Abstract
Brain signals refer to electrical signals or metabolic changes that occur as a consequence of brain cell activity. Among the various non-invasive measurement methods, electroencephalogram (EEG) stands out as a widely employed technique, providing valuable insights into brain patterns. The deviations observed in EEG reading serve as indicators of abnormal brain activity, which is associated with neurological diseases. Brain‒computer interface (BCI) systems enable the direct extraction and transmission of information from the human brain, facilitating interaction with external devices. Notably, the emergence of artificial intelligence (AI) has had a profound impact on the enhancement of precision and accuracy in BCI technology, thereby broadening the scope of research in this field. AI techniques, encompassing machine learning (ML) and deep learning (DL) models, have demonstrated remarkable success in classifying and predicting various brain diseases. This comprehensive review investigates the application of AI in EEG-based brain disease diagnosis, highlighting advancements in AI algorithms.
Keywords: Brain disease, Electroencephalography, Brain computer interface, Artificial intelligence
Abstract
大脑信号反映了脑细胞活动引起的电信号变化或代谢变化。在各种非侵入性测量方法中,脑电图作为一种广泛应用的技术可以帮助了解大脑模式。脑电图中的异常读数可作为与神经系统疾病相关的脑活动的指标。脑机接口(BCI)系统能够直接从人脑提取和传输信息,从而实现与外部设备的交互。人工智能(AI)的出现极大地提高了BCI技术精度和准确性,并拓宽了该领域的研究范围。AI技术包括机器学习(ML)和深度学习(DL)模型,可以利用脑信号对各种脑部疾病进行分类和预测。本文综述了AI在基于脑电图的脑部疾病诊断中的应用,特别是AI算法在该领域应用的进展。
Keywords: 脑部疾病, 脑电图, 脑机接口, 人工智能
1. Introduction
Brain–computer interface (BCI) technology represents a significant advancement within the field of neuroscience and biomedical engineering, as it facilitates a direct interface between the human brain and external devices. This technology has been used in various domains, including neurorehabilitation (Kwak et al., 2015), disabled motor assistance (Huang et al., 2012), and virtual reality (Lotte et al., 2012). Notably, BCI holds immense potential in brain disease diagnosis, offering a promising alternative to conventional diagnostic approaches that frequently necessitate invasive procedures for biomarker collection or costly imaging techniques. Moreover, these methods encounter the obstacles of real-time monitoring and early detection capabilities. Ever since Hans Berger successfully recorded the first human electroencephalogram (EEG) in 1924, EEG has evolved into an indispensable tool for identifying brain diseases. By continuously capturing the brain’s electrical activity, EEG surpasses other BCI modalities in terms of being non-invasive and cost-effective and possessing a high temporal resolution. EEG technology can provide detailed information about brain function, and the variations in EEG readings serve as indicators of abnormal brain activity associated with neurological disorders.
Additionally, computer-aided diagnosis (CAD) has emerged as a potentially effective detection tool. CAD utilizes artificial intelligence (AI) to automatically process and analyze medical data. The integration of machine learning (ML) and deep learning (DL) methodologies enables the identification of intricate patterns within input data, enhancing the accuracy and reliability of BCI systems and furnishing healthcare practitioners with valuable diagnostic insights. AI techniques, such as support vector machine (SVM), convolutional neural network (CNN), recurrent neural network (RNN), and long short-term memory network (LSTM), have been extensively applied to EEG data for detecting and predicting brain disorders.
Currently, AI-powered EEG diagnostic systems are being developed and refined to enhance their clinical applicability. These systems are capable of real-time monitoring and early detection, which are crucial for timely intervention and treatment. The prospects for AI-based EEG diagnostics are promising. Deepening AI algorithms and increasing computational power are expected to further improve the sensitivity and specificity of these diagnostic tools. Moreover, the development of portable and wearable EEG devices integrated with AI technology is likely to facilitate widespread application in clinical and home settings, enabling continuous monitoring and personalized healthcare.
In this review, we present a comprehensive examination of the diverse ML/DL techniques employed in the diagnosis of brain diseases using EEG data. Specifically, we investigate seven brain disorders, namely epilepsy, schizophrenia, depression, Parkinson’s disease (PD), Alzheimer’s disease (AD), brain stroke, and autism spectrum disorder (ASD).
2. Human EEG
2.1. Invasive EEG and non-invasive EEG
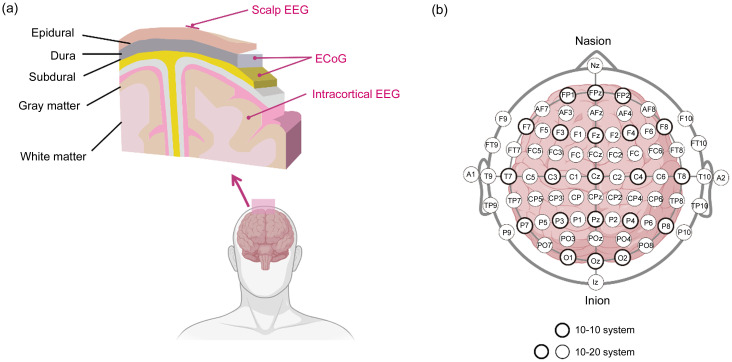
EEG acquisition can be classified into invasive and non-invasive techniques, primarily determined by the placement of electrodes. Invasive EEG entails the insertion of electrodes beneath the skull, including intracortical EEG (iEEG) and electrocorticogram (ECoG). iEEG captures signals from a specific implanted location that represents a restricted subset of neurons. Conversely, ECoG, although less invasive than iEEG, provides a higher signal-to-noise ratio (SNR), wider bandwidth, and more information. Nevertheless, it still entails potential surgical risks. Non-invasive EEG collects signals externally on the scalp, commonly referred to as scalp EEG. This technique provides a high temporal resolution but a relatively low spatial resolution. In Fig. 1a, the gathered scalp EEG, iEEG, and ECoG are depicted, each originating from distinct regions of the cortex.
Fig. 1. Electroencephalogram (EEG) signal types and EEG recording system. (a) Locations of different EEG signals relative to the brain. (b) The 10-10 and 10-20 electrode placement systems. ECoG: electrocorticogram.
2.2. EEG recording system
The EEG recording system employs electrode placement on the scalp, with the international 10-20 and 10-10 systems being the most commonly utilized configurations (Malmivuo and Plonsey, 1995). The former partitions the scalp into intervals of 10% and 20%, resulting in a total of 21 locations for electrode placement, while the latter employs 10% intervals and offers 75 sites for electrode placement. Electrodes are identified by a letter-number code denoting the lobe, hemisphere, and scalp position. Even numbers are indicative of the right hemisphere, while odd numbers denote the left hemisphere. The letters F, T, C, P, and O correspond to the frontal, temporal, central, parietal, and occipital lobes, respectively. A visual representation of the electrode location is shown in Fig. 1b.
In BCI systems, various types of electrodes are employed, namely wet, dry, and semi-dry electrodes. Wet electrodes, typically composed of silver chloride, are utilized for the purpose of achieving stable EEG recordings. However, their usage necessitates the aplication of gel, which entails drawbacks such as prolonged preparation time and discomfort (Ferree et al., 2001). Conversely, dry electrodes, devoid of conducting gel, offer quicker application and better comfort, albeit potentially compromising signal stability. Semi-dry electrodes combine wet and dry technologies, using a minimal quantity of gel to enhance conductivity without incurring the disadvantages associated with wet electrodes (Mota et al., 2013).
2.3. State-dependent EEG signals
EEG signals can be classified into distinct bands known as delta (0.5–4.0 Hz), theta (4–8 Hz), alpha (8–12 Hz), beta (12–30 Hz), and gamma (above 30 Hz). These bands correspond to various states of brain activity, encompassing wakefulness as well as diverse sleep patterns. Each frequency band exhibits unique characteristics and patterns that are closely associated with behavioral states (Roohi-Azizi et al., 2017). Notably, alterations in power intensity within different frequency bands have been observed in neurological disorders. Specifically, individuals with AD demonstrate slower EEG rhythms, accompanied by heightened energy in the delta and theta bands and diminished energy in the alpha band (Brenner et al., 1988). Moreover, abnormalities in the alpha band have been associated with the initial phase of autism, wherein children with ASD exhibit diminished alpha energy, indicative of heightened anxiety and reduced relaxation (Hoole et al., 2012). PD patients lacking emotional impairment display diminished intra-hemispheric asymmetry in alpha power. Additionally, they manifest decreased overall power in the delta, theta, alpha, and beta bands across various emotional states (Yuvaraj et al., 2014).
2.4. Spontaneous EEG and evoked potential
EEG can be divided into spontaneous EEG and evoked potential (EP) based on the presence or absence of external stimulation during the measurement process. The utilization of spontaneous EEG as a valuable tool in scientific and clinical contexts, such as the diagnosis and evaluation of epilepsy and PD, is facilitated by its ability to reveal the inherent neural activity pattern. However, spontaneous EEG exhibits low SNR and significant inter-subject variability, necessitating additional efforts in data processing. The introduction of external stimuli, such as visual or auditory stimulation, can induce specific deviations in brain waves, leading to the acquisition of EP. Certain characteristics observed in EP have the potential to elucidate disparities in brain function between individuals with brain disorders and healthy controls. EP signals are more robust among subjects and can be further divided into event-related evoked potential (EVP) and steady-state evoked potential (SSEP) according to the mode of external stimuli (Norcia et al., 2015). The detection of EPs aids in the diagnosis and monitoring of brain disorders like schizophrenia and AD.
3. Artificial intelligence
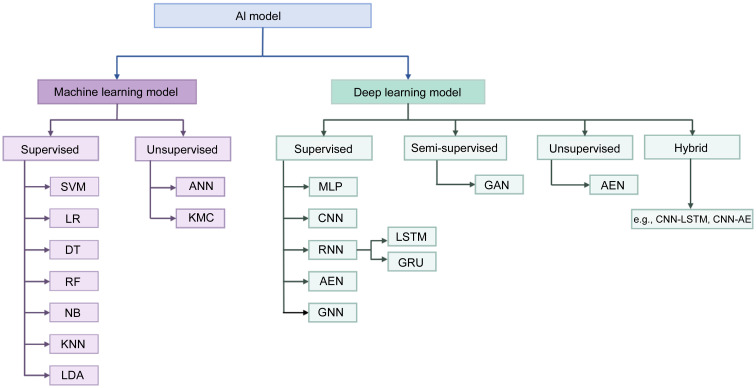
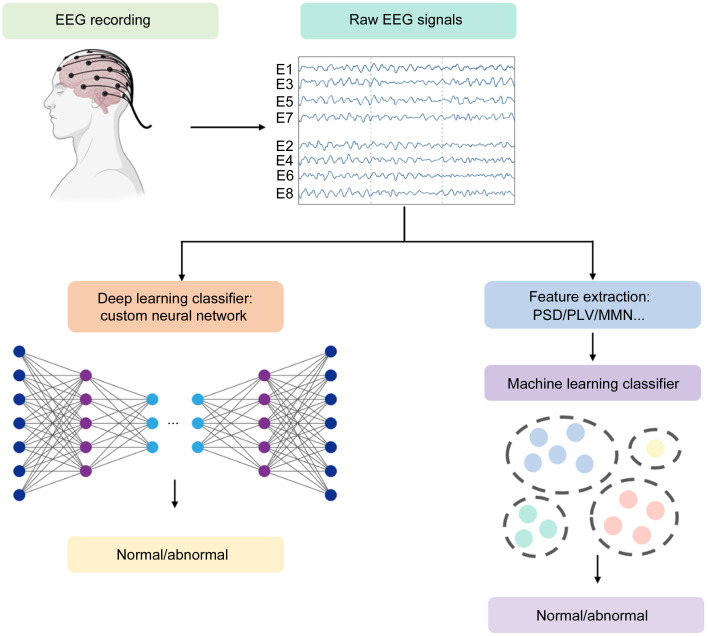
AI has emerged as a transformative force in the field of biomedical engineering, specifically in the domain of disease diagnosis. Over the preceding decades, AI technologies have significantly enhanced diagnostic accuracy and facilitated novel avenues for comprehending intricate brain disorders. Conventional ML and DL techniques both belong to the subfield of AI. ML primarily concentrates on the development of algorithms and models that empower computers to learn and make predictions or decisions without being explicitly programmed (Mohri et al., 2018). SVM, logistic regression (LR), decision tree (DT), random forest (RF), naive Bayes (NB), k-nearest neighbor (KNN), linear discriminant analysis (LDA), and other commonly utilized ML models are frequently employed. In brain disease detection tasks, feature extraction is an important step that requires manually extracting features and performing feature selection. DL, a more sophisticated branch of ML, distinguished itself by its capacity to automatically extract, analyze, and comprehend intrinsic information from raw data (LeCun et al., 2015; Schmidhuber, 2015; Goodfellow et al., 2016). DL includes various types of classifiers, including CNN, RNN, LSTM, autoencoder network (AEN), and graph neural network (GNN). The common ML and DL methods for EEG signals to detect brain diseases can be categorized as depicted in Fig. 2. Additionally, Fig. 3 illustrates a representative AI-based BCI system employed for the classification of normal and abnormal EEG signals. Fig. 4 shows a general summary diagram of AI-auxiliary EEG diagnosis of brain diseases.
Fig. 2. Common machine learning (ML) and deep learning (DL) methods. AI: artificial intelligence; SVM: support vector machine; LR: logistic regression; DT: decision tree; RF: random forest; NB: naive Bayes; KNN: k-nearest neighbor; LDA: linear discriminant analysis; ANN: artificial neural network; KMC: k-means clustering; MLP: multilayer perceptron; CNN: convolutional neural network; RNN: recurrent neural network; AEN: autoencoder network; GNN: graph neural network; LSTM: long short-term memory network; GRU: gated-recurrent unit; GAN: generative adversarial network; AE: autoencoder.
Fig. 3. Typical flow diagram of electroencephalogram (EEG)-based disease diagnosis. PSD: power spectral density; PLV: phase-locked value; MMN: mismatch negativity.
Fig. 4. General diagram of artificial intelligence (AI)-auxiliary electroencephalogram (EEG) for diagnosis of brain diseases. SVM: support vector machine; RF: random forest; KNN: k-nearest neighbor; CNN: convolutional neural network; LSTM: long short-term memory network; PD: Parkinson's disease; AD: Alzheimer's disease; ASD: autism spectrum disorder; AUC: area under the curve; ROC: receiver operating characteristic; FPR: false prediction rate.
4. Contemporary AI approaches for brain disease diagnosis using EEG signals
4.1. Epilepsy
Epilepsy is a neurological disorder characterized by the occurrence of recurrent seizures, which can be manifested in various symptoms such as convulsions, loss of consciousness, and atypical sensations or movements (Krook-Magnuson and Soltesz, 2015). The global prevalence of epilepsy exceeds 50 million individuals, with a continued upward trend. Although the precise etiology of epilepsy remains elusive, potential triggers include brain trauma, infections, and genetic factors. The impact of epilepsy on quality of life can include consequences such as physical injury, cognitive impairment, and social isolation. Therefore, timely and effective diagnosis and treatment of epilepsy are necessary. Clinically, the brain activity of individuals with epilepsy can be categorized into the following four states based on EEG readings: preictal, referring to the period immediately preceding a seizure; ictal, representing the time during which a seizure occurs; postictal, denoting the period following a seizure; and interictal, corresponding to the state between seizures (Jemal et al., 2021). Several studies emphasize the potential of AI-based techniques in predicting epileptic seizures using EEG data, which could yield substantial clinical advancement. These advancements are outlined in Table 1.
Table 1.
Summary of electroencephalogram (EEG)-based epilepsy diagnosis
| Study | Database | Classifier/detector | Performance |
|---|---|---|---|
| Alickovic et al., 2018 | Freiburg, CHB-MIT | ANN, KNN, SVM, RF | 99.77% accuracy |
| Kiral-Kornek et al., 2018 | Cook et al., 2013 | MLP | 69% sensitivity |
| Antoniades et al., 2018 | Local | CNN | 68% accuracy |
| Acharya et al., 2018b | Bonn | CNN | 88.67% accuracy, 90.00% specificity,95.00% sensitivity |
| Truong et al., 2018 | Freiburg, CHB-MIT, Kaggle | CNN | 81.4% (Freiburg), 81.2% (CHB-MIT), and75.0% (Kaggle) sensitivity |
| Tsiouris et al., 2018 | CHB-MIT | LSTM | 0.02–0.11 h-1 FPR |
| Hussein et al., 2019 | Bonn | LSTM | 100% accuracy, 100% sensitivity, 100% specificity |
| Li Y et al., 2019 | Bonn, Hauz Khas | SVM | 100.00%, 99.30%, and 99.30% accuracy |
| Zhang et al., 2020 | CHB-MIT | CNN | 92.2% sensitivity, 0.12 h-1 FPR |
| Daoud and Bayoumi, 2019 | CHB-MIT | MLP, DCNN, DCNN+BiLSTM, DCAE+BiLSTM | 99.6% accuracy, 0.004 h-1 FPR |
| Emami et al., 2019 | Local | AE | 100% sensitivity |
| Liang et al., 2020 | CHB-MIT | LRCN | 84% sensitivity, 99% specificity, 99% accuracy,0.2 h-1 FPR |
| Geng et al., 2020 | Freiburg | BiLSTM | 98.09% sensitivity/98.69% specificity (segment-based), 96.3% sensitivity/0.24 h-1 FPR (event-based) |
| Savadkoohi et al., 2020 | Bonn | SVM, KNN | 100% (SVM) and 99.5% (KNN) accuracy |
| Usman et al., 2021 | CHB-MIT, Kaggle | SVM, CNN, LSTM | 96.28% sensitivity/95.65% specificity (CHB-MIT), 94.2% sensitivity/95.8% specificity (Kaggle) |
| Shoeibi et al., 2021b | Bonn | ML/CNN | 99.53% (two-classes) and 98.67% (multi-classes) accuracy |
| Gramacki and Gramacki, 2022 | Local | CNN | 96%–97% accuracy |
| Tveit et al., 2023 | Local | CNN | 88.3% accuracy |
CHB: Boston Children's Hospital; MIT: Massachusetts Institute of Technology; ANN: artificial neural network; KNN: k-nearest neighbor; SVM: support vector machine; RF: random forest; MLP: multilayer perceptron; CNN: convolutional neural network; LSTM: long short-term memory network; DCNN: deep convolutional neural network; BiLSTM: bidirectional LSTM; DCAE: deep convolutional autoencoder; AE: autoencoder; LRCN: long-term recurrent convolutional network; ML: machine learning; FPR: false prediction rate.
ML methods have been employed in classification tasks where feature selection is necessary to identify the most informative features. Alickovic et al. (2018) developed an automated model for detecting and predicting seizure onset, applying ANN, KNN, SVM, and RF ML algorithms to Freiburg and CHB-MIT datasets (Shoeb, 2009; Ihle et al., 2012). The presented model successfully differentiated between interictal, preictal, and ictal EEGs with an accuracy of 99.77%. In a study conducted by Li Y et al. (2019), a high-resolution time-frequency image was initially generated using the multiscale radial basis function (MRBF)-modified particle swarm optimization (MPSO)-orthogonal least squares (OLS) algorithm. From each sub-band image, gray-level co-occurrence matrix (GLCM) descriptors and the Fisher vector (FV) were extracted. Prior to being inputted to the SVM classifier, dimensionality reduction was performed using the t-test statistical tool. The effectiveness of the proposed method was assessed using the datasets of Bonn and Hauz Khas, and the results demonstrated its capability to accurately differentiate between ictal signals and healthy brain activities (Andrzejak et al., 2001; Swami et al., 2016). However, the proposed framework used single-channel EEG segments, which may limit the optimal classification. Daoud and Bayoumi (2019) used a channel selection algorithm to extract the suitable channel and reduce the computational complexity, making the system suitable for real-time application. Autoencoder (AE) is an unsupervised ML model for which the input is the same as the output. Emami et al. (2019) employed AE to identify seizures as anomalous occurrences within interictal EEG data. The findings indicated that the AE error served as a viable feature for seizure detection, achieving a maximum sensitivity of 100% in distinguishing between seizure and non-seizure states. Savadkoohi et al. (2020) employed t-test and sequential forward floating selection (SFFS) to determine the optimal and statistically significant features from the Bonn dataset. These selected features were fed into SVM and KNN classifiers. The outcomes demonstrated a 99.5% accuracy in the time domain, while the frequency domain and time-frequency domain exhibited the highest accuracy of 100%.
The clinical feasibility of seizure prediction has been established, but certain limitations persist, such as uncertainty regarding the occurrence of seizures among individuals with epilepsy. In order to address this, wearable devices have been developed to enable continuous monitoring. Kiral-Kornek et al. (2018) developed an automated, patient-tunable epileptic seizure prediction system based on DL and deployed the system on a low-power neuromorphic chip. A DL model was trained using annotated iEEG data from Cook et al. (2013)’s dataset to distinguish between preictal and interictal signals and deployed onto the neuromorphic TrueNorth chip. The system allowed for instantaneous and easy adjustment, which meant that patients or clinicians could prioritize high sensitivity or low warning time depending on different needs or circumstances. The prediction system achieved a mean sensitivity of 69% using iEEG data. Although iEEG data improve data training due to their high SNR, iEEG’s invasive recording is impractical. Scalp EEG data represent a more convenient alternative.
To ensure the acquisition of high-quality data comparable to iEEG, Antoniades et al. (2018) proposed an ensemble DL architecture. This approach nonlinearly maps scalp EEGs to pseudo-intracranial EEGs, which are subsequently fed into a CNN model. Remarkably, the authors achieved an accuracy of 68% in automatically discriminating intracranial epileptic discharges (IEDs). Acharya et al. (2018b) constructed a 13-layer-deep CNN architecture to detect normal, preictal, and seizure classes using the Bonn dataset and achieved an accuracy, specificity, and sensitivity of 88.67%, 90.00%, and 95.00%, respectively. Nevertheless, large DL models involve a large number of parameters, which may impose additional computational burden. Truong et al. (2018) applied a simple and shallow CNN model with only three convolutional blocks. Information from both frequency and time domains was extracted using a short-time Fourier transform, and then the optimized features were automatically generated in an image-like format. This approach achieved sensitivity of 81.4%, 81.2%, and 75.0%, respectively, in classifying preictal and interictal segments using Freiburg, Boston Children’s Hospital (CHB)-Massachusetts Institute of Technology (MIT), and Kaggle datasets (Brinkmann et al., 2016). Zhang et al. (2020) also used the CHB-MIT dataset for seizure prediction but a data augmentation was implemented to obtain a diversity of data to solve the trial imbalance problem. A spatial filtering algorithm was adopted to extract the component signal that could best transduce the cerebral activity in seizure prediction, and a shallow CNN was utilized. Finally, the proposed approach achieved a sensitivity of 92.2% and a false prediction rate (FPR) of 0.12 h-1.
CNNs are capable of extracting spatial features from input data, but they tend to overlook temporal features, particularly in time series data like EEG. Epilepsy detection using EEG signals involves temporal aspects. In order to enhance the accuracy of detection, certain recurrent DL architectures have been implemented. Tsiouris et al. (2018) designed a two-layer LSTM network for seizure prediction. Time and frequency domain features, cross-correlation between channels, and graph theoretic features were fed into the model. An evaluation was performed on the CHB-MIT dataset, and the result showed a low FPR of 0.02–0.11 h-1, depending on the length of preictal windows. Instead of choosing features manually, Hussein et al. (2019) exploited an LSTM network to learn high-level representations of EEG signals, and a fully connected (FC) layer was applied to extract the most robust EEG features. They tested the approach on the Bonn dataset and achieved 100% classification accuracy, 100% sensitivity, and 100% specificity in both two-class and multi-class tasks using clean EEG signals that were free of artifacts and noise. They also evaluated the model on EEG signals without artifact removal and achieved an accuracy of over 90% in multi-class classification, which demonstrated its ability to be deployed in real-life conditions. Compared to LSTM’s single-time direction processing, bidirectional LSTM (BiLSTM) could process time series in both directions simultaneously. Geng et al. (2020) presented a BiLSTM algorithm that could automatically select features from the time-frequency matrix obtained by S-transform and perform classification tasks. The proposed system was evaluated on the Freiburg dataset. Results showed that segment-based classification achieved a sensitivity of 98.09% and a specificity of 98.69%. The event-based classification obtained a sensitivity of 96.3% and an FPR of 0.24 h-1.
However, it should be noted that training a DL model necessitates a substantial amount of data for effective learning. On the other hand, handcrafted features are computationally efficient and suitable for implementation. Shoeibi et al. (2021a, 2021b) employed handcrafted features for DL models. They first compared 50 different handcrafted features in terms of importance and computational complexity. The best features selected by the Fisher algorithm were then combined with CNN-AE. The performance of this approach was assessed using the Bonn dataset, revealing an accuracy of 99.53% in two-classes classification and 98.67% in multi-classes classification. Because of the efficient combination capabilities of the multi-networks method, Daoud and Bayoumi (2019) introduced a fusion DL model for automatic seizure prediction, where raw EEGs were input directly into the models without any handcrafted feature extraction. Initially, they implemented a deep convolutional autoencoder (DCAE) architecture to pre-train the model front-end in an unsupervised manner. Transfer learning was employed to enhance the generalization capability of the DCAE. Subsequently, the pre-trained encoder was integrated with a BiLSTM network for classification. The CHB-MIT EEG dataset achieved a performance with 99.6% accuracy and a 0.004 h-1 FPR.
Long-term recurrent convolutional network (LRCN) is a spatiotemporal model that combines characteristics of both CNN and LSTM. Liang et al. (2020) proposed an 18-layer LRCN to realize automatic seizure detection and epileptogenic zone localization. The model was trained and evaluated using the CHB-MIT database. The resulting classifier demonstrated a sensitivity of 84%, specificity of 99%, accuracy of 99%, and a relatively low FPR of 0.2 h-1. Nevertheless, it is important to acknowledge the potential limitation of computational intensity and the risk of overfitting due to the limited dataset availability. To address these concerns, Usman et al. (2021) employed ensemble learning of multiple classifiers, including SVM, CNN, and LSTM. Model-agnostic meta-learning (MAML) was used by feeding the output probabilities of each classifier to give the final decision of whether it was a preictal or interictal state. In this way, they trained the classifier with a smaller number of training examples without overfitting. The model achieved an average sensitivity of 96.28% and a specificity of 95.65% with an average anticipation time of 33 min on the CHB-MIT dataset and an average sensitivity of 94.2% and a specificity of 95.8% on the Kaggle dataset.
To improve the reproducibility of different datasets, Gramacki and Gramacki (2022) proposed a complete framework for EEG-based seizure detection using DL techniques. They have also proposed a sliding window design to generate fully balanced training data. Based on their open-access code, researchers could analyze their own EEG datasets with only minor modifications.
Beyond distinguishing abnormal from normal or identifying epileptiform activity, Tveit et al. (2023) reported a new CNN model, standardized computer-based organized reporting of EEG (SCORE)-AI, which could classify abnormal recordings into the major categories that were most relevant for decisions involving patients. These included epileptiform-focal, epileptiform-generalized, nonepileptiform-focal, and nonepileptiform-diffuse abnormalities. SCORE-AI had an area under the receiver operating characteristic (ROC) curve between 0.89 and 0.96 for the different categories of EEG abnormalities. This performance was comparable to the consensus of human experts.
4.2. Schizophrenia
Schizophrenia is a chronic neurodevelopmental disorder with a global prevalence of 1% (Saha et al., 2005). While the underlying mechanism remains unclear, numerous studies have established a correlation between schizophrenia and structural and functional abnormalities in the brain. Individuals suffering from schizophrenia typically present a heterogeneous combination of psychotic symptoms, including hallucinations and delusions, as well as behavioral and cognitive impairments (McCutcheon et al., 2020) that significantly impact their daily lives. Timely diagnosis of schizophrenia is essential to patients so that pharmacological treatments can be used to relieve the symptoms. Recent research has demonstrated the significance of EEG patterns in the identification of schizophrenia. Various ML techniques have been employed to achieve automated and accurate classification outcomes. Additionally, the potential of neural networks in the scope of schizophrenia diagnosis has been under investigation (Barros et al., 2021). Table 2 provides an overview of studies conducted on the diagnosis of schizophrenia using EEG signals.
Table 2.
Summary of electroencephalogram (EEG)-based schizophrenia diagnosis
| Study | Database | Classifier/detector | Performance |
|---|---|---|---|
| Johannesen et al., 2016 | Local | SVM | 87% accuracy |
| Shim et al., 2016 | Local | SVM | 88.24% accuracy |
| Oh et al., 2019 | Institute of Psychiatry and Neurology in Warsaw, Poland | CNN | 98.07% and 81.26% accuracy |
| Jahmunah et al., 2019 | Institute of Psychiatry and Neurology in Warsaw, Poland | SVM-RBF | 92.91% accuracy |
| Devia et al., 2019 | Local | LDA | 71% accuracy |
| Li FL et al., 2019 | Local | SVM/LDA | 90.48% accuracy |
| Phang et al., 2020 | Lomonosov Moscow State University, Russia | Multi-domain connectome CNN (MDC-CNN) | 91.69% accuracy |
| Goshvarpour and Goshvarpour, 2020 | RepOD | PNN | 100% accuracy |
| Ahmedt-Aristizabal et al., 2021 | Local | CNN-LSTM | 89.98% accuracy |
| Chang et al., 2021 | Local | SVM/GNN | 81.33% accuracy |
| Yin et al., 2023 | Local | GCN | 90.01% accuracy |
| Saadatinia and Salimi-Badr, 2024 | Local | CNN | 99.0% accuracy |
SVM: support vector machine; CNN: convolutional neural network; RBF: radial-basis-function; LDA: linear discriminant analysis; PNN: probabilistic neural network; LSTM: long short-term memory network; GNN: graph neural network; GCN: graph convolutional network; RepOD: Repository for Open Data.
Numerous experimental approaches have been introduced to investigate the EEG biomarkers in schizophrenia patients. Studies on the diagnosis of schizophrenia have examined the event-related potential (ERP) components of patients through the collection of EEG data during working memory tasks or visual and auditory processing tasks, which allowed the evaluation of brain response due to the result of specific sensory cognitive features. Johannesen et al. (2016) conducted EEG recordings while subjects were completing a Sternberg working memory task (SWMT), which consisted of four processing stages: baseline, encoding, retention, and retrieval. The utilization of an SVM classifier resulted in an accuracy of 87%. Additionally, frontal gamma at the encoding stage was identified by SVM as the most highly weighted feature, which coincided well with early studies about the gamma band’s role in memory encoding. Researchers have also recognized the potential utility of P300 and mismatch negativity (MMN) as biomarkers for assessing schizophrenia, which can be elicited through auditory oddball tasks. These tasks involve the presentation of infrequent deviant stimuli amidst frequent standard tones. Shim et al. (2016) conducted a study in which they collected EEG data while participants were engaged in a passive auditory oddball paradigm. They combined P300 peak data from 62 electrodes as sensor-level features with cortical current density as source-level EEG features. By utilizing an SVM classifier, they were able to achieve a classification accuracy of 88.24%. In a separate study, Chang et al. (2021) utilized MMN signals from ERPs and employed SVM to extract graph-theoretic features. These features were then used to classify individuals into three groups: first-episode schizophrenia, chronic schizophrenia, and healthy control. This classification approach was based on the understanding of the neural mechanism underlying MMN damage in different stages of schizophrenia.
However, these tasks were often goal-directed, which posed challenges for schizophrenia patients with cognitive impairments and distorted perception. In order to address this issue, Devia et al. (2019) employed a straightforward visual task-free paradigm to record EEG while participants viewed natural scenes. By examining the average EEG activity synchronized with the onset of the images, the researchers identified differences between patients with schizophrenia and healthy controls. Specifically, the patients exhibited a diminished occipital ERP amplitude of approximately 500 ms following the presentation of the images in comparison to the control group. Subsequently, these differences were employed for the categorization of individuals diagnosed with schizophrenia. The evaluation encompassed three classifiers: LDA, a rule-based classifier, and a combination of the posterior probability of two LDAs. Remarkably, the classifier based on the comparison of the posterior probability, employing occipital ERPs, exhibited the most notable sensitivity at 81% and accuracy at 71%.
Furthermore, alternative methodologies centered on resting-state EEG data were explored, and these data were suggested to reflect intrinsic neural activity (Ciprian et al., 2021). A publicly available database at the Repository for Open Data (RepOD) contains EEG data at eyes-closed resting state (Olejarczyk and Jernajczyk, 2017; Jahmunah et al., 2019). Multiple classifiers were evaluated, including DT, LDA, KNN, probabilistic neural network (PNN), and SVM. The SVM with radial-basis-function (SVM-RBF) classifier yielded the highest accuracy of 92.91%. Based on the same dataset, Goshvarpour and Goshvarpour (2020) implemented a feature-level fusion strategy to combine extracted features, resulting in improved accuracy compared to separate feature stacks. Classification was performed with PNN to separate the EEG features of schizophrenic patients and healthy controls.
According to certain studies, the absence of task-related elements in data may lead to a deficiency of comprehensive information for discrimination. However, incorporating features derived from both rest and task states can provide distinct insights into brain deficits associated with schizophrenia. Inspired by this theory, Li FL et al. (2019) conducted an experiment in which they trained LDA and SVM classifiers using concatenated inherent spatial pattern of network (SPN) features from both resting and task P300 EEGs. As a result, they achieved an accuracy of 90.48%.
When examining the DL method, most studies have utilized resting-state data as they allow for the automatic extraction of abstract and high-level features from raw data. Among the various DL models employed in the detection of schizophrenia, CNN stands out as one of the most extensively utilized. Oh et al. (2019) proposed an eleven-layered CNN model to distinguish schizophrenia patients and obtained an accuracy of 81.26% and 98.07% for the non-subject-based testing and subject-based testing, respectively. However, it is important to note that the study’s primary constraint lies in the limited sample size, which may not provide adequate training for a DL model. Phang et al. (2020) used multi-channel EEG as training data to enlarge the dataset. They designed a multi-domain connectome CNN (MDC-CNN) with a combined connectivity feature, which was suitable for spatial proximity modeling. The algorithm outperformed single-domain CNNs using individual features, achieving an accuracy of 91.69%. As well as enlarging the dataset, Saadatinia and Salimi-Badr (2024) generated synthetic datasets to solve the limited training data and overfitting problem. They used Wasserstein generative adversarial network (GAN) with gradient penalty (WGAN-GP) and variational autoencoder (VAE) for data augmentation, with the VAE improving accuracy by 3.0%, reaching 99.0%. Spectrograms were extracted from raw signals to utilize time-frequency features, and CNN was selected for initial diagnosis. They also employed the local interpretable model-agnostic explanation (LIME) algorithm to explain the most important frequencies in the diagnosis process, thus solving the trust issues.
However, it is possible that CNNs do not adequately represent the graph structure of brain networks. As a result, models that generalize CNNs to graphs (e.g., GNN and graph convolutional network (GCN)) have been explored. Chang et al. (2021) leveraged a GNN model to successfully learn the topological structure of brain networks, leading to superior improvements in classifying schizophrenia patients and healthy controls. Similarly, Yin et al. (2023) developed a GCN-based automatic recognition model for detecting schizophrenia using resting-state EEG. The feature matrix consisted of time-frequency features of the EEG signal and the local features of the brain network. The result showed that the parietal lobe was the most significant brain region contributing to the classification. They also compared the results using other models (CNN-2, CNN-5, and SVM). The highest accuracy achieved by the proposed model in identifying first-episode schizophrenia patients was 90.01%.
Recent studies have focused on distinguishing between people with schizophrenia and healthy controls. However, fewer studies have been conducted on people at risk of schizophrenia. The situation is more complicated because brain abnormalities are less evident among people at risk. Ahmedt-Aristizabal et al. (2021) identified children at risk of schizophrenia by exploring several ML and DL techniques. They collected MMN signals from children with a positive family history of schizophrenia and/or children who exhibit antecedents of schizophrenia. In the proposed DL architecture, they not only used CNN but also added recurrent layers (LSTM). The results showed improved performance of the hybrid network 2D-CNN-LSTM with an average accuracy of 89.98%.
4.3. Depression
Depression is a common psychiatric disorder. According to the World Health Organization (WHO), about 340 million people suffer from different degrees of depression worldwide. The neurological mechanism and pathological principle of depression still remain unclear. People with depression have severe psychological disorders and negative emotions characterized by grief, fatigue, despair, and even suicidal thoughts. They also have physical symptoms such as headaches, constipation, weight changes, and insomnia (Simon et al., 1999). The most commonly used international diagnostic criterion is the Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition (DSM-5), set by the United States (American Psychiatric Association, 2013). However, such a questionnaire-based approach is prone to doctor and patient subjectivity. With that in mind, studies have explored the use of EEG to find depression biomarkers that can be used as an objective indication. Researchers also made efforts to develop ML/DL-based techniques for depression recognition. A summary of the presented studies is provided in Table 3.
Table 3.
Summary of electroencephalogram (EEG)-based depression diagnosis
| Study | Dataset | Classifier/detector | Performance |
|---|---|---|---|
| Li et al., 2016 | Local | ML (BN/SVM/LR/KNN/RF) | >91% accuracy |
| Mumtaz et al., 2017 | Local | LR/SVM/NB | 98.4% accuracy |
| Acharya et al., 2018a | Local | CNN | 93.54%/95.49% (left/right hemisphere EEG data) accuracy |
| Shim et al., 2019 | Local | SVM | 70.34% accuracy |
| Ay et al., 2019 | Acharya et al., 2018a | CNN-LSTM | 99.12% accuracy |
| Li XW et al., 2019 | Local | SVM/CNN | 89.02% (SVM)/84.75% (CNN) accuracy |
| Shah et al., 2019 | Local | SNN | 68.18% (eyes-closed)/72.13% (eyes-open) accuracy |
| Cai et al., 2020 | Local | KNN/DT/SVM | 86.98% accuracy (KNN) |
| Sharma et al., 2021 | Psychology Department, University of Arizona, USA | CNN-LSTM | 99.10% accuracy |
| Sarkar et al., 2022 | Kaggle | MLP/CNN/RNN/LSTM/SVM/LR | 96.50% accuracy (RNN) |
| Wang et al., 2022 | MODMA dataset, University of New Mexico, USA | CNN | 93.97% accuracy |
ML: machine learning; BN: BayesNet; SVM: support vector machine; LR: logistic regression; KNN: k-nearest neighbor; RF: random forest; NB: naive Bayes; CNN: convolutional neural network; LSTM: long short-term memory network; SNN: spiking neural network; DT: decision tree; MLP: multilayer perceptron; RNN: recurrent neural network; MODMA: multi-modal open dataset for mental-disorder analysis.
Regarding biomarkers to detect depression, a review highlights that some features of EEG, such as alpha asymmetry, are promising (Yasin et al., 2021). Mumtaz et al. (2017) compared EEG alpha interhemispheric asymmetry between major depressive disorder (MDD) patients and healthy controls. Their findings revealed that depressed individuals showed greater anterior EEG activity. The most significant features were selected from candidate features (power computation at different frequency bands and EEG alpha asymmetry) according to a criterion named ROC. Features with substantial weights were used for training and testing classifier models, namely LR, SVM, and NB. Among these models, SVM demonstrated the highest performance (accuracy=98.4%, sensitivity=96.66%, specificity=100%).
Several studies have been conducted using ERP. Li et al. (2016) performed an experiment involving a facial expression-viewing task, building upon previous findings that depression can affect the ability to recognize different facial emotions. EEG signals were analyzed, specifically focusing on the theta, alpha, and beta frequency bands. Multiple classifiers and feature selection methods were employed in the analysis. The highest accuracy of 98% was achieved through the utilization of the feature selection method Greedy Stepwise (GSW) based on correlation features selection and the KNN classifier for the beta frequency band. It has been established that the ERP component P300 correlates with depression (Mumtaz et al., 2015). Shim et al. (2019) conducted a study to investigate the potential effectiveness of utilizing P300 features at both the sensor and source levels for the classification of post-traumatic stress disorder (PTSD) and MDD. During the auditory oddball tasks, they collected P300 signals and extracted the amplitude and latency of P300 as sensor-level features and the current source density (CSD) of P300 components as source-level feature. A classification accuracy of 70.34% for the discrimination between PTSD and MDD was obtained using a two-class linear SVM classifier.
As traditional ML classification methods require greater computational resources and process more feature dimensions, an increasing number of DL-based approaches have been proposed for classifying EEG data of both depressed and healthy individuals. Li XW et al. (2019) incorporated spatial information obtained from EEG caps to enhance the analysis of original features, namely power spectral density (PSD) in the frequency domain and activity in the time domain. These features were transformed into images that included spatial information, and a CNN was subsequently employed to recognize these feature images. The method achieved a maximum accuracy of 84.75%. Acharya et al. (2018a) employed a 13-layer-deep CNN model and achieved a higher accuracy of 95.49% when analyzing right hemisphere EEG signals. This finding aligns with previous research indicating a correlation between depression and heightened activity in the right hemisphere. Using the same dataset, Ay et al. (2019) improved the classification performance by employing a novel hybrid model that combined CNN and LSTM architectures. Their findings align with those of previous studies, indicating a superior performance of right-hemisphere compared to left-hemisphere EEG signals, achieving an accuracy of 99.12%. However, the aforementioned models entail significant computational demands. In contrast, Sharma et al. (2021) proposed a CNN-LSTM hybrid neural network model for depression screening that exhibits reduced time and computational complexity. CNN was used in temporal learning, and LSTM was used in sequence learning. The proposed model yielded a classification accuracy of 99.10%. Sarkar et al. (2022) comprehensively compared DL and ML techniques for depression detection using EEG. RNN achieved the highest accuracy, reaching 97.50% during training and 96.50% during testing.
Nevertheless, it is worth noting that the aforementioned studies solely employed a single pattern recognition technique, lacking the capability to acquire more profound diagnostic insights. The multi-modal technique has gained popularity in recent times due to its ability to offer more comprehensive information. Li XW et al. (2019) conducted an experiment utilizing an emotional face stimulus task and extracted the PSD and original activities as features. They adopted ensemble learning and SVM to process and classify these features, acquiring the best accuracy of 89.02%. Cai et al. (2020) proposed a multi-modal depression discrimination model that integrated EEG data from three modalities collected during neutral, negative, and positive audio stimuli, respectively. Linear and nonlinear features were extracted from each individual modality, followed by a feature-level fusion method to linearly combine the feature matrices of the three modalities. Several ML classifiers were employed, with the KNN classifier achieving the highest accuracy of 86.98%. Both studies shared the limitation of small sample sizes. To address this issue, Wang et al. (2022) employed multi-channel data fusion and clipping augmentation. Previous studies have consistently demonstrated the close association between the frontal lobe and depression as well as psychological activities (Shi et al., 2020). Therefore, they only utilized three electrode signals, namely Fp1, Fp2, and Fpz, originating from the prefrontal lobe section. They fused the 3-channel EEG signals and transformed them into 2D images. This approach improved depression diagnosis accuracy and clustering effects while maintaining low complexity and robust signal processing.
In addition to the utilization of deep neural network (DNN), some biologically realistic models have been employed, such as the spiking neural network (SNN). The SNN model is inspired by the spike activities observed in the human brain (Maass, 1997). Shah et al. (2019) employed the NeuCube SNN architecture to analyze data from a depression case study. The obtained accuracy was 68.18% for eyes-closed data and 72.13% for eyes-open data. Nevertheless, SNN was still inferior to the DL method in terms of accuracy, as DL models employ backpropagation to reduce the error. Future research endeavors may consider extracting features using CNN or other DL models and subsequently integrating them into the SNN framework to improve performance.
4.4. Parkinson’s disease
PD is a progressive neurological disorder that arises with the loss of neurons responsible for producing dopamine. It is the second most prevalent neurodegenerative disorder to increase with age after AD. People with PD encounter symptoms such as rest tremors, slow movements (bradykinesia), and muscular rigidity (Jankovic, 2008). PD diagnosis is mostly based on motor symptoms such as speech defects or freezing of gait. However, autopsy and neuroimaging studies indicate that the emergence of motor symptoms is a late manifestation, occurring when there is widespread degeneration of dopaminergic neurons (Beitz, 2014). Therefore, early identification of PD is vital for effective neuroprotective treatments. The application of ML/DL techniques in EEG has revealed another method for PD diagnosis with a low error rate, low cost, and low invasiveness. The summary of the selected studies is shown in Table 4.
Table 4.
Summary of electroencephalogram (EEG)-based Parkinson's disease (PD) diagnosis
| Study | Database | Classifier/detector | Performance |
|---|---|---|---|
| Chaturvedi et al., 2017 | Local | ML | 78% accuracy |
| Saikia et al., 2019 | Local | ANN | 98.8% classification rate |
| Oh et al., 2020 | Local | CNN | 88.25% accuracy, 84.71% sensitivity, 91.77% specificity |
| Shah et al., 2020 | PRED+CT data repository | DGHNet | 99.22% accuracy |
| Khare et al., 2021 | OpenNeuro, Hospital Universiti Kebangsaan Malaysia | CNN | 100% and 99.97% accuracy |
| Lee et al., 2021 | Local | ML/CRNN | 99.2% accuracy |
| Qiu et al., 2022 | OpenNeuro, Iowa dataset | SVM/MCNN | 0.999 AUC (MCNN) |
| Suuronen et al., 2023 | University of Turku, Finland | ML | 76% accuracy |
| Bdaqli et al., 2024 | UCSD | CNN-LSTM | 99.51% (binary) and 99.75% (multi-class) accuracy |
ML: machine learning; ANN: artificial neural network; CNN: convolutional neural network; PRED+CT: patient repository for EEG data+computational tools; DGHNet: dynamical system generated hybrid network; CRNN: convolutional recurrent neural network; SVM: support vector machine; MCNN: multi-scale CNN; AUC: area under the curve; UCSD: University of California, San Diego; LSTM: long short-term memory network.
The investigation of reliable and quantitative PD biomarkers derived from EEG is currently a thriving research field. In the majority of studies, EEG recordings are obtained during the resting state. Chaturvedi et al. (2017) employed ML techniques to identify a subset of features from quantitative EEG (QEEG) parameters. They calculated relative power in different frequency bands across different brain regions. Median and peak frequencies as well as alpha/theta ratios were calculated. The RF and the least absolute shrinkage and selection operator (LASSO) methods were employed for feature selection. Extracted features indicated that the alpha/theta spectral ratio in the central left region could be an influential feature for discriminating between PD patients and healthy controls. The discrimination method finally achieved an accuracy of 78%.
However, Shah et al. (2020) believed that the acquired EEG signals exhibited chaotic characteristics, yet neurodegenerative disorders such as PD progressively resulted in alteration in the dynamical properties of EEG. In accordance with this theory, they chose beta frequency synchronization and phase amplitude coupling (PAC) in the EEG signals as potential biomarkers. To emphasize the PD-associated alterations in the dynamical properties of the EEG data, an embedding reconstruction technique derived from chaos theory was employed on the EEG data sourced from the patient repository for EEG data+computational tools (PRED+CT) data repository (Cavanagh et al., 2017).
The utilization of DL has been shown to mitigate the necessity for feature engineering. Oh et al. (2020) designed a 13-layer CNN architecture to classify PD and healthy individuals, achieving an accuracy of 88.25%, a sensitivity of 84.71%, and a specificity of 91.77%. Khare et al. (2021) implemented an automatic PD detection approach named PD CNN (PDCNNet), wherein they transformed 1D signals into 2D time-frequency plots. Smoothed pseudo Wigner Ville distribution (SPWVD) was used to obtain time-frequency representation from EEG signals. Then, the 2D plots were fed to a CNN. The model achieved 100% and 99.97% accuracy for two public datasets, respectively.
In the context of multi-modal techniques, the combination of EEG and electromyography (EMG) was utilized to detect cognitive decline and muscle conditions in PD. Saikia et al. (2019) conducted a study in which EEG signals were extracted from the frontal and temporal brain, along with muscle activities associated with wrist flexion and extension. ANNs were employed for classification, and the result showed that the combination of EEG and EMG features resulted in the highest classification rate of 98.8%.
Recent research has focused on reducing the number of electrodes, as high-density EEG montages are not commonly utilized in hospitals. Suuronen et al. (2023) conducted an investigation on the impact of electrode placement on classification performance. They employed a budget-based search algorithm and successfully developed a unique channel-wise feature selection method. The result demonstrated that a mere five channels positioned at right-frontal, left-temporal, and midline-occipital sites were adequate for detecting PD with an accuracy of 76%.
However, the majority of participants in the aforementioned studies were taking medication, thus introducing potential confounding effects. Several studies have been conducted to classify patients in on-medication vs. off-medication conditions. Shah et al. (2020) proposed a DNN architecture named dynamical system generated hybrid network (DGHNet) for the classification of PD patients in on-medication vs. off-medication states. The network had a convolutional layer as the input layer to extract spectral features and an LSTM layer as the output layer, resulting in an accuracy of 99.22%. In a separate study, Lee et al. (2021) recorded two EEG sessions during off- and on-medication states, respectively. They employed a convolutional RNN (CRNN) model consisting of CNN and RNN, which achieved 99.2% accuracy, 98.9% precision, and 99.4% recall in classifying PD. Furthermore, the proposed model demonstrated its ability to detect the impact of dopaminergic medication by utilizing phase information. Phase-related features were also used in Qiu et al. (2022)’s study. They implemented a multi-pattern analysis that incorporated PSD and phase-locked value (PLV) features from EEG data. The results showed significant differences in PSD and PLV between healthy controls and PD patients (including PD_OFF and PD_ON), particularly within the beta and gamma frequency bands. Their proposed multi-scale CNN (MCNN) model exhibited exceptional performance.
In Bdaqli et al. (2024)’s work, a novel 1D CNN-LSTM architecture was introduced to extract salient features from the EEG signals. For classification, three algorithms were used, and the results indicated that SVM performed best, achieving a high accuracy of 99.51% in binary classification and 99.75% in multi-class classification. To enhance model interpretability, t-distributed stochastic neighbor embedding (t-SNE) was used as an explainable AI (XAI) method in the post-processing stage.
4.5. Alzheimer’s disease
AD is the most common cause of dementia and holds the seventh position as a leading cause of mortality on a global scale. It is a neurodegenerative disease with a high incidence among people over the age of 65 years, characterized by its hidden progression and devastating impact on cognitive function. Common pathological features of the disease include beta-amyloid plaques, dystrophic neurites associated with plaques, and neurofibrillary tangles within nerve cells (Vickers et al., 2000). The early and accurate diagnosis of AD is crucial for timely intervention and treatment strategies. The diagnosis of AD involves a range of clinical procedures, including neuropsychological tests, cognitive function evaluation, biochemical analysis, and brain imaging (Bateman et al., 2012). The most common cerebrospinal fluid (CSF) biomarkers used for AD diagnosis are amyloid beta and tau (Anoop et al., 2010). In the early clinical stages, AD often manifests as mild cognitive impairment (MCI) (Albert et al., 2011), which is a transitional stage between normal aging (NA) and more severe cognitive decline. During this stage, patients often exhibit cognitive alterations that surpass the expected performance for their age, while still maintaining a degree of functional independence. The distinguishing factor is whether the patient has specific cognitive deficits beyond typical age-related changes. Accurately diagnosing AD, especially in the MCI stage, poses a significant challenge in AD detection. Brain imaging techniques, such as functional magnetic resonance imaging (fMRI), computed tomography (CT), and positron emission tomography (PET), require exhaustive procedures and high cost. In the past decade, EEG signal analysis has emerged as an alternative method due to its non-invasive and cost-effective features.
The classification problems of identifying EEG for AD, MCI, and healthy control can be categorized into two or three categories. ML and DL can help robustly learn abstract representations from EEG so that better generalization can be achieved. In Table 5, the most recent advancements in diagnosing AD through the integration of EEG and AI are presented.
Table 5.
Summary of electroencephalogram (EEG)-based Alzheimer's disease (AD) diagnosis
| Study | Size of dataset | Classifier/detector | Performance |
|---|---|---|---|
| McBride et al., 2014 | 15 HC, 16 MCI, 17 AD | SVM | 85.4% accuracy |
| Cassani et al., 2017 | 85 HC, 99 AD | SVM | 91.4% (20-channel) and 77.3% (7-channel) accuracy |
| Ieracitano et al., 2019 | 63 HC, 63 MCI, 63 AD | CNN | 89.8% (AD vs. HC) and 83.3%(AD vs. MCI vs. HC) accuracy |
| Bi and Wang, 2019 | 4 HC, 4 MCI, 4 AD | CDBM | 95.04% accuracy |
| You et al., 2020 | 17 HC, 35 MCI, 35 AD | GCN | 91.07% accuracy |
| Vecchio et al., 2020 | 120 HC, 175 AD | SVM | 95% accuracy |
| Alsharabi et al., 2022 | 35 HC, 31 mild AD,22 moderate AD | LDA, QDA, SVM, NB, KNN, DT, ELM, ANN, RF | 99.98% accuracy (KNN) |
| Rad et al., 2021 | 17 HC, 10 AD | CNN | 97.5% (recall stage) and 99.0% (stimulation stage) accuracy |
| Xia et al., 2023 | 14 HC, 37 MCI, 49 AD | CNN | 97.10% accuracy |
| Dogan et al., 2023 | 11 HC, 12 AD | KNN | 100% (tenfold cross-validation) and 92.01% (LOSO cross-validation) accuracy |
| Han et al., 2024 | 21 NA, 29 NCD | SVM | 92.69% accuracy |
| Boudaya et al., 2024 | 13 MCI, 11 HC | Hybrid ML (SVM, RF, GB) | 93.86% accuracy, 93.87% sensitivity, 93.53% specificity |
HC: healthy control; MCI: mild cognitive impairment; NA: normal aging; SVM: support vector machine; CNN: convolutional neural network; CDBM: convolutional deep Boltzmann machine; GCN: graph convolutional network; LDA: linear discriminant analysis; QDA: quadratic discriminant analysis; NB: naive Bayes; KNN: k-nearest neighbor; DT: decision tree; ELM: extreme learning machine; ANN: artificial neural network; RF: random forest; NCD: neurocognitive disorder; ML: machine learning; GB: gradient boosting.
Several works have utilized ML technique to distinguish individuals with AD from healthy. For instance, Cassani et al. (2017) proposed an automatic EEG-based AD diagnosis system based on automated artifact removal (AAR) and a 7-channel EEG setup, which provided an opportunity for portable and affordable AD detection. EEG data from healthy controls and AD patients were analyzed, including spectral power, coherence, and amplitude-modulation. SVM was utilized as classifier and the system was evaluated on two different datasets, the 7-channel and the 20-channel datasets, with the accuracy of 77.3% and 91.4%, respectively. Vecchio et al. (2020) explored the automatic classification of healthy aging and AD based on EEG connectivity using SVM. Brain connectivity was computed using graph theory analysis on 84 regions of interest (ROIs) from both the left and right hemispheres. The results offered high accuracy (area under the curve (AUC) of 0.97), indicating that EEG connectivity analysis combined with source/connectivity biomarkers could be a promising method for identifying AD patients. Additionally, Alsharabi et al. (2022) developed a band-pass elliptic digital filter to remove interference, followed by discrete wavelet transform (DWT) to extract EEG signal features. Multiple features were integrated into the DWT to enhance diagnostic capabilities. Nine ML methods were evaluated for classifying EEG features. Among them, KNN classifier achieved the highest accuracy (99.98%).
Traditional EEG-based automatic systems aimed at early diagnosis of MCI and AD have heavily emphasized feature engineering. Unlike previous studies, an automated AD detection model was explored, using a directed graph for local texture feature extraction from EEG signals (Dogan et al., 2023). The graph is constructed based on a primate brain pattern (PBP) derived from neuronal pathways associated with visual object recognition and motor response. The model combined PBP with a tunable q-factor wavelet transform for multilevel feature extraction, achieving 100% and 92.01% accuracy using the KNN classifier with tenfold and LOSO cross-validation, respectively.
In addition to resting EEG, ERPs have shown promise in AD diagnosis. A study demonstrated that the amplitude and latency of the P300 component vary with age and the progression of AD following stimulation (Rad et al., 2021). This study employed four task states, namely closed-eye, open-eye, recall, and stimulation EEGs. Three channels (frontal zero (Fz), parietal zero (Pz), and central zero (Cz)) were used to record brain activity. LDA, Elman neural network, and CNN were used for classification. Notably, the Pz channel exhibited better features, achieving 97.5% and 99.0% accuracy in the recall and stimulation stages, respectively, when employing CNN for classification.
The two categories are less complex than the three categories, but the latter is more significant in research. The diagnosis of MCI can help in prevention and treatment of AD. McBride et al. (2014) used scalp EEG to detect cognitive impairments in older adults, specifically amnestic MCI (aMCI) and early AD. Spectral and complexity features were extracted and SVM model was used to distinguish between groups. The highest classification accuracy of 85.4% was achieved when analyzing EEG data during eyes closed conditions. One limitation of the study was the small sample size of participants. Ieracitano et al. (2019) expanded the dataset and proposed a method that involves analyzing PSDs of EEG traces and representing them as 2D grayscale images (PSD-images). A customized CNN is designed to extract features from these PSD-images for both binary and three-way classification tasks. The CNN outperforms traditional methods, achieving an average accuracy of 89.8% for binary classification and 83.3% for three-way classification.
However, the accuracy of the three-class classification task was diminished due to the minimal distinctions observed in the PSD between AD and MCI. Multi-task learning has recently been explored to address the risk of overfitting of deep models, which often occurs with small sample sizes. Bi and Wang (2019) introduced a multi-task learning approach using a discriminative convolutional high-order Boltzmann machine with hybrid feature maps. The deep Boltzmann machine (DBM) and its variants have proven to be a potentially powerful alternative tool for feature extraction. To further improve the model, a label layer was introduced to bridge the gap between feature learning and classification. In this study, EEG signals were transformed into a 2D image format to preserve spatial structure, while multiple color channels were employed to represent the spectral dimension. Ultimately, the model successfully classified EEG spectral images into one of three classes (healthy control, MCI, and AD) with an accuracy of 95.04%. Nevertheless, the process of converting 1D signals into images required complicated data processing, and the dataset was insufficient in size. To overcome the problem of limited data and improve the generalization ability of deep models, Xia et al. (2023) employed overlapping sliding windows to augment the EEG data. They also designed a deep pyramid CNN (DPCNN) to discriminate the resting-state EEGs of AD, MCI, and healthy controls. The model achieved an accuracy of 97.10% in classifying the augmented datasets into three distinct categories.
In addition to utilizing solely EEG signals, You et al. (2020) combined both gait and EEG data to achieve classification of AD. To this end, they proposed a two-step cascade neural network approach. The first step involved the implementation of attention-based spatial temporal graph convolutional network (AST-GCN) on gait data to distinguish between healthy controls and patients. The second step utilized spatial-temporal convolutional networks on EEG data to classify patients as either MCI or AD. Experimental results with three-way classification showed outperformance of 91.07% compared to other methods. Apart from the recorded signals, omics maps can also be used to aid in the diagnosis of brain diseases. Researchers collected fecal samples and urine exosomes from NA and neurocognitive disorder (NCD) seniors to identify multi-omics signatures and metabolic pathways (Han et al., 2024). SVM achieved 92.69% accuracy in classifying the NA and NCD groups based on fused EEG data and multi-omics profiles. Boudaya et al. (2024) incorporated EEG and heart rate variability (HRV) to classify MCI patients and healthy control while subjects were taking the cognitive examinations. A hybrid ML model (SVM, RF, and gradient boosting (GB)) with a voting system was proposed to improve accuracy. The hybrid model finally achieved an average accuracy of 93.86%, sensitivity of 93.87%, and specificity of 93.53%.
4.6. Brain stroke
Stroke is a devastating disease and has become one of the leading causes of death globally (Feigin et al., 2014). There are two main types of brain strokes: ischemic and hemorrhagic. Acute ischemic stroke (AIS) is a prevalent form of cerebral infarction, accounting for over 80% of cases. It arises from an interruption in the cerebral blood flow (CBF) to a specific region, resulting in diminished oxygen and nutrient supply neurons. The consequences of a brain stroke can be severe, as they can affect various cognitive, motor, and sensory functions. Common symptoms include sudden weakness or numbness on one side of the body, speech impairment, impaired coordination, intense headache, and confusion. Early prognosis is necessary to mitigate the negative impact for patients. MRI and CT are commonly diagnostic techniques for AIS, but they are not suitable for long-term monitoring. Recently, EEG has emerged as a promising tool for both diagnosis and prognosis of AIS (Jordan, 2004; Foreman and Claassen, 2012). The observation of an increase in slow-wave (delta) activity compared to faster (alpha/beta) activity following the stroke has been well-documented (Finnigan et al., 2016). Here we collected several representative studies in EEG-based stroke detection and the summary is provided in Table 6.
Table 6.
Summary of electroencephalogram (EEG)-based brain stroke diagnosis
| Study | Detection type | Classifier/detector | Performance |
|---|---|---|---|
| Li et al., 2014 | Stroke | MKL-SVM | 89% (0-back) and 80% (1-back) accuracy |
| Giri et al., 2016 | Stroke | CNN | 86% accuracy |
| Qureshi et al., 2018 | Stroke | MLP | 95% accuracy |
| Li et al., 2020 | Ischemic/hemorrhagic stroke | SVM, DT, RF | 98.51% accuracy |
| Wilkinson et al., 2020 | Stroke type/stroke severity | RF | 76% accuracy |
| Dewi et al., 2020 | Stroke type/stroke severity | CNN | 97.3% accuracy |
| Guntari et al., 2020 | Stroke type/stroke severity | RNN | 90.00% accuracy |
| Singh et al., 2024 | Stroke type/stroke severity/affected artery | FCNN | 97.74% accuracy (stroke type)/2.1955 RMSE (stroke severity)/95.7% accuracy (affected artery) |
MKL: multiple kernel learning; SVM: support vector machine; CNN: convolutional neural network; MLP: multilayer perceptron; DT: decision tree; RF: random forest; RNN: recurrent neural network; FCNN: fully connected neural network; RMSE: root mean square error.
Currently, there are limitations in brain stroke detection using EEG as most of the datasets were obtained after the stroke onset. However, CBF reperfusion and restoration may change the EEG characteristics compared with the hyper-acute phase of stroke. Most studies focused on investigating EEG as a prognostic tool to identify the severity of post-stroke patients. Dewi et al. (2020) proposed a method that employs the PSD of EEG recordings as inputs and employs CNN for the automatic extraction of features, which achieved 97.3% accuracy in distinguishing normal, mild, moderate, and severe stroke. Guntari et al. (2020) proposed a novel approach to optimize EEG channels for evaluating post-stroke patient. Genetic algorithms optimized channel combinations, followed by RNN to classify the signals into three categories: stroke-free, minor stroke, and moderate stroke. The proposed method demonstrated an accuracy of 90.00% when using only 12 channels, surpassing the 72.22% accuracy when using all channels.
Other studies have explored the feasibility and utility of using portable and low-density EEG systems in time-sensitive scenarios such as ambulances or emergency rooms for stroke detection. Giri et al. (2016) used 2-channel EEG (C3 and OZ), along with 2-electrooculography (EOG) channels (left EOG and right EOG) to classify stroke data from health controls. It achieved an average accuracy of 86% using 1D convolutional neural network (1DCNN) with batch normalization, which outperformed other classifiers with simple feature extraction. Qureshi et al. (2018) used wearable EEG devices and ML for ischemic stroke detection. EEG data were collected from only six channels. MLP and bootstrap models obtained 95% accuracy and a ROC curve area of 0.85. Wilkinson et al. (2020) designed an affordable and portable EEG system consisting of seven electrodes that could detect stroke and stroke severity. They investigated ratios like delta/alpha ratio (DAR) and (delta+theta)/(alpha+beta) ratio (DTABR), along with head movement data. RF was employed for subgroup differentiation. Moderate/severe stroke from minor stroke/controls could be distinguished with 63% sensitivity, 86% specificity, and 76% accuracy.
The aforementioned research used QEEG measures as stroke biomarkers, involving numerical analysis of the EEG signal spectrum. ERP was also used as an assessment feature to assess the extent of motor impairment in stroke patients. Li et al. (2014) conducted cognitive tasks involving working memory to collect ERP data, wherein participants were presented with a random sequence of 4-digit numbers on a monitor and tasked with determining if a specific digit appeared on the screen (0-back task) or if the current number had been previously shown (1-back task). Health controls and stroke patients could be classified using the multiple kernel learning (MKL)-SVM algorithm with over 89% accuracy for the 0-back task and 80% accuracy for the 1-back task.
Besides stroke and stroke severity detection, research has also been conducted on the classification of ischemic stroke and hemorrhagic stroke. Li et al. (2020) presented a novel method for classifying EEG signals obtained from stroke patients with cerebral infarction and cerebral hemorrhage. A feature extraction method based on the fusion feature of wavelet packet energy and hierarchical entropy feature was introduced. SVM, DT, and RF were employed as classifiers. Results showed that the RF using the proposed fusion method achieved the best accuracy of 98.51%. Singh et al. (2024) proposed a diagnostic tool that could quickly obtain multiple stroke information using one-minute EEG data. Three different fully connected neural network (FCNN) models were developed for each task: classifying stroke type, identifying the affected artery, and assessing stroke severity. The models achieved impressive results, including an accuracy of 97.74% for stroke type classification, a root mean square error (RMSE) of 2.1955 for stroke severity assessment, and an accuracy of 95.7% for identifying the affected artery.
4.7. Autism spectrum disorder
ASD is a complex neurodevelopmental condition characterized by disorders in social interaction and communication, along with restricted or repetitive behaviors (Lord et al., 2018). The etiology of its onset remains incompletely elucidated, yet it is postulated to result from a combination of genetic and environmental factors impacting brain development. ASD is referred to as a spectrum disorder as it manifests a wide range of symptoms that vary greatly among individuals. The association between epilepsy and ASD has been recognized due to the higher prevalence of epilepsy among individuals with ASD (Amiet et al., 2008). The most common protocol used to diagnose ASD involves a qualitative behavioral assessment conducted by experts following internationally established criteria such as the DSM-5. Early intervention and therapies for behavior and speech are crucial in improving communication skills and overall quality of life for individuals with ASD. Some research has indicated a U-shaped profile EEG pattern from patients with ASD, where the activity of high-frequency bands (beta and gamma) and low-frequency bands (delta and theta) abnormally increases, and the activity in the middle frequencies appears reduced (Wang et al., 2013). The summary of the related studies is presented in Table 7.
Table 7.
Summary of electroencephalogram (EEG)-based autism spectrum disorder (ASD) diagnosis
| Study | Detection type | Classifier/detector | Performance |
|---|---|---|---|
| Bosl et al., 2011 | HRA | SVM | Over 80% accuracy |
| Jamal et al., 2014 | ASD | SVM | 94.7% accuracy |
| Grossi et al., 2017 | ASD | ML | 92.8% accuracy (RF) |
| Bosl et al., 2018 | LRC vs. HRA vs. ASD | SVM | 99% specificity, 82% sensitivity,97% positive predictive values (three-month infants) |
| Ibrahim et al., 2018 | Epilepsy vs. ASD vs. normal | ANN, KNN, SVM, LDA | 94.6% accuracy |
| Thapaliya et al., 2018 | ASD | NB, LR | 100% accuracy |
| Abdolzadegan et al., 2020 | ASD | SVM | 99.91% sensitivity, 90.57% accuracy |
| Bouallegue et al., 2020 | ASD | CNN | 99.5% accuracy |
| Abou-Abbas et al., 2021 | HRA without ASD vs. HRA with ASD | SVM, KNN | 88.44% accuracy |
| Magboo and Magboo, 2022 | ASD | SVM, LR, AdaBoost | 99%–100% accuracy |
HRA: high risk for autism; SVM: support vector machine; ML: machine learning; RF: random forest; LRC: low-risk control; ANN: artificial neural network; KNN: k-nearest neighbor; LDA: linear discriminant analysis; NB: naive Bayes; LR: logistic regression; CNN: convolutional neural network.
Multiscale entropy (MSE) is designed to differentiate various levels of complexity and information within a biological signal. Bosl et al. (2011) used modified MSE derived from resting-state EEG data to distinguish between typically developing children and those who were at high risk for autism (HRA) based on older siblings’ ASD diagnoses. A multiclass SVM algorithm was employed for classification within different age groups (6 to 24 months). Infants were classified with over 80% accuracy into control and HRA groups at nine months of age. This research indicated that EEG complexity could be a potential biomarker for ASD at very early ages before the onset of symptoms. However, children at high risk do not necessarily develop ASD; typically developing children can also show developmental problems later on. Grossi et al. (2017) collected EEG data from a group of 15 children with proven ASD and 10 proven typically developing children. They also introduced an advanced processing algorithm called multi-scale ranked organizing map coupled with implicit function as squashing time (MS-ROM/I-FAST) algorithm, extracting features from EEG data without pre-processing. Results showed consistent 100% accuracy in distinguishing autistic cases using various ML systems in a training-testing protocol. The Leave One Out protocol yielded the highest accuracy of 92.8% using the RF classifier. However, due to the limitation of the small sample size, it was not enough to derive definitive conclusions. Later, Bosl et al. (2018) improved their previous study by extending the EEG measurement period. EEG data from 99 infants with ASD-risk siblings (high-risk) and 89 low-risk controls were collected from 3 to 36 months of age. Then, participants were classified into three outcome groups: low-risk control (LRC)—infants from the low-risk group without ASD; HRA—infants from the high-risk group without ASD; ASD—infants from either group who received an ASD diagnosis. Classification and prediction were computed using the SVM, which and achieved accurate diagnosis of ASD or not ASD from as early as the age of three months, with high specificity (99%), sensitivity (82%), and positive predictive values (97%).
However, recording accurate EEG signals from restless children with autism is challenging. Commonly, recordings are taken during sleep with medication, which may alter the EEG and complicate the analysis. Abdolzadegan et al. (2020) introduced a new protocol to record EEG without sedation as well as extract reliable features. EEG was acquired during cartoon video playback, and density-based spatial clustering of applications with noise (DBSCAN) was used to remove the artifacts. SVM classifiers achieved a classification accuracy of 90.57% and a notably high sensitivity of 99.91%. Besides solely focusing on EEG, Thapaliya et al. (2018) combined EEG and eye-tracking data to identify ASD. Data retrieval was performed while watching video clips. Multiple ML classifiers were employed, and 100% accuracy was obtained using NB and LR classifiers.
Studies have investigated impairments in emotion perception in ASD using ERPs recorded in response to different face stimuli. Jamal et al. (2014) extracted EEG signals during face processing tasks when three types of emotional face stimuli were presented, including fearful, neutral, and happy facial expression. Complex network parameters were used as features, and SVM was used as the classifier; the research achieved an accuracy of 94.7%. Abou-Abbas et al. (2021) explored visual ERPs and ML to classify high-risk infants who later developed ASD. ERP data were collected from infants reacting to changing gaze directions in faces (Elsabbagh et al., 2012). They used the empirical mode decomposition (EMD) technique to decompose ERPs into a set of intrinsic mode functions. Features extracted via IMFs were input into SVM and KNN for classification. The highest accuracy the study reached was 88.44% using SVM.
Ibrahim et al. (2018) investigated EEG classification techniques for epilepsy and ASD automated diagnosis. DWT and cross-correlation (measuring synchronization between EEG channels) were used to extract features from the EEG segment. Several ML methods, including ANN, KNN, SVM, and LDA, were used for classification. The ASD dataset was provided by King Abdulaziz University (KAU) in Saudi Arabia with ten normal controls and nine ASD patients (Alhaddad et al., 2012). The study finally obtained an overall accuracy of 94.6% on the three-class classification problem (normal vs. epilepsy vs. autism). Based on the same dataset, Bouallegue et al. (2020) proposed an approach involving a DL process. They used finite impulse response (FIR) and infinite impulse response (IIR) filters with a gated-recurrent unit (GRU) RNN to preprocess informative sub-bands, followed by independent component analysis (ICA). CNN was employed in the classification. The system attained an accuracy of 99.5% for autism diagnosis. Magboo and Magboo (2022) applied several ML models, including SVM, LR, and AdaBoost, to a publicly available child ASD dataset. The best models could achieve 99%–100% accuracy. The LIME algorithm provided model explainability, helping physicians understand the model’s predictions.
5. Discussion
5.1. Types of EEG dataset and challenges in brain disease diagnosis
Different types of EEG signals, including resting-state EEG and ERP, should be taken into account depending on the disease. It is important to determine which type of biomarker from an EEG signal is the most informative for a certain situation. For instance, in schizophrenia detection, ERPs such as the MMN component were used to study auditory processing deficits in individuals (Shim et al., 2016; Chang et al., 2021). The P300 component was frequently utilized to investigate cognitive and memory functions in individuals with depression and AD (Shim et al., 2019; Rad et al., 2021). Greater anterior EEG activity, particularly the EEG alpha asymmetry and beta frequency band, has shown promise for depression detection (Li et al., 2016; Mumtaz et al., 2017). In the context of PD diagnosis, alpha/theta spectral ratio, alterations in beta frequency synchronization, and PAC have been emphasized as potential biomarkers (Chaturvedi et al., 2017; Shah et al., 2020). Additionally, time-frequency plots from EEG signals have also been used as EEG biomarkers. Some studies have successfully converted 1D EEG signals into 2D feature images, a transformation that has improved the applicability of image-based models such as CNN and GNN (Truong et al., 2018; Bi and Wang, 2019; Ieracitano et al., 2019; Li XW et al., 2019; Li Y et al., 2019; Khare et al., 2021; Wang et al., 2022). This advancement allows for the capture of both spatial and hierarchical patterns. However, computational intensity should be considered at the same time to strike a balance between improved representation and efficient processing.
When designing the architecture of the detection model, both the size and characteristics of the dataset should be considered, as AI is based on methods driven by data. Datasets in most of the studies accommodated a relatively small number of participants, which may lead to a lack of generalizability and pose limitations when translating into clinical utilities. Solutions include data augmentation, such as applying transformations to EEG signals or using GANs to create synthetic data, which can expand the dataset and improve model robustness (Zhang et al., 2020; Wang et al., 2022; Saadatinia and Salimi-Badr, 2024). Transfer learning, which uses a pre-trained model on a larger relevant dataset and fine-tunes it on a smaller target dataset, is also useful for solving overfitting problems (Daoud and Bayoumi, 2019). Regularization techniques, like dropout and weight regularization, can also help prevent overfitting by ensuring simpler models that generalize better. Data-sharing platforms can also be established among researchers to create larger, more diverse datasets.
In some mental diseases, such as depression, patients participating in the test were under medication, and the possible confounding effects of psychotropic medication may not be controlled. Even when participants are asked to be medication-free for a while before EEG recording, the effect of medication cannot be ruled out completely. Studies could explore strategies to minimize confounding variables or conduct research to assess the impact of specific psychotropic medications on EEG (Shah et al., 2020; Lee et al., 2021; Qiu et al., 2022).
Most published work on EEG-based brain disease diagnosis typically relied on EEG setups ranging among 16, 20, 32, 64, and 128 electrodes. In the future, it would be advantageous to focus on acquiring data with a reduced number of electrodes, thereby enabling the development of portable EEG devices capable of conducting long-term recordings in everyday settings.
5.2. Advancements and challenges of AI models
Before the advent of DL, many analyses from statistics, time, frequency, or time-frequency domains were used in various conventional ML algorithms. Comparing the classification performance achieved by different architecture designs across diverse brain diseases and EEG datasets poses challenges, but the complexity of EEG signals makes it difficult to achieve truly spectacular results. DL deployment stands out as having several advantages. First, DL models can achieve a deeper understanding of data due to their ability to learn hierarchical representations directly from raw data. This hierarchical learning enables the extraction of complex and abstract features that might be difficult to identify using traditional ML methods. Second, DL models facilitate automatic feature selection, reducing the reliance on hand-crafted features and domain-specific knowledge. In addition, hybrid algorithms, such as CNN-LSTM or CNN-AE, tend to outperform other individual algorithms in ablation experiments (Ay et al., 2019; Daoud and Bayoumi, 2019; Liang et al., 2020; Ahmedt-Aristizabal et al., 2021; Sharma et al., 2021). However, one of the problems of the DL models is their limited interpretability; because they involve intricate mathematical operations, clinicians may find it challenging to trust and adopt DL models for EEG data analysis in medical contexts. This can be improved through the development of techniques such as attention mechanisms or the use of XAI methods.
One limitation in determining the best model is the database used. Local datasets can introduce biases due to varying factors such as age and gender, potentially affecting the reported results. Conversely, using open-access databases makes research more comparable and helps identify the best methods. For instance, in schizophrenia detection research, studies by Oh et al. (2019) and Jahmunah et al. (2019) utilized the same dataset from the Institute of Psychiatry and Neurology in Warsaw, Poland. Their results demonstrated that the CNN model outperformed the traditional SVM model. However, even studies using the same model and dataset can yield different results due to minor differences in parameters and data processing methods. For example, in seizure detection research, Acharya et al. (2018b) and Shoeibi et al. (2021b) proposed CNN models based on the same dataset but achieved different performance levels. In the future, efforts should be made to standardize the reporting of AI technologies with standardized methods for describing and measuring results (Sounderajah et al., 2021).
In general, high accuracy often comes with low specificity compared to recall and accuracy, highlighting the challenge of achieving relative precision. Additionally, DL models, such as CNNs and LSTMs, are increasingly favored for their ability to automatically extract and learn complex features from high-dimensional data. CNNs automatically extract spatial features from EEG data and are efficient at parameter sharing. LSTMs can capture long-term temporal dependencies in EEG signals and alleviate vanishing gradient issues. Traditional ML models like SVMs remain valuable, especially when combined with hand-crafted features and used in ensemble approaches.
Integrating advanced AI-driven methodologies into the diagnosis and management of brain diseases also holds significant promise for enhancing diagnostic precision and improving patient outcomes. By leveraging natural language processing (NLP) and large language models like chat generative pre-trained transformer (ChatGPT), clinicians can screen clinical notes for risk factors and suggest potential diagnoses, thereby facilitating earlier intervention. Additionally, the future exploration of web-based diagnosis techniques has the potential to improve the efficiency of CAD systems. Pre-trained models can be deployed based on cloud computing, reducing the computational burden on local systems and enabling healthcare professionals to receive results in a timely manner.
5.3. Potential of multimodal EEG and neurophysiological data
The exploration of multimodal EEG holds potential for the future. For example, the simultaneous collection of EEG and EOG or eye-tracking data helps us to understand how visual attention and processing are reflected in both brain activity and eye movement (Giri et al., 2016; Thapaliya et al., 2018). EMG and gait data provide insight into the motor conditions of PD and AD patients (Saikia et al., 2019; You et al., 2020). EMG can also aid in the elimination of muscle artifacts in EEG data. It has been indicated that changes in short-term heart rate recordings can be observed during a seizure preictal state (Moridani and Farhadi, 2017).
However, EEG has its own advantages and disadvantages. For instance, while EEG is non-invasive and relatively inexpensive and provides real-time brain activity data, it has limited spatial resolution compared to imaging modalities like MRI. Additionally, EEG is sensitive to noise and artifacts, which can complicate data interpretation. Apart from EEG data, combining multiple data sources such as medical records, imaging studies, and omics data can provide a comprehensive view of a patient’s condition. By integrating EEG and MRI data, it is possible to capture comprehensive information about both brain activity and structure, considering spatial and temporal aspects simultaneously. HRV can be further combined with EEG recordings to improve epilepsy prediction methods, as HRV offers valuable information about the autonomic nervous system’s functioning, reflecting the balance between sympathetic and parasympathetic activity. In the diagnosis of complex brain diseases such as AD, PD, and schizophrenia, omics data provide a comprehensive view of the molecular mechanisms underlying brain function and disease. By exploring the interaction between neural oscillations and molecular pathways, clinicians can uncover biomarkers that are more specific and informative. Future research can investigate innovative techniques to fuse information from different modalities, thereby offering a more comprehensive understanding of brain diseases, leading to improved diagnostic precision and more personalized therapeutic strategies. To merge different modalities, several approaches can be explored. For example, DL methods, such as multimodal neural networks and attention mechanisms, dynamically integrate multiple data sources. Graph-based methods, like GNN, model interactions between modalities, while canonical correlation analysis (CCA) finds shared patterns between them.
6. Conclusions
In this article, a review of EEG-based diagnosis of brain disease is presented with a focus on AI technology. AI has shown significant promise in the diagnosis of brain diseases using EEG signals. By leveraging both traditional ML and advanced DL techniques, AI systems can enhance diagnostic accuracy and offer insights into complex brain disorders. The studies reviewed highlight the effectiveness of various AI models, such as SVM and CNN, and hybrid models, like CNN-LSTM, in classifying and predicting conditions such as epilepsy, schizophrenia, depression, PD, AD, brain stroke, and ASD. Key considerations include the selection of the most informative type of EEG signal for specific diseases, the balance between improved representation and computational efficiency, and the need for larger and more balanced datasets to improve generalizability. Future research should integrate multimodal data, including other types of EEG, such as EOG and EMG, or combine EEG with other brain imaging techniques, like MRI and HRV, to provide a more comprehensive understanding of brain diseases. Additionally, enhancing the interpretability of DL models and exploring web-based diagnostic systems using pre-trained models on cloud platforms could further improve the efficiency and accessibility of AI-driven diagnostic tools.
Acknowledgments
This work was supported by the National Key Research and Development Project of China (No. 2021ZD0200405) and the National Natural Science Foundation of China (Nos. 62271443, 32250008, and 82330064).
Author contributions
Ping WANG, Liujing ZHUANG, and Hong ZHANG designed the manuscript. Shunuo SHANG wrote the manuscript. Yingqian SHI completed the drawing of all the figures. Yajie ZHANG and Mengxue LIU proofread the article. All authors have read and approved the final version.
Compliance with ethics guidelines
Liujing ZHUANG is a Young Scientist Committee Member for Journal of Zhejiang University-SCIENCE B (Biomedicine & Biotechnology) and was not involved in the editorial review or the decision to publish this article. Shunuo SHANG, Yingqian SHI, Yajie ZHANG, Mengxue LIU, Hong ZHANG, Ping WANG, and Liujing ZHUANG declare that they have no conflict of interest.
This review does not contain any studies with human or animal subjects performed by any of the authors.
References
- Abdolzadegan D, Moattar MH, Ghoshuni M, 2020. A robust method for early diagnosis of autism spectrum disorder from EEG signals based on feature selection and DBSCAN method. Biocybern Biomed Eng, 40(1): 482-493. 10.1016/j.bbe.2020.01.008 [DOI] [Google Scholar]
- Abou-Abbas L, van Noordt S, Desjardins JA, et al. , 2021. Use of empirical mode decomposition in ERP analysis to classify familial risk and diagnostic outcomes for autism spectrum disorder. Brain Sci, 11(4): 409. 10.3390/brainsci11040409 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acharya UR, Oh SL, Hagiwara Y, et al. , 2018a. Automated EEG-based screening of depression using deep convolutional neural network. Comput Methods Programs Biomed, 161: 103-113. 10.1016/j.cmpb.2018.04.012 [DOI] [PubMed] [Google Scholar]
- Acharya UR, Oh SL, Hagiwara Y, et al. , 2018b. Deep convolutional neural network for the automated detection and diagnosis of seizure using EEG signals. Comput Biol Med, 100: 270-278. 10.1016/j.compbiomed.2017.09.017 [DOI] [PubMed] [Google Scholar]
- Ahmedt-Aristizabal D, Fernando T, Denman S, et al. , 2021. Identification of children at risk of schizophrenia via deep learning and EEG responses. IEEE J Biomed Health Inform, 25(1): 69-76. 10.1109/JBHI.2020.2984238 [DOI] [PubMed] [Google Scholar]
- Albert MS, Dekosky ST, Dickson D, et al. , 2011. The diagnosis of mild cognitive impairment due to Alzheimer’s disease: Recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement, 7(3): 270-279. 10.1016/j.jalz.2011.03.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alhaddad MJ, Kamel MI, Malibary HM, et al. , 2012. Diagnosis autism by fisher linear discriminant analysis FLDA via EEG. Int J Bio-Sci Bio-Technol, 4(2): 45-54. [Google Scholar]
- Alickovic E, Kevric J, Subasi A, 2018. Performance evaluation of empirical mode decomposition, discrete wavelet transform, and wavelet packed decomposition for automated epileptic seizure detection and prediction. Biomed Signal Process Control, 39: 94-102. 10.1016/j.bspc.2017.07.022 [DOI] [Google Scholar]
- Alsharabi K, Salamah YB, Abdurraqeeb AM, et al. , 2022. EEG signal processing for Alzheimer’s disorders using discrete wavelet transform and machine learning approaches. IEEE Access, 10: 89781-89797. 10.1109/ACCESS.2022.3198988 [DOI] [Google Scholar]
- American Psychiatric Association , 2013. Diagnostic and Statistical Manual of Mental Disorders, Fifth Edition. American Psychiatric Publishing, Washington, USA. 10.1176/appi.books.9780890425596 [DOI] [Google Scholar]
- Amiet C, Gourfinkel-An I, Bouzamondo A, et al. , 2008. Epilepsy in autism is associated with intellectual disability and gender: evidence from a meta-analysis. Biol Psychiatry, 64(7): 577-582. 10.1016/j.biopsych.2008.04.030 [DOI] [PubMed] [Google Scholar]
- Andrzejak RG, Lehnertz K, Mormann F, et al. , 2001. Indications of nonlinear deterministic and finite-dimensional structures in time series of brain electrical activity: dependence on recording region and brain state. Phys Rev E, 64(6): 061907. 10.1103/PhysRevE.64.061907 [DOI] [PubMed] [Google Scholar]
- Anoop A, Singh PK, Jacob RS, et al. , 2010. CSF biomarkers for Alzheimer’s disease diagnosis. Int J Alzheimers Dis, 2010: 606802. 10.4061/2010/606802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antoniades A, Spyrou L, Martin-Lopez D, et al. , 2018. Deep neural architectures for mapping scalp to intracranial EEG. Int J Neur Syst, 28(8): 1850009. 10.1142/S0129065718500090 [DOI] [PubMed] [Google Scholar]
- Ay B, Yildirim O, Talo M, et al. , 2019. Automated depression detection using deep representation and sequence learning with EEG signals. J Med Syst, 43: 205. 10.1007/s10916-019-1345-y [DOI] [PubMed] [Google Scholar]
- Barros C, Silva CA, Pinheiro AP, 2021. Advanced EEG-based learning approaches to predict schizophrenia: promises and pitfalls. Artif Intell Med, 114: 102039. 10.1016/j.artmed.2021.102039 [DOI] [PubMed] [Google Scholar]
- Bateman RJ, Xiong CJ, Benzinger TLS, et al. , 2012. Clinical and biomarker changes in dominantly inherited Alzheimer’s disease. N Engl J Med, 367(9): 795-804. 10.1056/NEJMoa1202753 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bdaqli M, Shoeibi A, Moridian P, et al. , 2024. Diagnosis of Parkinson disease from EEG signals using a CNN-LSTM model and explainable AI. In: Ferrández Vicente JM, Val Calvo M, Adeli H (Eds.), Artificial Intelligence for Neuroscience and Emotional Systems. IWINAC 2024. Lecture Notes in Computer Science, Vol. 14674. Springer, Cham. 10.1007/978-3-031-61140-7_13 [DOI] [Google Scholar]
- Beitz JM, 2014. Parkinson’s disease: a review. Front Biosci, 6(1): 65-74. 10.2741/s415 [DOI] [PubMed] [Google Scholar]
- Bi XJ, Wang HB, 2019. Early Alzheimer’s disease diagnosis based on EEG spectral images using deep learning. Neural Netw, 114: 119-135. 10.1016/j.neunet.2019.02.005 [DOI] [PubMed] [Google Scholar]
- Bosl W, Tierney A, Tager-Flusberg H, et al. , 2011. EEG complexity as a biomarker for autism spectrum disorder risk. BMC Med, 9: 18. 10.1186/1741-7015-9-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bosl WJ, Tager-Flusberg H, Nelson CA, 2018. EEG analytics for early detection of autism spectrum disorder: a data-driven approach. Sci Rep, 8: 6828. 10.1038/s41598-018-24318-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouallegue G, Djemal R, Alshebeili SA, et al. , 2020. A dynamic filtering DF-RNN deep-learning-based approach for EEG-based neurological disorders diagnosis. IEEE Access, 8: 206992-207007. 10.1109/ACCESS.2020.3037995 [DOI] [Google Scholar]
- Boudaya A, Chaabene S, Bouaziz B, et al. , 2024. Mild cognitive impairment detection based on EEG and HRV data. Digital Signal Process, 147: 104399. 10.1016/j.dsp.2024.104399 [DOI] [Google Scholar]
- Brenner RP, Reynolds CF III, Ulrich RF, 1988. Diagnostic efficacy of computerized spectral versus visual EEG analysis in elderly normal, demented and depressed subjects. Electroencephalogr Clin Neurophysiol, 69(2): 110-117. 10.1016/0013-4694(88)90206-4 [DOI] [PubMed] [Google Scholar]
- Brinkmann BH, Wagenaar J, Abbot D, et al. , 2016. Crowdsourcing reproducible seizure forecasting in human and canine epilepsy. Brain, 139(6): 1713-1722. 10.1093/brain/aww045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai HS, Qu ZD, Li Z, et al. , 2020. Feature-level fusion approaches based on multimodal EEG data for depression recognition. Inf Fusion, 59: 127-138. 10.1016/j.inffus.2020.01.008 [DOI] [Google Scholar]
- Cassani R, Falk TH, Fraga FJ, et al. , 2017. Towards automated electroencephalography-based Alzheimer’s disease diagnosis using portable low-density devices. Biomed Signal Process Control, 33: 261-271. 10.1016/j.bspc.2016.12.009 [DOI] [Google Scholar]
- Cavanagh JF, Napolitano A, Wu C, et al. , 2017. The patient repository for EEG data+computational tools (PRED+CT). Front Neuroinform, 11: 67. 10.3389/fninf.2017.00067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang Q, Li CC, Tian Q, et al. , 2021. Classification of first-episode schizophrenia, chronic schizophrenia and healthy control based on brain network of mismatch negativity by graph neural network. IEEE Trans Neural Syst Rehabil Eng, 29: 1784-1794. 10.1109/TNSRE.2021.3105669 [DOI] [PubMed] [Google Scholar]
- Chaturvedi M, Hatz F, Gschwandtner U, et al. , 2017. Quantitative EEG (QEEG) measures differentiate Parkinson’s disease (PD) patients from healthy controls (HC). Front Aging Neurosci, 9: 3. 10.3389/fnagi.2017.00003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciprian C, Masychev K, Ravan M, et al. , 2021. Diagnosing schizophrenia using effective connectivity of resting-state EEG data. Algorithms, 14(5): 139. 10.3390/a14050139 [DOI] [Google Scholar]
- Cook MJ, O'Brien TJ, Berkovic SF, et al. , 2013. Prediction of seizure likelihood with a long-term, implanted seizure advisory system in patients with drug-resistant epilepsy: a first-in-man study. Lancet Neurol, 12(6): 563-571. 10.1016/s1474-4422(13)70075-9 [DOI] [PubMed] [Google Scholar]
- Daoud H, Bayoumi MA, 2019. Efficient epileptic seizure prediction based on deep learning. IEEE Trans Biomed Circuits Syst, 13(5): 804-813. 10.1109/TBCAS.2019.2929053 [DOI] [PubMed] [Google Scholar]
- Devia C, Mayol-Troncoso R, Parrini J, et al. , 2019. EEG classification during scene free-viewing for schizophrenia detection. IEEE Trans Neural Syst Rehabil Eng, 27(6): 1193-1199. 10.1109/TNSRE.2019.2913799 [DOI] [PubMed] [Google Scholar]
- Dewi FY, Faza A, Prajitno P, et al. , 2020. Stroke severity classification based on EEG signals using 1D convolutional neural network. J Phys Conf Ser, 1528: 012006. 10.1088/1742-6596/1528/1/012006 [DOI] [Google Scholar]
- Dogan S, Baygin M, Tasci B, et al. , 2023. Primate brain pattern-based automated Alzheimer’s disease detection model using EEG signals. Cogn Neurodyn, 17(3): 647-659. 10.1007/s11571-022-09859-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elsabbagh M, Mercure E, Hudry K, et al. , 2012. Infant neural sensitivity to dynamic eye gaze is associated with later emerging autism. Curr Biol, 22(4): 338-342. 10.1016/j.cub.2011.12.056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emami A, Kunii N, Matsuo T, et al. , 2019. Autoencoding of long-term scalp electroencephalogram to detect epileptic seizure for diagnosis support system. Comput Biol Med, 110: 227-233. 10.1016/j.compbiomed.2019.05.025 [DOI] [PubMed] [Google Scholar]
- Feigin VL, Forouzanfar MH, Krishnamurthi R, et al. , 2014. Global and regional burden of stroke during 1990‒2010: findings from the global burden of disease study 2010. Lancet, 383(9913): 245-255. 10.1016/S0140-6736(13)61953-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferree TC, Luu P, Russell GS, et al. , 2001. Scalp electrode impedance, infection risk, and EEG data quality. Clin Neurophysiol, 112(3): 536-544. 10.1016/S1388-2457(00)00533-2 [DOI] [PubMed] [Google Scholar]
- Finnigan S, Wong A, Read S, 2016. Defining abnormal slow EEG activity in acute ischaemic stroke: delta/alpha ratio as an optimal QEEG index. Clin Neurophysiol, 127(2): 1452-1459. 10.1016/j.clinph.2015.07.014 [DOI] [PubMed] [Google Scholar]
- Foreman B, Claassen J, 2012. Quantitative EEG for the detection of brain ischemia. Crit Care, 16: 216. 10.1186/cc11230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geng MX, Zhou WD, Liu GY, et al. , 2020. Epileptic seizure detection based on stockwell transform and bidirectional long short-term memory. IEEE Trans Neural Syst Rehabil Eng, 28(3): 573-580. 10.1109/TNSRE.2020.2966290 [DOI] [PubMed] [Google Scholar]
- Giri EP, Fanany MI, Arymurthy AM, et al. , 2016. Ischemic stroke identification based on EEG and EOG using ID convolutional neural network and batch normalization. 2016 International Conference on Advanced Computer Science and Information Systems (ICACSIS), Malang, Indonesia, p.484-491. 10.1109/ICACSIS.2016.7872780 [DOI] [Google Scholar]
- Goodfellow I, Bengio Y, Courville A, 2016. Deep Learning. Translators, MIT Press, Cambridge, Massachusetts. [Google Scholar]
- Goshvarpour A, Goshvarpour A, 2020. Schizophrenia diagnosis using innovative EEG feature-level fusion schemes. Phys Eng Sci Med, 43(1): 227-238. 10.1007/s13246-019-00839-1 [DOI] [PubMed] [Google Scholar]
- Gramacki A, Gramacki J, 2022. A deep learning framework for epileptic seizure detection based on neonatal EEG signals. Sci Rep, 12: 13010. 10.1038/s41598-022-15830-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossi E, Olivieri C, Buscema M, 2017. Diagnosis of autism through EEG processed by advanced computational algorithms: a pilot study. Comput Methods Programs Biomed, 142: 73-79. 10.1016/j.cmpb.2017.02.002 [DOI] [PubMed] [Google Scholar]
- Guntari EW, Djamal EC, Nugraha F, et al. , 2020. Classification of post-stroke EEG signal using genetic algorithm and recurrent neural networks. 2020 7th International Conference on Electrical Engineering, Computer Sciences and Informatics (EECSI), Yogyakarta, Indonesia, p.156-161. 10.23919/EECSI50503.2020.9251296 [DOI] [Google Scholar]
- Han Y, Zeng XL, Hua L, et al. , 2024. The fusion of multi-omics profile and multimodal EEG data contributes to the personalized diagnostic strategy for neurocognitive disorders. Microbiome, 12: 12. 10.1186/s40168-023-01717-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoole P, Pirapaharan K, Basar SA, et al. , 2012. Autism, EEG and brain electromagnetics research. 2012 IEEE-EMBS Conference on Biomedical Engineering and Sciences, Langkawi, Malaysia, p.541-543. 10.1109/IECBES.2012.6498036 [DOI] [Google Scholar]
- Huang DD, Qian K, Fei DY, et al. , 2012. Electroencephalography (EEG)-based brain-computer interface (BCI): a 2-D virtual wheelchair control based on event-related desynchronization/synchronization and state control. IEEE Trans Neural Syst Rehabil Eng, 20(3): 379-388. 10.1109/TNSRE.2012.2190299 [DOI] [PubMed] [Google Scholar]
- Hussein R, Palangi H, Ward RK, et al. , 2019. Optimized deep neural network architecture for robust detection of epileptic seizures using EEG signals. Clin Neurophysiol, 130(1): 25-37. 10.1016/j.clinph.2018.10.010 [DOI] [PubMed] [Google Scholar]
- Ibrahim S, Djemal R, Alsuwailem A, 2018. Electroencephalography (EEG) signal processing for epilepsy and autism spectrum disorder diagnosis. Biocybern Biomed Eng, 38(1): 16-26. 10.1016/j.bbe.2017.08.006 [DOI] [Google Scholar]
- Ieracitano C, Mammone N, Bramanti A, et al. , 2019. A Convolutional Neural Network approach for classification of dementia stages based on 2D-spectral representation of EEG recordings. Neurocomputing, 323: 96-107. 10.1016/j.neucom.2018.09.071 [DOI] [Google Scholar]
- Ihle M, Feldwisch-Drentrup H, Teixeira CA, et al. , 2012. EPILEPSIAE—A European epilepsy database. Comput Methods Programs Biomed, 106(3): 127-138. 10.1016/j.cmpb.2010.08.011 [DOI] [PubMed] [Google Scholar]
- Jahmunah V, Oh SL, Rajinikanth V, et al. , 2019. Automated detection of schizophrenia using nonlinear signal processing methods. Artif Intell Med, 100: 101698. 10.1016/j.artmed.2019.07.006 [DOI] [PubMed] [Google Scholar]
- Jamal W, Das S, Oprescu IA, et al. , 2014. Classification of autism spectrum disorder using supervised learning of brain connectivity measures extracted from synchrostates. J Neural Eng, 11(4): 046019. 10.1088/1741-2560/11/4/046019 [DOI] [PubMed] [Google Scholar]
- Jankovic J, 2008. Parkinson’s disease: clinical features and diagnosis. J Neurol Neurosurg Psychiatry, 79(4): 368-376. 10.1136/jnnp.2007.131045 [DOI] [PubMed] [Google Scholar]
- Jemal I, Mitiche A, Mezghani N, 2021. A study of EEG feature complexity in epileptic seizure prediction. Appl Sci, 11(4): 1579. 10.3390/app11041579 [DOI] [Google Scholar]
- Johannesen JK, Bi JB, Jiang RH, et al. , 2016. Machine learning identification of EEG features predicting working memory performance in schizophrenia and healthy adults. Neuropsychiatr Electrophysiol, 2: 3. 10.1186/s40810-016-0017-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jordan KG, 2004. Emergency EEG and continuous EEG monitoring in acute ischemic stroke. J Clin Neurophysiol, 21(5): 341-352. 10.1097/01.WNP.0000145005.59766.D2 [DOI] [PubMed] [Google Scholar]
- Khare SK, Bajaj V, Acharya UR, 2021. PDCNNet: an automatic framework for the detection of Parkinson’s disease using EEG signals. IEEE Sensors J, 21(15): 17017-17024. 10.1109/JSEN.2021.3080135 [DOI] [Google Scholar]
- Kiral-Kornek I, Roy S, Nurse E, et al. , 2018. Epileptic seizure prediction using big data and deep learning: toward a mobile system. eBioMedicine, 27: 103-111. 10.1016/j.ebiom.2017.11.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krook-Magnuson E, Soltesz I, 2015. Beyond the hammer and the scalpel: selective circuit control for the epilepsies. Nat Neurosci, 18(3): 331-338. 10.1038/nn.3943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak NS, Müller KR, Lee SW, 2015. A lower limb exoskeleton control system based on steady state visual evoked potentials. J Neural Eng, 12(5): 056009. 10.1088/1741-2560/12/5/056009 [DOI] [PubMed] [Google Scholar]
- LeCun Y, Bengio Y, Hinton G, 2015. Deep learning. Nature, 521(7553): 436-444. 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- Lee S, Hussein R, Ward R, et al. , 2021. A convolutional-recurrent neural network approach to resting-state EEG classification in Parkinson’s disease. J Neurosci Methods, 361: 109282. 10.1016/j.jneumeth.2021.109282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li FL, Wang JJ, Liao YY, et al. , 2019. Differentiation of schizophrenia by combining the spatial EEG brain network patterns of rest and task P300. IEEE Trans Neural Syst Rehabil Eng, 27(4): 594-602. 10.1109/TNSRE.2019.2900725 [DOI] [PubMed] [Google Scholar]
- Li FL, Fan YZ, Zhang XY, et al. , 2020. Multi-feature fusion method based on EEG signal and its application in stroke classification. J Med Syst, 44: 39. 10.1007/s10916-019-1517-9 [DOI] [PubMed] [Google Scholar]
- Li XO, Chen X, Yan YN, et al. , 2014. Classification of EEG signals using a multiple kernel learning support vector machine. Sensors, 14(7): 12784-12802. 10.3390/s140712784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XW, Hu B, Sun ST, et al. , 2016. EEG-based mild depressive detection using feature selection methods and classifiers. Comput Methods Programs Biomed, 136: 151-161. 10.1016/j.cmpb.2016.08.010 [DOI] [PubMed] [Google Scholar]
- Li XW, Zhang X, Zhu J, et al. , 2019. Depression recognition using machine learning methods with different feature generation strategies. Artif Intell Med, 99: 101696. 10.1016/j.artmed.2019.07.004 [DOI] [PubMed] [Google Scholar]
- Li Y, Cui WG, Huang H, et al. , 2019. Epileptic seizure detection in EEG signals using sparse multiscale radial basis function networks and the Fisher vector approach. Knowl-Based Syst, 164: 96-106. 10.1016/j.knosys.2018.10.029 [DOI] [Google Scholar]
- Liang WX, Pei HJ, Cai QL, et al. , 2020. Scalp EEG epileptogenic zone recognition and localization based on long-term recurrent convolutional network. Neurocomputing, 396: 569-576. 10.1016/j.neucom.2018.10.108 [DOI] [Google Scholar]
- Lord C, Elsabbagh M, Baird G, et al. , 2018. Autism spectrum disorder. Lancet, 392(10146): 508-520. 10.1016/S0140-6736(18)31129-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lotte F, Faller J, Guger C, et al. , 2012. Combining BCI with virtual reality: towards new applications and improved BCI. In: Allison BZ, Dunne S, Leeb R, (Eds.), Towards Practical Brain-Computer Interfaces. Biological and Medical Physics, Biomedical Engineering. Springer, Berlin, Heidelberg, p.197-220. 10.1007/978-3-642-29746-5_10 [DOI] [Google Scholar]
- Maass W, 1997. Networks of spiking neurons: the third generation of neural network models. Neural Netw, 10(9): 1659-1671. 10.1016/S0893-6080(97)00011-7 [DOI] [Google Scholar]
- Magboo MSA, Magboo VPC, 2022. Explainable AI for autism classification in children. In: Jezic G, Chen-Burger YHJ, Kusek M, (Eds.), Agents and Multi-Agent Systems: Technologies and Applications 2022. Smart Innovation, Systems and Technologies, Vol. 306. Springer, Singapore, p.195-205. 10.1007/978-981-19-3359-2_17 [DOI] [Google Scholar]
- Malmivuo J, Plonsey R, 1995. Bioelectromagnetism: Principles and Applications of Bioelectric and Biomagnetic Fields. Translators, Oxford University Press, New York, USA. [Google Scholar]
- McBride JC, Zhao XP, Munro NB, et al. , 2014. Spectral and complexity analysis of scalp EEG characteristics for mild cognitive impairment and early Alzheimer’s disease. Comput Methods Programs Biomed, 114(2): 153-163. 10.1016/j.cmpb.2014.01.019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCutcheon RA, Marques TR, Howes OD, 2020. Schizophrenia—an overview. JAMA Psychiatry, 77(2): 201-210. 10.1001/jamapsychiatry.2019.3360 [DOI] [PubMed] [Google Scholar]
- Mohri M, Rostamizadeh A, Talwalkar A, 2018. Foundations of Machine Learning, 2nd Ed. Translators, MIT Press, Cambridge, USA. [Google Scholar]
- Moridani MK, Farhadi H, 2017. Heart rate variability as a biomarker for epilepsy seizure prediction. Bratisl Lek Listy, 118(1): 3-8. 10.4149/BLL_2017_001 [DOI] [PubMed] [Google Scholar]
- Mota AR, Duarte L, Rodrigues D, et al. , 2013. Development of a quasi-dry electrode for EEG recording. Sens Actuators A Phys, 199: 310-317. 10.1016/j.sna.2013.06.013 [DOI] [Google Scholar]
- Mumtaz W, Malik AS, Yasin MAM, et al. , 2015. Review on EEG and ERP predictive biomarkers for major depressive disorder. Biomed Signal Process Control, 22: 85-98. 10.1016/j.bspc.2015.07.003 [DOI] [Google Scholar]
- Mumtaz W, Xia LK, Ali SSA, et al. , 2017. Electroencephalogram (EEG)-based computer-aided technique to diagnose major depressive disorder (MDD). Biomed Signal Process Control, 31: 108-115. 10.1016/j.bspc.2016.07.006 [DOI] [Google Scholar]
- Norcia AM, Appelbaum LG, Ales JM, et al. , 2015. The steady-state visual evoked potential in vision research: a review. J Vis, 15(6): 4. 10.1167/15.6.4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SL, Vicnesh J, Ciaccio EJ, et al. , 2019. Deep convolutional neural network model for automated diagnosis of schizophrenia using EEG signals. Appl Sci, 9(14): 2870. 10.3390/app9142870 [DOI] [Google Scholar]
- Oh SL, Hagiwara Y, Raghavendra U, et al. , 2020. A deep learning approach for Parkinson’s disease diagnosis from EEG signals. Neural Comput Appl, 32(15): 10927-10933. 10.1007/s00521-018-3689-5 [DOI] [Google Scholar]
- Olejarczyk E, Jernajczyk W, 2017. EEG in schizophrenia. RepOD, V1. 10.18150/repod.0107441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phang CR, Noman F, Hussain H, et al. , 2020. A multi-domain connectome convolutional neural network for identifying schizophrenia from EEG connectivity patterns. IEEE J Biomed Health Inform, 24(5): 1333-1343. 10.1109/JBHI.2019.2941222 [DOI] [PubMed] [Google Scholar]
- Qiu LN, Li JP, Pan JH, 2022. Parkinson’s disease detection based on multi-pattern analysis and multi-scale convolutional neural networks. Front Neurosci, 16: 957181. 10.3389/fnins.2022.957181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi AA, Zhang CX, Zheng R, et al. , 2018. Ischemic stroke detection using EEG signals. CASCON '18: Proceedings of the 28th Annual International Conference on Computer Science and Software Engineering, p.301-308. [Google Scholar]
- Rad EM, Azarnoosh M, Ghoshuni M, et al. , 2021. Diagnosis of mild Alzheimer’s disease by EEG and ERP signals using linear and nonlinear classifiers. Biomed Signal Process Control, 70: 103049. 10.1016/j.bspc.2021.103049 [DOI] [Google Scholar]
- Roohi-Azizi M, Azimi L, Heysieattalab S, et al. , 2017. Changes of the brain’s bioelectrical activity in cognition, consciousness, and some mental disorders. Med J Islam Republic Iran, 31(1): 307-312. 10.14196/mjiri.31.53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saadatinia M, Salimi-Badr A, 2024. An explainable deep learning-based method for schizophrenia diagnosis using generative data-augmentation. IEEE Access, 12: 98379-98392. 10.1109/ACCESS.2024.3428847 [DOI] [Google Scholar]
- Saha S, Chant D, Welham J, et al. , 2005. A systematic review of the prevalence of schizophrenia. PLoS Med, 2(5): e141. 10.1371/journal.pmed.0020141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saikia A, Hussain M, Barua AR, et al. , 2019. EEG-EMG correlation for Parkinson’s disease. Int J Eng Adv Technol, 8(6): 1179-1185. 10.35940/ijeat.F8360.088619 [DOI] [Google Scholar]
- Sarkar A, Singh A, Chakraborty R, 2022. A deep learning-based comparative study to track mental depression from EEG data. Neurosci Inf, 2(4): 100039. 10.1016/j.neuri.2022.100039 [DOI] [Google Scholar]
- Savadkoohi M, Oladunni T, Thompson L, 2020. A machine learning approach to epileptic seizure prediction using Electroencephalogram (EEG) Signal. Biocybern Biomed Eng, 40(3): 1328-1341. 10.1016/j.bbe.2020.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidhuber J, 2015. Deep learning in neural networks: an overview. Neural Netw, 61: 85-117. 10.1016/j.neunet.2014.09.003 [DOI] [PubMed] [Google Scholar]
- Shah D, Wang GY, Doborjeh M, et al. , 2019. Deep learning of EEG data in the NeuCube brain-inspired spiking neural network architecture for a better understanding of depression. In: Gedeon T, Wong K, Lee M (Eds.), Neural Information Processing. ICONIP 2019. Lecture Notes in Computer Science, Vol. 11955. Springer, Cham, p.195-206. 10.1007/978-3-030-36718-3_17 [DOI] [Google Scholar]
- Shah SAA, Zhang L, Bais A, 2020. Dynamical system based compact deep hybrid network for classification of Parkinson disease related EEG signals. Neural Netw, 130: 75-84. 10.1016/j.neunet.2020.06.018 [DOI] [PubMed] [Google Scholar]
- Sharma G, Parashar A, Joshi AM, 2021. DepHNN: a novel hybrid neural network for electroencephalogram (EEG)-based screening of depression. Biom Signal Process Control, 66: 102393. 10.1016/j.bspc.2020.102393 [DOI] [Google Scholar]
- Shi QX, Liu A, Chen RY, et al. , 2020. Depression detection using resting state three-channel EEG signal. Electrical Engineering and Systems Science, Signal Processing. arXiv: 2002.09175. 10.48550/arXiv.2002.09175 [DOI] [Google Scholar]
- Shim M, Hwang HJ, Kim DW, et al. , 2016. Machine-learning-based diagnosis of schizophrenia using combined sensor-level and source-level EEG features. Schizophr Res, 176(2-3): 314-319. 10.1016/j.schres.2016.05.007 [DOI] [PubMed] [Google Scholar]
- Shim M, Jin MJ, Im CH, et al. , 2019. Machine-learning-based classification between post-traumatic stress disorder and major depressive disorder using P300 features. NeuroImage Clin, 24: 102001. 10.1016/j.nicl.2019.102001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shoeb AH, 2009. Application of Machine Learning to Epileptic Seizure Onset Detection and Treatment. PhD Dissemination, Massachusetts Institute of Technology, Cambridge, USA. [Google Scholar]
- Shoeibi A, Ghassemi N, Alizadehsani R, et al. , 2021a. A comprehensive comparison of handcrafted features and convolutional autoencoders for epileptic seizures detection in EEG signals. Expert Syst Appl, 163: 113788. 10.1016/j.eswa.2020.113788 [DOI] [Google Scholar]
- Shoeibi A, Khodatars M, Ghassemi N, et al. , 2021b. Epileptic seizures detection using deep learning techniques: a review. Int J Environ Res Public Health, 18(11): 5780. 10.3390/ijerph18115780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simon GE, Vonkorff M, Piccinelli M, et al. , 1999. An international study of the relation between somatic symptoms and depression. N Engl J Med, 341(18): 1329-1335. 10.1056/NEJM199910283411801 [DOI] [PubMed] [Google Scholar]
- Singh S, Dawar D, Mehmood E, et al. , 2024. Determining diagnostic utility of EEG for assessing stroke severity using deep learning models. Biomed Eng Adv, 7: 100121. 10.1016/j.bea.2024.100121 [DOI] [Google Scholar]
- Sounderajah V, Ashrafian H, Golub RM, et al. , 2021. Developing a reporting guideline for artificial intelligence-centred diagnostic test accuracy studies: the STARD-AI protocol. BMJ Open, 11(6): e047709. 10.1136/bmjopen-2020-047709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suuronen I, Airola A, Pahikkala T, et al. , 2023. Budget-based classification of Parkinson’s disease from resting state EEG. IEEE J Biomed Health Inform, 27(8): 3740-3747. 10.1109/JBHI.2023.3235040 [DOI] [PubMed] [Google Scholar]
- Swami P, Gandhi TK, Panigrahi BK, et al. , 2016. A novel robust diagnostic model to detect seizures in electroencephalography. Expert Syst Appl, 56: 116-130. 10.1016/j.eswa.2016.02.040 [DOI] [Google Scholar]
- Thapaliya S, Jayarathna S, Jaime M, 2018. Evaluating the EEG and eye movements for autism spectrum disorder. 2018 IEEE International Conference on Big Data (Big Data), Seattle, WA, USA, p.2328-2336. 10.1109/BigData.2018.8622501 [DOI] [Google Scholar]
- Truong ND, Nguyen AD, Kuhlmann L, et al. , 2018. Convolutional neural networks for seizure prediction using intracranial and scalp electroencephalogram. Neural Netw, 105: 104-111. 10.1016/j.neunet.2018.04.018 [DOI] [PubMed] [Google Scholar]
- Tsiouris KM, Pezoulas VC, Zervakis M, et al. , 2018. A Long Short-Term Memory deep learning network for the prediction of epileptic seizures using EEG signals. Comput Biol Med, 99: 24-37. 10.1016/j.compbiomed.2018.05.019 [DOI] [PubMed] [Google Scholar]
- Tveit J, Aurlien H, Plis S, et al. , 2023. Automated interpretation of clinical electroencephalograms using artificial intelligence. JAMA Neurol, 80(8): 805-812. 10.1001/jamaneurol.2023.1645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Usman SM, Khalid S, Bashir S, 2021. A deep learning based ensemble learning method for epileptic seizure prediction. Comput Biol Med, 136: 104710. 10.1016/j.compbiomed.2021.104710 [DOI] [PubMed] [Google Scholar]
- Vecchio F, Miraglia F, Alù F, et al. , 2020. Classification of Alzheimer’s disease with respect to physiological aging with innovative EEG biomarkers in a machine learning implementation. J Alzheimers Dis, 75(4): 1253-1261. 10.3233/JAD-200171 [DOI] [PubMed] [Google Scholar]
- Vickers JC, Dickson TC, Adlard PA, et al. , 2000. The cause of neuronal degeneration in Alzheimer’s disease. Prog Neurobiol, 60(2): 139-165. 10.1016/S0301-0082(99)00023-4 [DOI] [PubMed] [Google Scholar]
- Wang BY, Kang YY, Huo DY, et al. , 2022. EEG diagnosis of depression based on multi-channel data fusion and clipping augmentation and convolutional neural network. Front Physiol, 13: 1029298. 10.3389/fphys.2022.1029298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Barstein J, Ethridge LE, et al. , 2013. Resting state EEG abnormalities in autism spectrum disorders. J Neurodev Disord, 5: 24. 10.1186/1866-1955-5-24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson CM, Burrell JI, Kuziek JWP, et al. , 2020. Predicting stroke severity with a 3-min recording from the Muse portable EEG system for rapid diagnosis of stroke. Sci Rep, 10: 18465. 10.1038/s41598-020-75379-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia W, Zhang R, Zhang X, et al. , 2023. A novel method for diagnosing Alzheimer’s disease using deep pyramid CNN based on EEG signals. Heliyon, 9(4): e14858. 10.1016/j.heliyon.2023.e14858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yasin S, Hussain SA, Aslan S, et al. , 2021. EEG based Major Depressive disorder and Bipolar disorder detection using Neural Networks: a review. Comput Methods Programs Biomed, 202: 106007. 10.1016/j.cmpb.2021.106007 [DOI] [PubMed] [Google Scholar]
- Yin GM, Chang Y, Zhao YL, et al. , 2023. Automatic recognition of schizophrenia from brain-network features using graph convolutional neural network. Asian J Psychiatr, 87: 103687. 10.1016/j.ajp.2023.103687 [DOI] [PubMed] [Google Scholar]
- You Z, Zeng RH, Lan XY, et al. , 2020. Alzheimer’s disease classification with a cascade neural network. Front Public Health, 8: 584387. 10.3389/fpubh.2020.584387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj R, Murugappan M, Mohamed Ibrahim N, et al. , 2014. On the analysis of EEG power, frequency and asymmetry in Parkinson’s disease during emotion processing. Behav Brain Funct, 10: 12. 10.1186/1744-9081-10-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Guo Y, Yang P, et al. , 2020. Epilepsy seizure prediction on EEG using common spatial pattern and convolutional neural network. IEEE J Biomed Health Inform, 24(2): 465-474. 10.1109/JBHI.2019.2933046 [DOI] [PubMed] [Google Scholar]