-analysis of genetic variations across every protein encoded by M. tuberculosis. We focused only on protein-coding genes since the intended application of this dataset was to allow investigation of the presence of target-based non-synonymous changes in circulating clinical isolates of M. tuberculosis. This means that genetic differences in ribosomal RNA genes including rrs and rrl, linked to resistance to streptomycin and linezolid, respectively, are not included in this analysis. A total of 1,063,811 non-synonymous changes across 694,579 sites were observed in 4029 protein coding genes. We identified an average of 173 changes per gene, and when normalised to protein length, implicated an average of 49% of sites containing a polymorphism. The conservation at sites was generally quite high, with mean gene conservation values of 99.93%. We also isolated a smaller representative dataset of 5844 samples that better reflected the underlying genetic diversity. Compared to this representative dataset, our full dataset had similar distribution of drug susceptibility (Fig. 1A). As expected, for the full dataset, samples were mainly deposited by countries with lower TB incidence but higher whole genome sequencing capacity (Fig. 1B). Indeed, most of underrepresentation was in high burden countries in the global south especially Asia (China, the Philippines, India, Indonesia). Our full dataset was slightly over-represented for lineage 4 samples and underrepresented for lineage 2 as well as lineages 5–9 (Fig. 1C). Whilst accurate metadata of the date of collection was not available for most of the clinical samples, all the sample data collated in our dataset was deposited between 2010 and 2023 (Fig. 1D). This final dataset represents a comprehensive catalogue of genetic variance from clinical isolates for every protein in M. tuberculosis.
Fig. 1.
Overview of the data collection and analysis–Comparison of actual or predicted drug responsiveness for the (A) total dataset (outer ring) compared to the representative sample set (inner ring) and (B) based on the originating country for the total dataset. (C) Lineage of the total dataset (outer ring) compared with the representative dataset (inner ring). (D) Histogram of date when clinical isolate was deposited.
To make this extensive resource more accessible to the TB drug discovery research community, we established a user-friendly interface for data interrogation (https://www.lshtm.ac.uk/research/centres-projects-groups/satellite-centre-for-global-health-discovery#genetic-diversity). This dashboard can be used to investigate the genetic variance of any protein of interest in M. tuberculosis whilst also providing metrics such as conservation between closely related Mycobacterium species.
Genetic diversity and species conservation of genes correlate with gene vulnerability
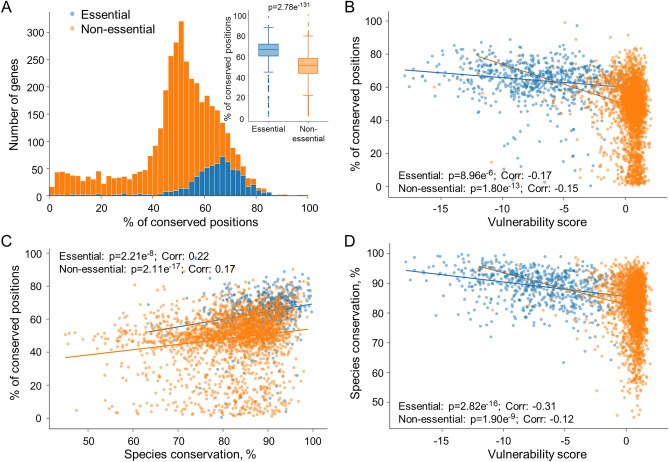
To compare essentiality and conservation on a global scale, we used our extensive database to compare genetic variance between clinical isolates against gene vulnerability scores, a measure of gene essentiality, identified through a genome-wide CRISPRi-mediated essentiality screen8 (Fig. 2A,B). These relationships were investigated separately for genes classed as essential and non-essential (based on the CRISPRi-mediated essentiality screen8) as these groups formed distinct clusters within the feature space. Firstly, this showed a statistical difference in the level of genetic variance of coding regions between genes classed as either essential or non-essential genes (Fig. 2A). Secondly, essential genes were more likely to have a higher number of conserved positions compared to non-essential genes. Indeed, there was a clear correlation between the vulnerability score, which uses Bayesian modelling to quantify the vulnerability of each gene, and the genetic variance between clinical isolates (Fig. 2B).
Fig. 2.
Genome-wide comparison of gene vulnerability, genetic diversity and species conservation–(A) Histogram of genes per percentage of polymorphic positions. Insert shows direct comparison between genes predicted to be essential verses non-essential. (B) Comparison of genetic diversity (% of positions that are completely conserved amongst all isolates) and gene vulnerability score8. (C) Comparison of genetic diversity and species conservation (between mycobacterium species). (D) Comparison of gene vulnerability score and species conservation. Genes are colour-coded based on essentiality.
Aside from genetic variance within a species, there is also evidence that genetic conservation between bacterial species is directly linked to gene essentiality4. The genus of Mycobacterium contains over 190 species, with the most commonly known members including M. tuberculosis and M. leprae, the causative agent of leprosy. Members of the genus are identified by their waxy, lipid rich cell walls consisting of mycolic acid. Within our dashboard, we have included the amino acid sequences of six species of this genus including M. abscessus, M. marinum and Mycolicibacterium smegmatis, together containing more than 24,000 protein sequences. Species conservation was then scored based on the average sequence identity between M. tuberculosis and the other species. As with essentiality and vulnerability, there was a statistically significant association between the genetic variance among clinical isolates and the species conservation when analysed via linear ordinary least squares regression (Fig. 2C). This supports previous work that suggests essential genes are more conserved between species in bacteria than non-essential genes. Finally, comparison of the species conservation and the vulnerability scores also revealed a statistically relevant correlation (Fig. 2D; Supplemental Table S1). Taken together this data suggests there is a relationship between the genetic conservation, both in clinical isolates and different species, and the vulnerability and essentiality of M. tuberculosis proteins (Supplemental Table S1).
Identifying inherent drug resistance in new antitubercular drug targets
The main application of this genetic diversity dataset is to provide a baseline for genetic variance of any drug target of new or future clinical compounds to measure future population dynamics. This would identify both inherent resistance within the population but also provide an indication of whether target-based mutations observed in lab-generated resistance strains could be viable in clinical isolates. As demonstration of the utility of our dashboard, we focused on the respective drug targets of four compounds undergoing stage II clinical trials for the treatment of TB: (i) SQ109, an 1,2-ethylenediamine, that targets the mycolic acid transporter, MmpL39; (ii) GSK070, an oxaborole derivative that inhibits leucyl tRNA synthetase (LeuS)10; (iii) BTZ-043, a benzothiazinone, shown to inhibit DprE111 and; (iv) Q203 (Telacebec) an imidazopyridine amide, known to target the cytochrome bc1 complex, specifically QcrB12.
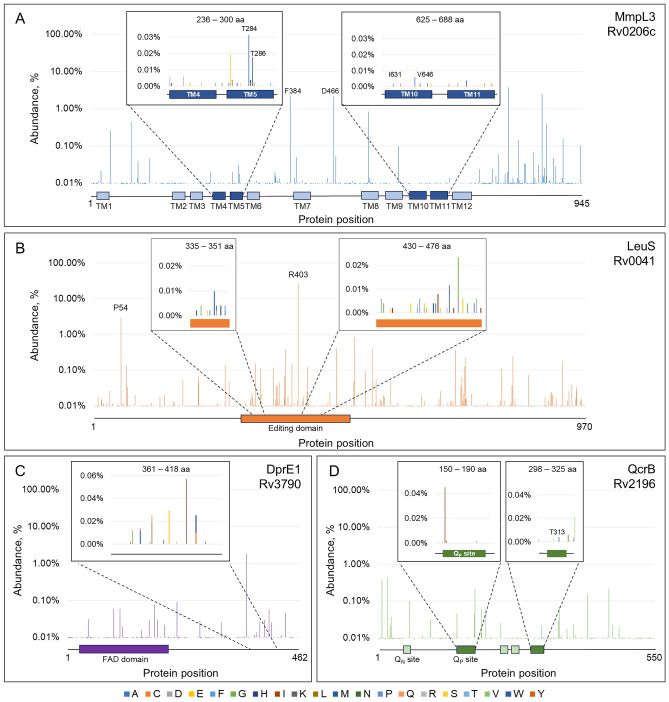
MmpL3 (Rv0206c) belongs to the Resistance, Nodulation and Division (RND) superfamily and transports trehalose monomycolate for cell wall biogenesis. While SQ109 is the most advanced compound to target MmpL3, there are multiple classes of compounds shown to inhibit this promiscuous drug target. During the development of these compounds, 136 unique amino acid changes have been identified in 83 different positions within the MmpL3 protein from a range of Mycobacterium species, predominately M. tuberculosis13. While many of these mutations have not been associated directly with SQ109 resistance, it is conceivable that several will lead to cross-resistance. We analysed our genetic diversity dataset to identify mutations in the MmpL3 coding sequence (Fig. 3A). SQ109 is predicted to interact with transmembrane domains (TMs) 4–5 and 10–11 (236–300 and 625–688 aa) of MmpL3, however mutations have been identified covering the whole protein13. Two mutations, unconnected to drug resistance, F384I and D466E, were prominent in our dataset and further investigation revealed that these mutations occurred almost exclusively in samples from lineage 6 and animal associated lineages. This suggests the mutations originated from single acquisition events and likely evolved under neutral evolution. We next compared the genetic diversity of MmpL3 with in vitro lab-generated mutations known to provide resistance to inhibitors of this target13. Our analysis identified genetic variance at 10 amino acid positions that can also maintain resistance-conferring mutations (Table 1; Fig. 3A). Two amino acid positions were particularly enriched with T284A occurring in 17 isolates from L4.5 originating mostly in China and Vietnam and T286M occurring in 8 isolates from L3 mostly with unknown origin as well as two isolates from the United Kingdom. In the absence of selective pressure, this would suggest that these mutations have minimal impact on bacterial growth and could be selected under drug pressure.
Fig. 3.
Genetic diversity of next generation drug targets in current development–Gene-wide genetic variation in (A) MmpL3, (B) LeuS, (C) DprE1 and (D) QcrB. See Supplemental Figs. S1 and S2 for sequence alignment and predicted drug binding regions for LeuS and DprE1. Inserts include regions predicted to interact with inhibitors and where mutations have been identified from lab-adapted resistance strains.
Table 1.
Genetic diversity of clinical isolates compared with known resistance-conferring mutations.
| Target | Gene ID | Mtb position | Mutation observed | Genetic diversity | Location of clinical isolates |
|---|---|---|---|---|---|
| Mmpl3 | Rv0206c | Q40 | Q40R, Q40H | Q 99.98%; R < 0.01%; − 0.02% | Portugal |
| T284 | T284A | T 99.95%; A 0.03%; − 0.02% | China, Vietnam | ||
| T286 | T286K (+ L320P M. bovis) | T 99.97%; K < 0.01%; M 0.02%; − 0.01% | UK, Thailand | ||
| N365 | N365S (+ V581A, V681I) | N 99.99%; K < 0.01%; S < 0.01%; − < 0.01% | India | ||
| M393 | M384I (M. abscessus) | M 99.99%; I < 0.01%; − < 0.01% | India | ||
| M492 | M492T (+ V564A, V681I) | M 99.99%; T < 0.01%; −0.01% | Malawi | ||
| I631 | I616F (+M313I M. abscessus) | I > 99.99%; F < 0.01%; − < 0.01% | |||
| V646 | V646M (+ F255L) | V 99.99%; M < 0.01%; − < 0.01% | UK | ||
| A700 | A700T | A 99.99%; T < 0.01%; −0.01% | Pakistan | ||
| V713 | V713M | V > 99.99%; L < 0.01%; M < 0.01% | Ethiopia, Netherlands | ||
| LeuS | Rv0041 | V482 | V468L (M. abscessus) | V 99.98%; L 0.01%; −0.01% | Belgium, Germany, Malawi |
| K516 | K502E (M. abscessus) | K 99.99%; E < 0.01%; −0.01% | India | ||
| QcrB | Rv2196 | T313 | T313A | T 99.99%; A < 0.01%; − < 0.01% | |
| M342 | M342V | M 99.98%; V 0.01%; −0.01% | Ghana, Italy, UK |
A: alanine; C: cysteine; D: aspartic acid; E: glutamic acid; F: phenylalanine; G: glycine; H: histidine; I: isoleucine; K: lysine; L: leucine; M: methionine; N: asparagine; P: proline; Q: glutamine; R: arginine; S: serine; T: threonine; V: valine; W: tryptophan; Y: tyrosine.
The oxaborole derivative, GSK070, is the most advanced of several chemical series targeting LeuS (Rv0041). The compounds are predicted to bind within the editing domain and multiple target-based mutations have been identified in in vitro experiments, predominately in the related pathogen, M. abscessus14–20 (Fig. 3B; Supplemental Fig. S1; Supplemental Table S2). The most prominent genetic variations, not connected to drug resistance, was seen at positions P54 (L4.2 associated) and R403 (L2.2 associated). These mutations occurred mostly in a single clade and thus probably only originated in the absence of selective pressure. In terms of drug resistance, two mutations–V468L and K502E (equivalent to V482 and K516 in M. tuberculosis)—identified in lab-adapted M. abscessus resistant strains were observed in our genetic diversity dataset (Table 1). DprE1 (Rv3790) is also a promiscuous target in TB drug discovery with multiple compounds inhibiting this target including BTZ-043. Indeed, a myriad of mutations providing resistance to DprE1 inhibitors have been identified from lab-adapted resistance strains largely associated with the compound binding region11,21–26 (Supplemental Fig. S2; Supplemental Table S3). A significant enrichment of A356T was observed which was associated with isolates from L1.2.1.2 (Fig. 3C), however, no genetic variance was observed that correlated with known resistance-conferring mutations. Finally, the cytochrome bc1 complex, specifically the QcrB subunit (Rv2196), is the target of multiple compounds including Q203 and several resistance-conferring mutations have been identified in the Qp (or Qo) site where the compounds are predicted to bind12,27–36 (Supplemental Table S4). Whilst, there was no major enrichment of genetic variance in this target, two known resistance-conferring mutations—T313A and M342V–were observed (Table 1; Fig. 3D; Supplemental Table S4). It is noteworthy to mention that for all of the drug targets discussed here, the genetic variance was noticeably lower in the predicted drug binding sites compared to the surrounding protein sequence.
Conclusion
In this study, we curated an extensive repository of genetic variants derived from whole genome sequencing data obtained from > 50,000 clinical isolates of M. tuberculosis collected since 2010. This resource encompasses genetic diversity profiles for more than 4000 genes of M. tuberculosis, conveniently accessible to the research community through a dedicated dashboard. Our analysis of this dataset unveiled a notable pattern: essential genes in Mycobacteria exhibit higher levels of conservation compared to non-essential genes, evident both within clinical isolates and across closely related species. This dataset serves as a valuable tool for exploring the genetic landscape of potential drug targets or resistance determinants. To demonstrate its utility, we focused on several targets of compounds currently undergoing phase II clinical trials. Our investigation revealed that while significant resistance was not prevalent in clinical settings, instances of laboratory-induced resistance-conferring mutations were detected in clinical isolates. Importantly, our findings suggest that these mutations do not compromise bacterial survival in patients and could therefore potentially emerge under drug pressure.
Our analyses of species conservation and genetic diversity focused on the entire coding region rather than specific functional domains or regions involved in compound binding. It is conceivable that these domain regions exhibit higher conservation and may more accurately reflect the hypothesis that increased conservation correlates with greater essentiality. Additionally, gene vulnerability scores and essentiality predictions were determined under in vitro growth conditions, which may inaccurately classify certain genes as non-essential, such as those essential only during host infection. By examining published resistance-conferring mutations, we demonstrate the viability of some of these mutations within clinical populations, evidenced by the presence of several clinical isolates exhibiting inherent resistance. This underscores the considerable potential for these mutations, already established as viable in clinical settings, to be rapidly selected under drug pressure. However, our analysis is constrained by the availability of known resistance mutations, as it is highly probable that many resistance-conferring mutations remain unidentified, particularly for drug targets in early developmental stages. This highlights the urgent need for a comprehensive catalogue of resistance mutations prior to clinical deployment, which would serve as crucial resistance markers for future surveillance efforts. Furthermore, for several drugs in development, as well as those already in the clinic, the dominant mechanism of resistance are non-target based resistance mechanisms. For example, disruption of Rv0678 (MmpS5-MmpL5 efflux pump repressor) either by mutation, indel and/or frameshift leads to resistance against a broad spectrum of antibiotics including bedaquiline, clofazimine and Q20337. Finally, our dataset was assembled using available sequences mainly deposited in countries with low TB incidence but higher sequencing capacity. Whilst this is unavoidable, there is a potential that this distorts the true global distribution of clinical isolates. This is further compounded by the limited metadata attached to each isolate sequence, making it more challenging, for example, to identify ‘hotspots’ of potential resistance.
This comprehensive catalogue of genetic variance provides a baseline reference of M. tuberculosis genetic diversity on a single-gene level and can assist in future surveillance of drug resistance. Overall, the genetic baseline generated through our collated database and dashboard provides valuable information for understanding the genetic composition of M. tuberculosis populations in the clinic.
Methods
The dataset was assembled from publicly available published sequence data (Supplementary Table S5). Sequence data in fastq format was downloaded from the European Nucleotide Archive (ENA). Reads were trimmed using Trimmomatic38 (v0.39; parameters: LEADING:3 TRAILING:3 SLIDINGWINDOW:4:20 MINLEN:36). Trimmed reads were aligned to the H37Rv reference genome (accession: NC000962.3) using bwa39 (v0.1.17). Duplicate aligned reads were marked with Samtools40 markdup (v1.12). Variants were then called using freebayes41 (v3.1.6) for each sample. For each sample, the predicted protein sequence was inferred by substituting in the variant calls into the reference and translating all genomic coding sequences into amino acid sequences. A custom script using biopython (v1.84; github.com/jodyphelan/genetic-diversity-db) was used to substitute variants into the reference genome. Low depth regions from each sample were inferred using Samtools depth and was used to mask the genomic sequence. Separately, TB-Profiler42 (v4.4.0; database version: e25540b) was used to predict drug resistance profile based on the presence of known drug resistance mutations and to assign a strain type. Linked data such as country of collection and date of collection were extracted from associated publications. To reduce the effect of oversampling from certain strain types, a subset of the data representative of the genetic diversity was generated by randomly selecting up to 10 different sequences from 117 different sublineages found by TB-Profiler. The percentage of polymorphic positions per gene was defined as the percentage of positions in a gene that have a polymorphism.
Species based conservation was calculated using protein sequences from Mycobacterium tuberculosis (ASM19595v2), M. abscessus (ASM6918v1), M. avium (ASM1498v1), M. kansasii (ASM15789v2), M. marinum (ASM1834v1) and M. smegmatis (ASM1500v1). Orthologues were found using OrthoFinder43 (v2.2.5) and orthogroups which had a single M. tuberculosis sequence were aligned using mafft44 (v7.520). Conservation by site was calculated at the percentage of alleles that were identical to the majority call. Gene conservation was calculated as the average of conservation across all sites. Association between the percent polymorphic positions across M. tuberculosis, species conservation and vulnerability scores8 was investigated by performing linear ordinary least squares regression using the regression.linear_model.OLS function from the statsmodels45 python package (v0.14.1). Figures were created using the plotly python package (v5.20.0). Sequence alignment was generated using ESPript 3.x software46.
Data preparation and ETL (extract, transform, and load) for aligned sequences was done using python version 3.9.7 to obtain the desired level of aggregation and output format for Tableau. The dashboard was developed in Tableau Desktop version 2023.3 and published to Tableau Public. The dashboard was subsequently integrated to the LSHTM website (https://www.lshtm.ac.uk/research/centres-projects-groups/satellite-centre-for-global-health-discovery#genetic-diversity) through an embedding code from Tableau Embedding API.
Supplementary Information
Acknowledgements
The authors would like to thank Anne-Theres Henze (Akkodis Belgium) for medical writing support on behalf of Janssen Pharmaceutica NV. This work was funded by a grant from Janssen Pharmaceutica to the London School of Hygiene & Tropical Medicine.
Author contributions
R.J.W., D.A.L. and A.K. conceived the project; J.P. compiled and analysed the clinical isolate dataset; K.V.H. assembled the dashboard interface; S.M. and R.V. supervised the dashboard assembly; R.J.W. wrote the manuscript with input from other authors. All authors reviewed the manuscript and approved the final version for publication.
Data availability
The datasets used in this study are publicly available via the European Nucleotide Archive (ENA) and the accession numbers are provided in Supplementary Table S5. Access to the dashboard generated in this project is available from the LSHTM website (https://www.lshtm.ac.uk/research/centres-projects-groups/satellite-centre-for-global-health-discovery#genetic-diversity).
Competing interests
KVdH, SM, RV, DAL and AK were/are all full-time employees or external contractors of Janssen, a Johnson & Johnson company, and potential stockholders of Johnson & Johnson. The other authors declare no competing interests.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Anil Koul, Email: anil.koul@lshtm.ac.uk.
Richard J. Wall, Email: richard.wall@lshtm.ac.uk
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-024-75818-y.
References
- 1.World Health Organisation. Global tuberculosis report. https://www.who.int/teams/global-tuberculosis-programme/tb-reports/global-tuberculosis-report-2023/tb-disease-burden/1-2-tb-mortality (2023).
- 2.Dartois, V. A. & Rubin, E. J. Anti-tuberculosis treatment strategies and drug development: Challenges and priorities. Nat. Rev. Microbiol.20, 685–701. 10.1038/s41579-022-00731-y (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rocha, E. P. C. Inference and analysis of the relative stability of bacterial chromosomes. Mol. Biol. Evol.23, 513–522. 10.1093/molbev/msj052 (2005). [DOI] [PubMed] [Google Scholar]
- 4.Jordan, I. K., Rogozin, I. B., Wolf, Y. I. & Koonin, E. V. Essential genes are more evolutionarily conserved than are nonessential genes in bacteria. Genome Res.12, 962–968. 10.1101/gr.87702 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Pál, C., Papp, B. & Lercher, M. J. Adaptive evolution of bacterial metabolic networks by horizontal gene transfer. Nat. Genet.Bold">37, 1372–1375. 10.1038/ng1686 (2005). [DOI] [PubMed] [Google Scholar]
- 6.Luo, H., Gao, F. & Lin, Y. Evolutionary conservation analysis between the essential and nonessential genes in bacterial genomes. Sci. Rep.5, 13210. 10.1038/srep13210 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gong, X. et al. Comparative analysis of essential genes and nonessential genes in Escherichia coli K12. Mol. Genet. Genomics279, 87–94. 10.1007/s00438-007-0298-x (2008). [DOI] [PubMed] [Google Scholar]
- 8.Bosch, B. et al. Genome-wide gene expression tuning reveals diverse vulnerabilities of M. tuberculosis. Cell184, 4579-4592.e4524. 10.1016/j.cell.2021.06.033 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tahlan, K. et al. SQ109 targets MmpL3, a membrane transporter of trehalose monomycolate involved in mycolic acid donation to the cell wall core of Mycobacterium tuberculosis. Antimicrob. Agents Chemother.56, 1797–1809. 10.1128/aac.05708-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li, X. et al. Discovery of a potent and specific M. tuberculosis leucyl-tRNA synthetase inhibitor: (S)-3-(Aminomethyl)-4-chloro-7-(2-hydroxyethoxy)benzo[c][1,2]oxaborol-1(3H)-ol (GSK656). J. Med. Chem.60, 8011–8026. 10.1021/acs.jmedchem.7b00631 (2017). [DOI] [PubMed] [Google Scholar]
- 11.Makarov, V. et al. Benzothiazinones kill Mycobacterium tuberculosis by blocking Arabinan synthesis. Science324, 801–804. 10.1126/science.1171583 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pethe, K. et al. Discovery of Q203, a potent clinical candidate for the treatment of tuberculosis. Nat. Med.19, 1157–1160. 10.1038/nm.3262 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Adams, O. et al. Cryo-EM structure and resistance landscape of M. tuberculosis MmpL3: An emergent therapeutic target. Structure29, 1182-1191.e1184. 10.1016/j.str.2021.06.013 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nguyen, T. Q. et al. DS86760016, a Leucyl-tRNA Synthetase inhibitor, is active against Mycobacterium abscessus. Antimicrob. Agents Chemother.67, e01567-e1522. 10.1128/aac.01567-22 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu, W. et al. A novel leucyl-tRNA synthetase inhibitor, MRX-6038, expresses anti-Mycobacterium abscessus activity in vitro and in vivo. Antimicrob. Agents Chemother.66, e00601-00622. 10.1128/aac.00601-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ganapathy, U. S., Gengenbacher, M. & Dick, T. Epetraborole is active against Mycobacterium abscessus. Antimicrob. Agents Chemother.65, 1128. 10.1128/aac.01156-01121 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palencia, A. et al. Discovery of novel oral protein synthesis inhibitors of Mycobacterium tuberculosis that target leucyl-tRNA synthetase. Antimicrob. Agents Chemother.60, 6271–6280. 10.1128/aac.01339-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ganapathy, U. S. et al. A Leucyl-tRNA synthetase inhibitor with broad-spectrum anti-mycobacterial activity. Antimicrob. Agents Chemother.95, 1128. 10.1128/aac.02420-20 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sullivan, J. R. et al. Efficacy of epetraborole against Mycobacterium abscessus is increased with norvaline. PLOS Pathog.17, e1009965. 10.1371/journal.ppat.1009965 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hoffmann, G. et al. Adenosine-dependent activation mechanism of prodrugs targeting an aminoacyl-tRNA synthetase. J. Am. Chem. Soc.145, 800–810. 10.1021/jacs.2c04808 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Neres, J. et al. 2-Carboxyquinoxalines kill Mycobacterium tuberculosis through noncovalent inhibition of DprE1. ACS Chem. Biol.10, 705–714. 10.1021/cb5007163 (2015). [DOI] [PubMed] [Google Scholar]
- 22.Chen, X., Li, Y., Wang, B. & Lu, Y. Identification of mutations associated with Macozinone-resistant in Mycobacterium tuberculosis. Curr. Microbiol.79, 205. 10.1007/s00284-022-02881-x (2022). [DOI] [PubMed] [Google Scholar]
- 23.Sarathy, J. P., Zimmerman, M. D., Gengenbacher, M., Dartois, V. & Dick, T. Mycobacterium tuberculosis DprE1 inhibitor OPC-167832 is active against Mycobacterium abscessus in vitro. Antimicrob. Agents Chemother.66, e01237-e1222. 10.1128/aac.01237-22 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Foo, C.S.-Y. et al. Characterization of DprE1-mediated benzothiazinone resistance in Mycobacterium tuberculosis. Antimicrob. Agents Chemother.60, 6451–6459. 10.1128/aac.01523-16 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Makarov, V. et al. The 8-pyrrole-benzothiazinones are noncovalent inhibitors of DprE1 from Mycobacterium tuberculosis. Antimicrob. Agents Chemother.59, 4446–4452. 10.1128/aac.00778-15 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.de Jesus Lopes Ribeiro, A. L. et al. Analogous mechanisms of resistance to benzothiazinones and dinitrobenzamides in Mycobacterium smegmatis. PLoS ONE6, e26675. 10.1371/journal.pone.0026675 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Harrison, G. A. et al. Identification of 4-amino-thieno[2,3-d]pyrimidines as QcrB inhibitors in Mycobacterium tuberculosis. MSphere4, 1128. 10.1128/msphere.00606-00619 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lupien, A. et al. New 2-ethylthio-4-methylaminoquinazoline derivatives inhibiting two subunits of cytochrome bc1 in Mycobacterium tuberculosis. PLOS Pathog.16, e1008270. 10.1371/journal.ppat.1008270 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Foo, C. S. et al. Arylvinylpiperazine amides, a new class of potent inhibitors targeting QcrB of Mycobacterium tuberculosis. MBio9, 1128. 10.1128/mbio.01276-01218 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chandrasekera, N. S. et al. Improved phenoxyalkylbenzimidazoles with activity against Mycobacterium tuberculosis appear to target QcrB. ACS Infect. Dis.3, 898–916. 10.1021/acsinfecdis.7b00112 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Arora, K. et al. Respiratory flexibility in response to inhibition of cytochrome c oxidase in Mycobacterium tuberculosis. Antimicrob. Agents Chemother.58, 6962–6965. 10.1128/aac.03486-14 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Phummarin, N. et al. SAR and identification of 2-(quinolin-4-yloxy)acetamides as Mycobacterium tuberculosis cytochrome bc1 inhibitors. MedChemComm7, 2122–2127. 10.1039/C6MD00236F (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Subtil, F. T. et al. Activity of 2-(quinolin-4-yloxy)acetamides in Mycobacterium tuberculosis clinical isolates and identification of their molecular target by whole-genome sequencing. Int. J. Antimicrob. Agents51, 378–384. 10.1016/j.ijantimicag.2017.08.023 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Abrahams, K. A. et al. Identification of novel imidazo[1,2-a]pyridine inhibitors targeting M. tuberculosis QcrB. PLoS ONE7, e52951. 10.1371/journal.pone.0052951 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rybniker, J. et al. Lansoprazole is an antituberculous prodrug targeting cytochrome bc1. Nat. Commun.6, 7659. 10.1038/ncomms8659 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cleghorn, L. A. T. et al. Identification of Morpholino thiophenes as novel Mycobacterium tuberculosis inhibitors, targeting QcrB. J. Med. Chem.61, 6592–6608. 10.1021/acs.jmedchem.8b00172 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Waller, N. J. E., Cheung, C. Y., Cook, G. M. & McNeil, M. B. The evolution of antibiotic resistance is associated with collateral drug phenotypes in Mycobacterium tuberculosis. Nat. Commun.14, 1517. 10.1038/s41467-023-37184-7 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bolger, A. M., Lohse, M. & Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics30, 2114–2120. 10.1093/bioinformatics/btu170 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li, H. Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. arXiv:1303.3997 (2013).
- 40.Li, H. et al. The sequence alignment/map format and SAMtools. Bioinformatics25, 2078–2079. 10.1093/bioinformatics/btp352 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Garrison, E. & Marth, G. Haplotype-based variant detection from short-read sequencing. arXiv:1207.3907 (2012).
- 42.Phelan, J. E. et al. Integrating informatics tools and portable sequencing technology for rapid detection of resistance to anti-tuberculous drugs. Genome Med.11, 41. 10.1186/s13073-019-0650-x (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Emms, D. M. & Kelly, S. OrthoFinder: Phylogenetic orthology inference for comparative genomics. Genome Biol.20, 238. 10.1186/s13059-019-1832-y (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Katoh, K., Misawa, K., Kuma, K. I. & Miyata, T. MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucl. Acids Res.30, 3059–3066. 10.1093/nar/gkf436 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seabold, S. & Perktold, J. Statsmodels: Econometric and statistical modeling with Python (eds van der Walt S. & Millman J.) pp. 92–96.
- 46.Robert, X. & Gouet, P. Deciphering key features in protein structures with the new ENDscript server. Nucl. Acids Res.42, W320–W324. 10.1093/nar/gku316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used in this study are publicly available via the European Nucleotide Archive (ENA) and the accession numbers are provided in Supplementary Table S5. Access to the dashboard generated in this project is available from the LSHTM website (https://www.lshtm.ac.uk/research/centres-projects-groups/satellite-centre-for-global-health-discovery#genetic-diversity).