Abstract
Serine is essential to maintain maximal growth and proliferation of cancer cells by providing adequate intermediate metabolites and energy. Phosphoserine aminotransferase 1 (PSAT1) is a key enzyme in de novo serine synthesis. However, little is known about the mechanisms underlying PSAT1 degradation. We found that acetylation was the switch that regulated the degradation of PSAT1 in lung adenocarcinoma (LUAD). Deacetylation of PSAT1 on Lys51 by histone deacetylase 7 (HDAC7) enhanced the interaction between PSAT1 and the deubiquitinase ubiquitin-specific processing protease 14 (USP14), leading to the deubiquitination and stabilization of PSAT1; while acetylation of PSAT1 promoted its interaction with the E3 ligase ubiquitination factor E4B (UBE4B), leading to proteasomal degradation. Acetylation of PSAT1 on Lys51 regulated serine metabolism and tumor proliferation in LUAD. Thus, acetylation and ubiquitination cooperatively regulated the protein homeostasis of PSAT1. In conclusion, our study reveals a key regulatory mechanism for maintaining PSAT1 protein homeostasis in LUAD.
Subject terms: Metabolomics, Non-small-cell lung cancer, Post-translational modifications
Acetylation and ubiquitination cooperatively regulate the protein homeostasis of PSAT1 and contribute to serine synthesis and tumorigenesis of lung adenocarcinoma.
Introduction
Metabolic processes support the high proliferation rates of cancer cells by supplying a major source of energy and critical cellular components1. Serine is a necessary precursor for the de novo synthesis of many cellular components, including nucleotides, proteins, lipids, and other metabolites2. Serine can be obtained from various sources: it can be taken up directly from the extracellular environment, derived from intracellular protein via autophagy, produced by glycine conversion, or synthesized de novo via the serine synthesis pathway (SSP). In SSP, the glycolytic intermediate 3-phosphoglycerate (3-PG) is first converted to 3-phosphohydroxypyruvate (3-PHP) by phosphoglycerate dehydrogenase (PHGDH). Phosphoserine aminotransferase 1 (PSAT1) then converts 3-phosphohydroxypyruvate to 3-phosphoserine (3-PS), which is subsequently dephosphorylated to serine by phosphoserine phosphatase (PSPH)3,4. During this process, PSAT1 also converts glutamate to α-ketoglutarate, an anaplerotic intermediate that refuels the tricarboxylic acid (TCA) cycle and sustains cancer metabolism, thus playing a crucial role in cancer metabolism5.
As a pivotal metabolic enzyme, upregulation of PSAT1 has been observed in cancer cell lines, and elevated PSAT1 expression enables cancer cells to survive under serine starvation conditions and promote tumorigenesis6,7. The protein abundance is tightly controlled by transcriptional, post-transcriptional, translational, and post-translational regulatory mechanisms. PSAT1 can be regulated by multiple upstream proteins and signaling molecules. A key regulator of SSP gene expression is Activating transcription factor 4 (ATF4), which directly binds to and activates the promoters. Also, nuclear factor erythroid 2-related factor 2 (NRF2) controls the expression of PSAT1 via ATF4 to support glutathione and nucleotide production8,9. Moreover, a recent study demonstrated that c-Myc stimulated SSP activation by transcriptionally upregulating the expression of multiple SSP enzymes10. PSAT1 is also subject to epigenetic control. The histone H3 lysine 9 methyltransferases (G9A) can transcriptionally activate PSAT111,12. However, little is known about the roles of post-translational modification in regulating PSAT1.
In this study, we demonstrated that acetylation occurred on PSAT1 in lung adenocarcinoma (LUAD) cells and this modification affected its protein stability, further affecting serine metabolism and cell proliferation. Deacetylation of PSAT1 by histone deacetylase 7 (HDAC7) promoted the interaction between PSAT1 and ubiquitin-specific processing protease 14 (USP14), resulting in deubiquitination and protein stabilization. However, acetylated PSAT1 tended to interact with ubiquitination factor E4B (UBE4B), the E3 ligase that ubiquitinated and degraded PSAT1 in the proteasomal pathway. Interestingly, cisplatin (DDP) treatment could increase the expression of PSAT1 and decrease the acetylation and ubiquitination of PSAT1-WT rather than PSAT1-K51R. Knocking down PSAT1 increased the sensitivity of LUAD cells to cisplatin. Taken together, our findings clarified the role of acetylation and ubiquitination in regulating the protein stability of PSAT1 in LUAD.
Results
Acetylation affects the protein stability and ubiquitination of PSAT1
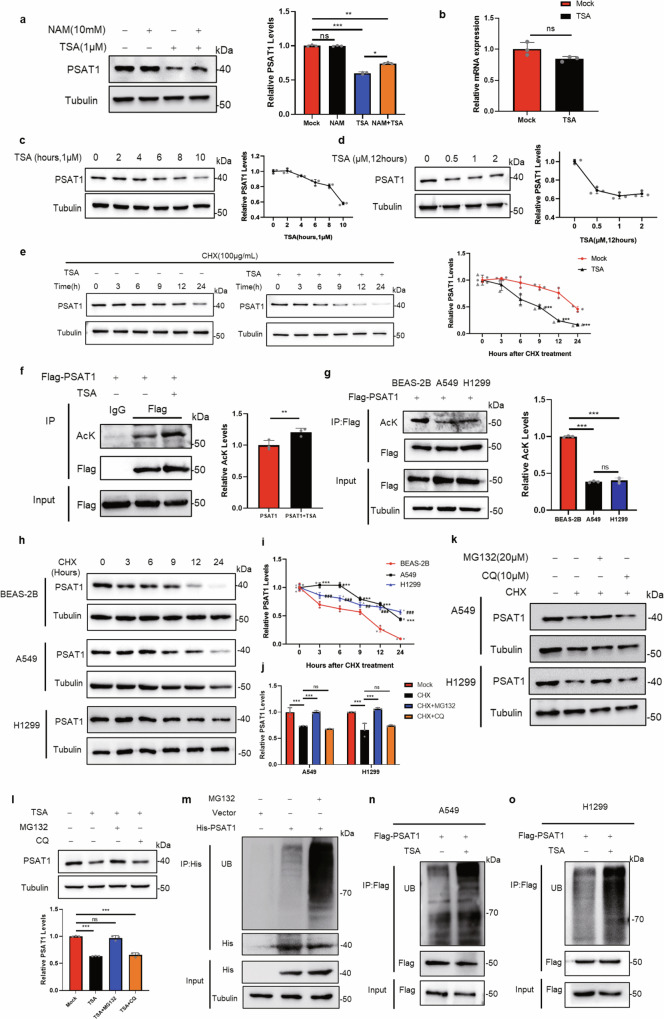
Acetylation is involved in various physiological processes, such as tumor development, signal transduction, and metabolism13. To clarify whether PSAT1 was regulated by acetylation in LUAD, we treated H1299 and A549 cells with trichostatin A (TSA, an inhibitor of histone deacetylase (HDAC) class I/II family deacetylases) and nicotinamide (NAM, an inhibitor of sirtuin (SIRT) family deacetylases). The treatment efficiencies of the inhibitors were pre-tested using the biomarkers proven by other studies (Supplementary Fig. 1a, b)14,15. The protein expression of PSAT1 was significantly decreased when treating cells with TSA but not NAM in lung cancer cells, however, these effects were not observed in breast cancer cells (MCF7) or hepatocellular carcinoma cells (HLE) (Fig. 1a and Supplementary Fig. 1c), suggesting that the phenomenon was specific to LUAD cells. Then, the mRNA level of PSAT1 was also examined when cells were treated with TSA in H1299 cells. Figure 1b showed that the mRNA level of PSAT1 was not significantly changed, indicating that the decreased expression of PSAT1 was not caused by transcriptional repression. To further validate the above results, we treated H1299 cells by adding different concentrations of TSA for 12 h or 1 μM TSA for the indicated time and detected PSAT1 expression. Figure 1c, d showed that the protein levels of PSAT1 decreased in a time or concentration-dependent manner. The cell viability was also determined under these conditions and Supplementary Fig. 1d, e showed that these treatments did not affect the cell viability significantly. Next, we performed cycloheximide (CHX)-chase assays to measure the protein stability of PSAT1 in physiological conditions or TSA treatment. The PSAT1 protein showed a significant decrease at 12 h in physiological conditions, while TSA treatment accelerated its degradation rate, promoting the degradation time to 6 h (Fig. 1e). These results indicated that TSA treatment decreased PSAT1 expression by accelerating its protein degradation.
Fig. 1. Acetylation affects the protein stability and ubiquitination of PSAT1.
a Western blot was used to detect PSAT1 protein levels after NAM (10 mM) and TSA (1 μM) treatment for 12 h in H1299. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001. b The mRNA levels of PSAT1 were detected after TSA treatment in H1299 by qPCR. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05. c Western blot was used to detect PSAT1 protein levels after TSA (1 μM) treatment at different time points. The relative PSAT1 expression compared with that of Tubulin was quantified. d TSA at different concentrations was added into H1299 cells for 12 h, then cells were collected and the protein level of PSAT1 was detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. e H1299 cells were experimented with CHX (100 μg/mL) for various time points and were treated with or without TSA (1 μM). The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ***P < 0.001. f H1299 cells were transfected with Flag-PSAT1 plasmid, and TSA (1 μM) was added 12 h before sample collection. Co-IP was used to detect the acetylation level of PSAT1. The relative AcK expression compared with that of Flag was quantified. Data represented as the average of three independent experiments (mean ± SD), **P < 0.01. g BEAS-2B, A549, and H1299 cells were transfected with Flag-PSAT1 plasmid, and Co-IP was used to detect the acetylation level of PSAT1. The relative AcK expression compared with that of Flag was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, ***P < 0.001. h, i BEAS-2B, A549, and H1299 cells were experimented with CHX (100 μg/mL) for various time points. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD). *, A549 versus BEAS-2B; #, H1299 versus BEAS-2B. ##: P < 0.01, *** or ###: P < 0.001. j, k CHX (100 μg/mL) was added into A549 and H1299 cells for 12 h with or without MG132 (20 μM) or CQ (10 μM). PSAT1 protein was detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, ***P < 0.001. l TSA (1 μM) was added into H1299 cells for 12 h with or without MG132 (20 μM) or CQ (10 μM). PSAT1 protein was detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, ***P < 0.001. m H1299 cells were transfected with His-PSAT1 plasmid, and MG132 (20 μM) was added 12 h before sample collection. Co-IP was used to detect the ubiquitination level of PSAT1. n, o A549 and H1299 cells were transfected with Flag-PSAT1 plasmid, and TSA (1 μM) was added 12 h before sample collection. Co-IP was used to detect the ubiquitination level of PSAT1.
Subsequently, the acetylation of PSAT1 was detected and we found that TSA treatment increased the acetylation of PSAT1 in H1299 and A549 cells (Fig. 1f and Supplementary Fig. 1f). In addition, we checked the acetylation levels of PSAT1 in LUAD cell lines (A549 and H1299) and normal bronchial epithelial cells BEAS-2B. Figure 1g showed that the acetylation levels of PSAT1 were downregulated in A549 and H1299 cells compared with BEAS-2B. We next examined the degradation rates of PSAT1 in these cells and found that the degradation of PSAT1 was attenuated in LUAD cells (Fig. 1h, i), suggesting that acetylation affected PSAT1 protein stability.
The main degradation pathways of intracellular protein are divided into two categories: ubiquitin–proteasome system and autophagy-lysosomal pathway16. Figure 1j, k showed that the protein degradation of PSAT1 induced by CHX treatment could be recovered by the proteasomal inhibitor MG132 but not the lysosomal inhibitor chloroquine (CQ). The treatment efficiency of CQ was pre-tested using the autophagy-related proteins by western blot (Supplementary Fig. 1g). MG132 treatment also reversed the decreased expression of PSAT1 caused by TSA treatment (Fig. 1l). This indicated that TSA treatment might accelerate PSAT1 protein degradation through ubiquitin–proteasome system. The ubiquitination of PSAT1 under MG132 treatment was also examined. The result showed that the ubiquitination of PSAT1 significantly increased by adding MG132 (Fig. 1m). We then treated LUAD cells with TSA and examined PSAT1 ubiquitination levels. Figure 1n, o demonstrated that TSA treatment induced PSAT1 ubiquitination. All the results demonstrate that acetylation affects the protein stability and ubiquitination of PSAT1.
HDAC7 is a key deacetylase that regulates PSAT1 acetylation and protein stability
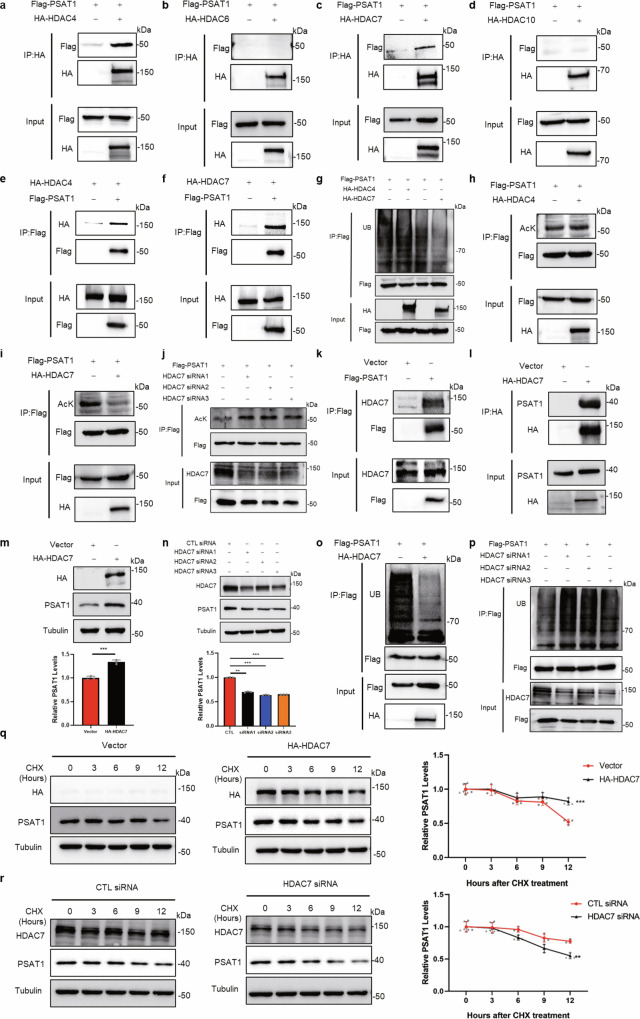
TSA is an inhibitor of HDAC class I/II family deacetylases and our results in Fig. 1 suggested that these HDAC proteins might be related to PSAT1 protein homeostasis. Because the PSAT1 protein is located in the cytoplasm, we selected several HDAC proteins which distributed in the cytoplasm for immunoprecipitation to detect their interactions with PSAT117. Figure 2a–f showed that PSAT1 interacted strongly with HDAC4 and HDAC7, but not with HDAC6 and HDAC10. Overexpressing HDAC7 reduced the ubiquitination and acetylation of PSAT1 in H1299 cells, while HDAC4 did not have these effects (Fig. 2g–i). Also, the reduction of ubiquitination and acetylation of PSAT1 was observed in A549 cells (Supplementary Fig. 2a, b). Then we detected the deacetylation efficiencies of some other proteins involved in serine synthesis by HDAC7 overexpression. Supplementary Fig. 2c–f showed that HDAC7 did not affect the acetylation levels of PHGDH, PSPH, or serine hydroxymethyltransferase1/2 (SHMT1/2). Knocking down HDAC7 increased the acetylation of PSAT1 (Fig. 2j). We further demonstrated the endogenous interaction between PSAT1 and HDAC7 in H1299 and A549 cells (Fig. 2k, l and Supplementary Fig. 2g). These data show that HDAC7 is the key deacetylase for PSAT1 in LUAD cells.
Fig. 2. HDAC7 is a key deacetylase that regulates PSAT1 acetylation and protein stability.
a–d H1299 cells were transfected with Flag-PSAT1 plasmid alone or co-transfected with HA-HDAC4 (a), HA-HDAC6 (b), HA-HDAC7 (c), or HA-HDAC10 (d). The immunoprecipitations were blotted with anti-HA or anti-Flag antibodies. e, f H1299 cells were transfected with HA-HDAC4 or HA-HDAC7 plasmid alone or co-transfected with Flag-PSAT1. The immunoprecipitations were blotted with anti-HA or anti-Flag antibodies. g In H1299 cells, Flag-PSAT1 and HA-HDAC4 or HA-HDAC7 plasmids were co-transfected. Co-IP was used to detect the ubiquitination level of PSAT1. h, i In H1299 cells, Flag-PSAT1 and HA-HDAC4 or HA-HDAC7 plasmids were co-transfected. The acetylation of PSAT1 was detected. j H1299 cells were transiently transfected with CTL siRNA or HDAC7 selective siRNAs. The acetylation of PSAT1 was detected. k, l In H1299 cells, endogenous interaction between PSAT1 and HDAC7 was tested using anti-Flag and anti-HA antibodies for Co-IP. The immunoprecipitations were blotted with anti-HDAC7 or anti-PSAT1 antibodies. m, n In H1299 cells, HDAC7 was overexpressed or knocked down and to detect PSAT1 protein by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), **P < 0.01, ***P < 0.001. o, p HDAC7 was overexpressed or knocked down in H1299 cells. Co-IP was used to detect the ubiquitination level of PSAT1. q, r H1299 cells were experimented with CHX (100 μg/mL) for various time points and were transfected with HA-HDAC7 or HDAC7 selective siRNA. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), **P < 0.01, ***P < 0.001.
To explore the role of HDAC7 in PSAT1 protein expression, we overexpressed HDAC7 in H1299 and A549 cells and PSAT1 expression was examined. The result showed that the protein expression of PSAT1 increased significantly (Fig. 2m and Supplementary Fig. 2h). To further validate the above results, we transfected the HDAC7-specific siRNAs into H1299 cells followed by detecting PSAT1 expression. Knocking down HDAC7 reduced PSAT1 expression (Fig. 2n). Supplementary Fig. 2i, j showed HDAC7 did not affect the transcription of PSAT1. Figure 2o, p demonstrated that overexpressing HDAC7 decreased the ubiquitination of PSAT1, while HDAC7 knockdown showed the opposite effect. Then, we tested whether HDAC7 affected the protein stability of PSAT1. The CHX-chase assays showed that the protein degradation rates of PSAT1 slowed down when overexpressing HDAC7 while knocking down HDAC7 accelerated the degradation of PSAT1 (Fig. 2q, r). These above results suggest that HDAC7 is the key deacetylase that regulates the acetylation and protein degradation of PSAT1.
Deubiquitinase USP14 stabilizes PSAT1 in lung adenocarcinoma
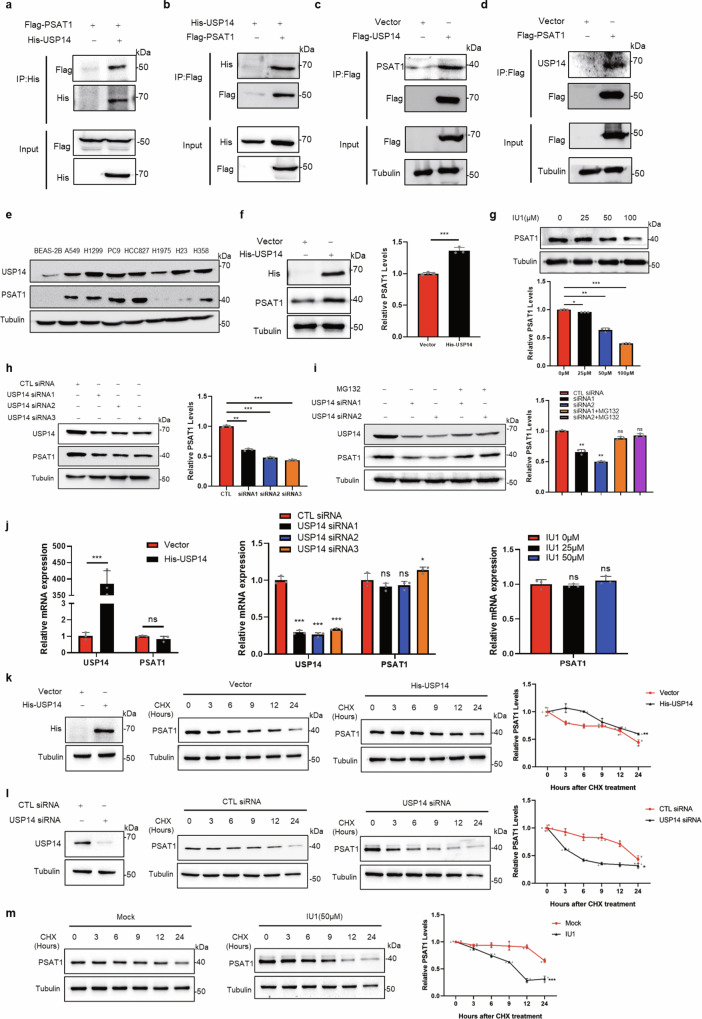
In the proteasomal degradation system, substrates were labeled with ubiquitin by E3 ligase and recruited on the proteasome for degradation, whereas deubiquitinases (DUBs) could stabilize the targeted proteins by inducing their deubiquitination18. To explore the potential E3 ligases or deubiquitinases for PSAT1, we performed mass spectrometry analysis. Ultimately, two proteins of DUBs, named USP39 and USP14, were identified as a putative PSAT1-interacting protein (Supplementary Data 1). Supplementary Fig. 3a–c showed that USP39 could not interact with PSAT1 and regulate the protein or ubiquitination levels of PSAT1. However, we found that USP14 interacted with PSAT1 by co-immunoprecipitations in H1299 and A549 cells (Fig. 3a–d and Supplementary Fig. 3d). To further validate their interaction, immunofluorescence assays were performed. The results showed that USP14 colocalized with PSAT1 in the cytoplasm (Supplementary Fig. 3e), indicating that PSAT1 interacted with USP14.
Fig. 3. Deubiquitinase USP14 stabilizes PSAT1 in lung adenocarcinoma.
a, b H1299 cells were co-transfected with His-USP14 and Flag-PSAT1. The immunoprecipitations were blotted with anti-His or anti-Flag antibodies. c, d In H1299 cells, endogenous interaction between PSAT1 and USP14 was tested using anti-Flag antibodies for Co-IP. The immunoprecipitations were blotted with anti-PSAT1 or anti-USP14 antibodies. e The protein levels of USP14 and PSAT1 were detected in lung cancer cell lines and normal bronchial epithelial cell BEAS-2B. f In H1299 cells, USP14 was overexpressed to detect PSAT1 protein by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ***P < 0.001. g H1299 cells were experimented with different concentrations of IU1. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), *P < 0.05, **P < 0.01, ***P < 0.001. h H1299 cells were transiently transfected with CTL siRNA or USP14 selective siRNAs. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), **P < 0.01, ***P < 0.001. i H1299 cells were transfected with USP14 selective siRNAs and MG132 (20 μM) was added 12 h before sample collection. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, **P < 0.01. j H1299 cells were transfected with control Vector or His-USP14 plasmid or transiently transfected with CTL siRNA or USP14 selective siRNAs or experimented with different concentrations of IU1. The mRNA levels of PSAT1 and USP14 were detected by qPCR. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, *P < 0.05, ***P < 0.001. k–m H1299 cells were experimented with CHX (100 μg/mL) for various time points and were transfected with His-USP14 or USP14 selective siRNA or treated with IU1. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), *P < 0.05, **P < 0.01, ***P < 0.001.
Interestingly, we found that the expression pattern between PSAT1 and USP14 showed a high consistency in LUAD cells (Fig. 3e). This indicated that USP14 might be related to PSAT1 expression. Indeed, overexpression of USP14 increased the protein level of PSAT1 in H1299 and A549 cells (Fig. 3f and Supplementary Fig. 3f). In contrast, the protein levels of PSAT1 were decreased when USP14 was inhibited by IU1, a specific inhibitor of USP14, or by USP14-specific siRNAs (Fig. 3g, h). The treatment efficiencies of IU1 were detected in Supplementary Fig. 3g using the USP14-target protein CyclinB119. At the same time, Fig. 3i demonstrated that MG132 treatment reversed the decreased expression of PSAT1 caused by USP14 inhibition, suggesting the proteasome degradation of PSAT1. The deubiquitinases could also bind to specific transcription factors or histone-associated proteins to regulate transcription20. To elucidate whether USP14 affected PSAT1 expression through transcriptional regulation, we examined the effect of USP14 on PSAT1 mRNA expression. Figure 3j showed that the mRNA levels of PSAT1 were not affected or only slightly changed when USP14 was overexpressed or inhibited. Then, we examined the degradation rates of PSAT1 under USP14 overexpression or inhibition conditions. The protein degradation rate of PSAT1 was slowed down when USP14 was overexpressed (Fig. 3k). Instead, USP14 knockdown or IU1 treatment accelerated the degradation rates of PSAT1 protein (Fig. 3l, m). Thus, these results demonstrated that USP14 interacts with PSAT1 and regulates its protein stability.
USP14 affects the protein stability of PSAT1 by regulating its ubiquitination
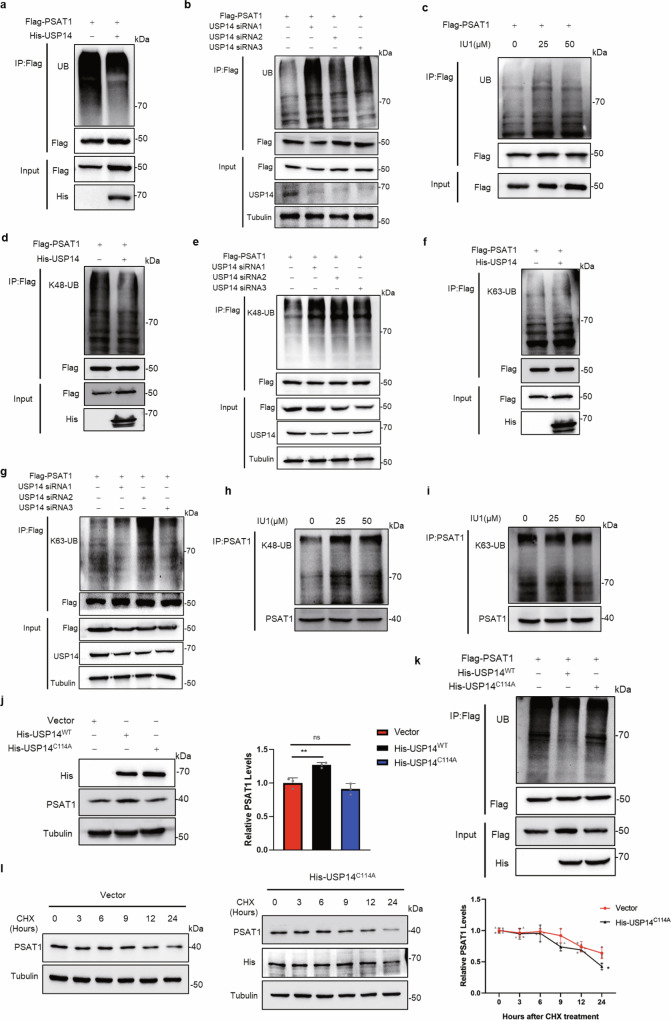
As a member of DUBs, USP14 played a central role in the release of ubiquitin to protect the substrate from degradation21. Our previous studies demonstrated that USP14 regulated protein homeostasis of cell cycle-related proteins by deubiquitination22,23. We next intended to clarify whether USP14 also regulated PSAT1 expression through deubiquitination. Consistent with expectations, overexpression of USP14 reduced the ubiquitination level of PSAT1, while either genetic or pharmacological inhibition of USP14 increased the ubiquitination of PSAT1 in H1299 cells (Fig. 4a–c). Besides, USP14 also regulated endogenous PSAT1 ubiquitination levels (Supplementary Fig. 3h–j), indicating that USP14 regulated the ubiquitination of PSAT1.
Fig. 4. USP14 regulates the K48-linked ubiquitination of PSAT1.
a, b In H1299 cells, Flag-PSAT1 and His-USP14 plasmids or USP14 selective siRNA were co-transfected. Co-IP was used to detect the ubiquitination level of PSAT1. c In H1299 cells, Flag-PSAT1 plasmid was transfected with different concentrations of IU1 treatment. Co-IP was used to detect the ubiquitination level of PSAT1. d, f In H1299 cells, Flag-PSAT1 and His-USP14 plasmids were co-transfected. Co-IP was used to detect the K48-linked or K63-linked ubiquitination level of PSAT1. e, g In H1299 cells, Flag-PSAT1 plasmid and USP14 selective siRNAs were co-transfected. Co-IP was used to detect the K48-linked or K63-linked ubiquitination level of PSAT1. h, i H1299 cells were treated with different concentrations of IU1. Co-IP was used to detect the K48-linked or K63-linked ubiquitination level of PSAT1. j H1299 cells were transfected with His-USP14WT or His-USP14C114A plasmids. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, **P < 0.01. k H1299 cells were transfected with Flag-PSAT1 plasmid alone or co-transfected with His-USP14WT or His-USP14C114A plasmids. Co-IP was used to detect the ubiquitination level of PSAT1. l H1299 cells were experimented with CHX (100 μg/mL) for various time points and were transfected with His-USP14C114A. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), *P < 0.05.
It is well known that ubiquitin chains may exist in several linkages, such as Lysine 48-linkage (K48) or Lysine 63-linkage (K63)24. We further detected the ubiquitin chain types using antibodies specific for K48-linked or K63-linked ubiquitination. As shown in Fig. 4d–g, overexpression or knockdown of USP14 regulated K48-linked ubiquitination, while K63-linked ubiquitination of PSAT1 was less changed in H1299 cells. Supplementary Fig. 3k–m showed the same effects in A549 cells. We further detected the ubiquitination chain types of endogenous PSAT1 following treatment with IU1. Figure 4h, i showed that IU1 treatment increased the K48-linked ubiquitination, but not K63-linkage. These data support the hypothesis that USP14 deubiquitinates PSAT1 with K48-linkage to escape from proteasomal degradation.
Since USP14 acted as a member of DUBs, we further confirmed whether the deubiquitinase activity of USP14 directly participated in PSAT1 regulation. It has been reported that the USP14-C114A mutant was incapable of hydrolyzing polyubiquitin chains25. Therefore, inactivated mutants USP14-C114A were constructed to detect its effect on protein homeostasis and ubiquitination of PSAT1. Figure 4j indicated that USP14-C114A did not increase PSAT1 expression as USP14 wild-type (USP14-WT). Also, overexpressing this mutation could not decrease the ubiquitination level of PSAT1 and stabilize the protein level of PSAT1 like USP14-WT (Fig. 4k, l). These results demonstrate that USP14 regulates the ubiquitination and protein stability of PSAT1.
Lysine 51 of PSAT1 is an essential site for the interaction with USP14
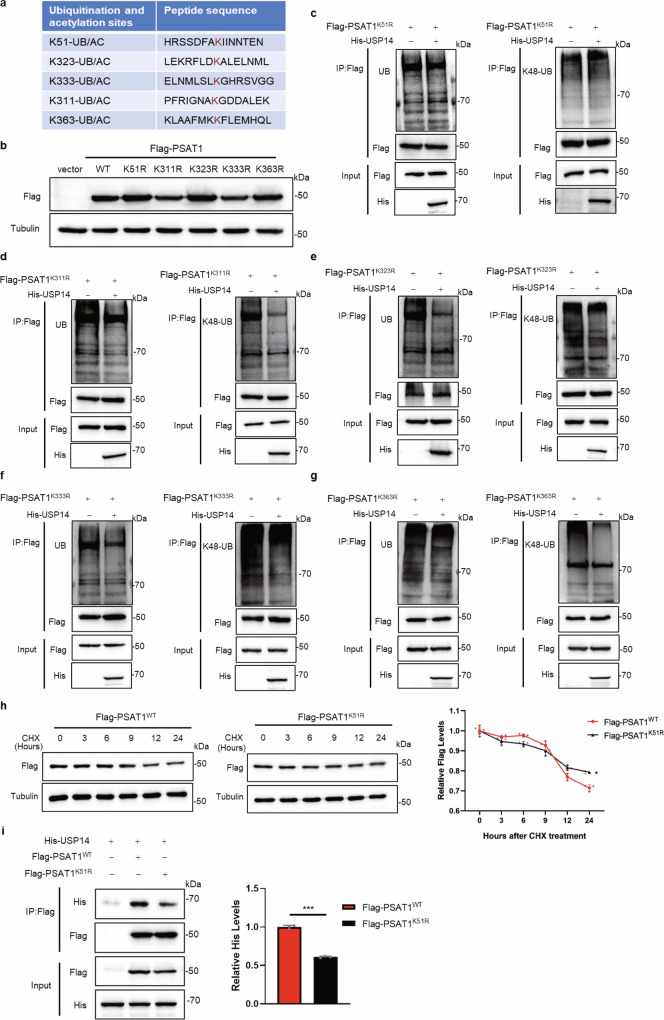
Above results revealed that both the ubiquitination and acetylation of PSAT1 affected its stability. To further clarify the regulatory mechanism on protein stability of PSAT1, we tried to identify the key sites of PSAT1 modified with ubiquitination and acetylation. Using PhosphoSitePlus, we found that the K51, K323, K333, K311 and K363 were identified as the putative ubiquitination and acetylation sites for PSAT1 (Fig. 5a). We tested the expression effects of the PSAT1 plasmids containing indicated mutations (K51R, K311R, K323R, K333R and K363R) (Fig. 5b) and examined whether these mutations affected the ubiquitination levels of PSAT1 regulated by USP14. We found that only the PSAT1-K51R mutant showed no significant change in the total and K48-linked ubiquitination of PSAT1 when overexpressing USP14 (Fig. 5c). Whereas ectopic expression of USP14 notably reduced the total and K48-linked ubiquitination levels of other PSAT1 mutants, including K323R, K333R, K311R and K363R (Fig. 5d–g). This indicated that PSAT1-K51 might be the key ubiquitination site for regulating its protein stability. However, the protein degradation rates of PSAT1 did not show a significant change in cells transfected with PSAT1-K51R compared with PSAT1-WT (Fig. 5h). This result contradicted our expectation that K51 was the key ubiquitination site. As mutation of this site abolished the deubiquitination effect of USP14 on PSAT1, we next examined if K51 mutation affected the interaction between USP14 and PSAT1. Figure 5i and Supplementary Fig. 4a showed that the interaction between USP14 and PSAT1-K51R was significantly attenuated compared with the interaction between USP14 and PSAT1-WT in H1299 and A549 cells. These results demonstrate that PSAT1-K51 is not the direct deubiquitination site for USP14, but an essential site for the interaction with USP14.
Fig. 5. Lysine 51 of PSAT1 is an essential site for the interaction with USP14.
a The co-modified sites with ubiquitination and acetylation of PSAT1 predicted by PhosphoSitePlus. b The expression effects of PSAT1 plasmids containing indicated mutations (K51R, K311R, K323R, K333R and K363R) were tested. c–g H1299 cells were co-transfected with His-USP14 and Flag-PSAT1K51R, Flag-PSAT1K311R, Flag-PSAT1K323R, Flag-PSAT1 K333R or Flag-PSAT1K363R plasmids. Co-IP was used to detect the total ubiquitination (left panel) and K48-linked ubiquitination (right panel) levels of PSAT1. h H1299 cells were experimented with CHX (100 μg/mL) for various time points and were transfected with Flag-PSAT1WT or Flag-PSAT1K51R. The Flag-PSAT1WT or Flag-PSAT1K51R protein expressions were detected by Western blot. The relative protein expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), *P < 0.05. i H1299 cells were co-transfected with His-USP14, Flag-PSAT1WT or Flag-PSAT1K51R. The immunoprecipitations were blotted with anti-His or anti-Flag antibodies. The relative His-USP14 expression compared with that of Flag was quantified. Data represented as the average of three independent experiments (mean ± SD), ***P < 0.001.
Lysine 51 is the key acetylation site regulating the protein stability of PSAT1
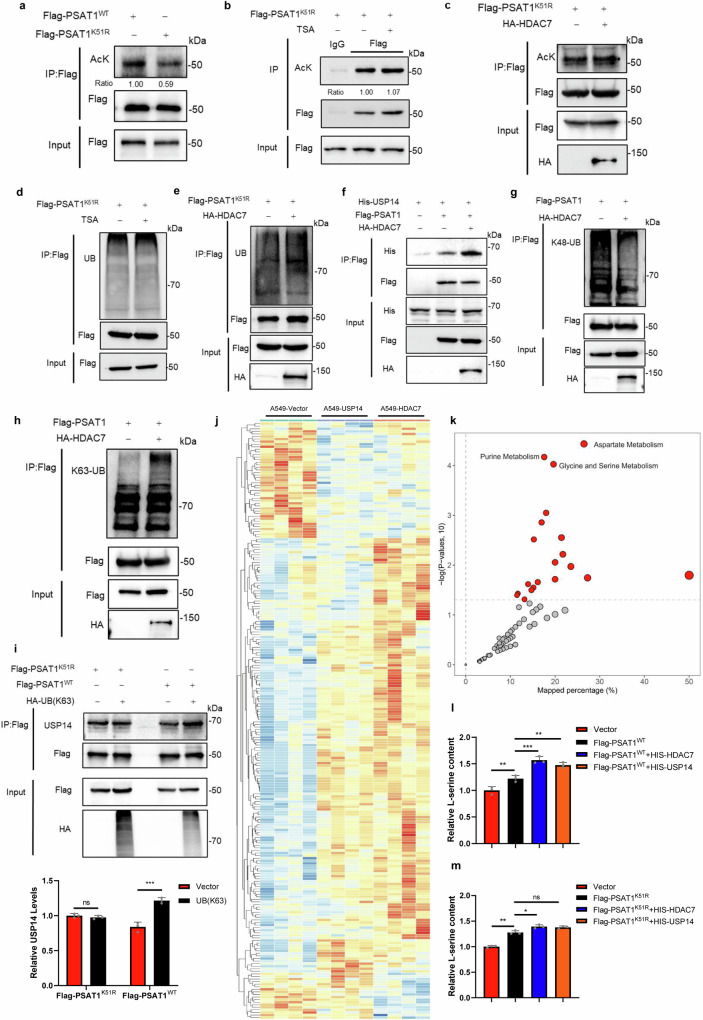
We next detected if K51 was an acetylation site. Figure 6a and Supplementary Fig. 4b showed that the acetylation level of PSAT1-K51R was reduced compared with PSAT1-WT in H1299 and A549 cells. Neither TSA treatment nor HDAC7 overexpression affected the acetylation of PSAT1-K51R, indicating that PSAT1-K51R might be the main acetylation site (Fig. 6b, c and Supplementary Fig. 4c). We also found that TSA treatment did not alter the ubiquitination of PSAT1-K51R (Fig. 6d). Similar effect was also observed in Fig. 6e that the ubiquitination of PSAT1-K51R was not changed when overexpressing HDAC7. These results demonstrated that PSAT1-K51 was the key acetylation site for regulating PSAT1 ubiquitination. Interestingly, overexpressing HDAC7 facilitated the interaction between PSAT1 and USP14 (Fig. 6f), indicating that deacetylation on PSAT1-K51 promoted the interaction between USP14 and PSAT1. This result seemed to contradict the result in Fig. 5i. Although both the mutation on PSAT1-K51 and HDAC7 overexpression showed reduced acetylation of PSAT1, the two experiments were different. PSAT1-K51R abolished all the modifications on Lys51, while deacetylation of PSAT1 on Lys51 by HDAC7 removed the acetyl residue at this site and provided an opportunity for other modifications. Interestingly, we discovered that HDAC7 overexpression decreased the K48-linked ubiquitination of PSAT1, but increased its K63-linked ubiquitination in H1299 and A549 cells (Fig. 6g, h and Supplementary Fig. 4d, e). As K63-linked ubiquitination regulated various functions of proteins except for protein degradation, we next wanted to explore if the K63-linked ubiquitination affected the interaction between USP14 and PSAT1. Figure 6i showed that overexpressing the Ub-K63 mutant (all the lysine residues were mutated to arginine except for Lys63) significantly increased the interaction between USP14 and PSAT1-WT. However, this effect was not observed in PSAT1-K51R. This indicated that deacetylation of PSAT1 on K51 promoted the K63-linked ubiquitination on this site, then USP14 tended to interact with PSAT1 with K63-linked ubiquitination on K51, which enhanced the interaction between USP14 and PSAT1. The immunofluorescence assay was performed and Supplementary Fig. 4f demonstrated that the localization of PSAT1-WT and PSAT1-K51R was consistent, both located in the cytoplasm. All these results demonstrate that Lys51 is the key acetylation site of PSAT1 that regulates its protein stability.
Fig. 6. Acetylation of PSAT1 on Lysine 51 regulates the protein stability of PSAT1 and serine metabolism.
a H1299 cells were transfected with Flag-PSAT1WT or Flag-PSAT1K51R. Co-IP was used to detect the acetylation level of PSAT1. The relative AcK expression compared with that of Flag was quantified. b, d H1299 cells were transfected with Flag-PSAT1K51R plasmid, and TSA (1 μM) was added 12 h before sample collection. Co-IP was used to detect the acetylation or ubiquitination level of PSAT1. c, e H1299 cells were transfected with Flag-PSAT1K51R plasmid alone or co-transfected with HA-HDAC7. Co-IP was used to detect the acetylation or ubiquitination level of PSAT1. f H1299 cells were co-transfected with His-USP14, Flag-PSAT1, and HA-HDAC7 or Vector. The immunoprecipitations were blotted with anti-His or anti-Flag antibodies. g, h In H1299 cells, Flag-PSAT1 and HA-HDAC7 plasmids were co-transfected. Co-IP was used to detect the K48-linked or K63-linked ubiquitination level of PSAT1. i H1299 cells were co-transfected with HA-UB(K63), Flag-PSAT1WT or Flag-PSAT1K51R. The immunoprecipitations were blotted with anti-USP14, anti-Flag, or anti-HA antibodies. The relative USP14 expression compared with that of Flag was quantified. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, ***P < 0.001. j The heat map of the metabolites identified from metabolomics analysis. k KEGG pathway analysis of the DEGs identified from (j). l, m The serine concentration in lysates of cells was measured using the DL-Serine Assay Kit. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, *P < 0.05, **P < 0.01, ***P < 0.001.
Acetylation of PSAT1 on Lysine 51 regulates serine metabolism and proliferation of lung adenocarcinoma
Metabolic reprogramming is one of the most distinctive features of tumorigenesis and progression. PSAT1 catalyzes 3-phosphohydroxylpyruvate and glutamate into 3-phosphoserine and α-ketoglutarate, integrating metabolic pathways such as glycolysis, de novo serine synthesis, citric acid cycle and one-carbon metabolism26. To investigate the effect of the HDAC7-USP14-PSAT1 axis on global tumor metabolism, we performed untargeted metabolomics in A549-Vector, A549-HDAC7, and A549-USP14 cells. A series of metabolites with differential contents between these groups were identified (Supplementary Data 2). Figure 6j and Supplementary Fig. 5 showed that the overexpression of HDAC7 or USP14 showed a similar trend in tumor metabolic regulation. We subsequently classified these differentially abundant metabolites using KEGG pathway analysis and found that the glycine and serine metabolic pathway was one of the most significantly changed metabolic pathways (Fig. 6k). This result was in accordance with the function of HDAC7-USP14-PSAT1 axis and further confirmed that HDAC7 and USP14 regulated the serine metabolism pathway by affecting PSAT1.
Serine provides major cellular metabolic precursors as anabolic and energic supports for a variety of biosynthetic pathways27. Next, we conducted a separate analysis of the metabolites which were closely related to the serine metabolic pathway using our metabolomic analysis. The results revealed that HDAC7 or USP14 overexpression decreased the upstream metabolite of serine synthesis: 3-phosphoglycerate, while increasing the production of α-ketoglutarate which is the direct metabolite of PSAT1 (Supplementary Fig. 6a, b). Overexpressing HDAC7 or USP14 increased GSH levels and reduced GSSG and NADP+, thus enhancing the antioxidant capacity of cancer cells (Supplementary Fig. 6c–e). Meanwhile, the purines and pyrimidines that required for RNA and DNA biosynthesis were also upregulated in HDAC7 or USP14 overexpressing cells (Supplementary Fig. 6f–h). Besides, the phosphatidylcholine and phosphatidyl ethanolamine levels were also increased (Supplementary Fig. 6i, j). To further provide direct evidence for the regulation of serine synthesis by the HDAC7-USP14-PSAT1 axis, we analyzed the serine levels. The data were congruent and showed that the cells with PSAT1 + HDAC7 or PSAT1 + USP14 produced a significantly higher level of serine than overexpressing PSAT1 alone (Fig. 6l). However, in PSAT1-K51R cells, this increase of serine levels was less significant (Fig. 6m). To demonstrate whether acetylation on the K51R site affected the proliferation of LUAD cells, we conducted cell proliferation experiments under specific conditions where the non-essential amino acids serine and glycine is deprived to exclude the influence of exogenous serine uptake. We found that the decreased proliferation caused by serine/glycine- depletion was recovered when PSAT1 overexpression, and overexpressing HDAC7 or USP14 further enhanced this effect (Supplementary Fig. 6k). However, overexpression HDAC7 or USP14 could not promote the proliferation in cells expressing PSAT1-K51R (Supplementary Fig. 6l). Taken together, our data demonstrated the potential impact of PSAT1 acetylation on serine metabolism and proliferation of LUAD cells.
Cisplatin (DDP) and paclitaxel (PTX) are two first-line chemotherapy drugs for the clinical treatment of non-small cell lung cancer (NSCLC)28. Mechanically, as a platinum chemotherapeutic agent, DDP can enter cells to cause DNA cross-linking, leading to DNA damage. Unlike DDP, PTX targets the microtubule cytoskeleton, inducing cell cycle arrest and apoptosis29. To investigate the effects of PSAT1 on chemotherapy treatment, we first measured the protein expression of PSAT1 when treating cells with different concentrations of DDP or PTX. Supplementary Fig. 7a, b showed that PSAT1 protein levels significantly increased in a DDP concentration-dependent manner, while the protein levels slightly decreased with PTX treatment. We subsequently tested whether these chemotherapy drugs regulated the acetylation and ubiquitination levels of PSAT1. Supplementary Fig. 7c–f showed that DDP treatment had no significant effect on the acetylation and ubiquitination levels of PSAT1-K51R cells, whereas it led to a remarkable decrease in PSAT1-WT cells. We also found that PTX treatment did not alter the acetylation and ubiquitination levels. These results indicated that DDP affected PSAT1 expression by regulating the interplay between acetylation and ubiquitination, while PTX did not have these effects. Then, we tested if PSAT1 expression affected the therapeutic effect of DDP or PTX on lung cancer cells. Knocking down PSAT1 increased the sensitivity of cancer cells to DDP treatment rather than PTX (Supplementary Fig. 7g, h). Thus, we speculated that the different anticancer mechanisms between DDP and PTX led to the unique effects of PSAT1. These results exhibit the potential application of PSAT1 as a therapeutic target for cancer treatment.
UBE4B ubiquitinates the acetylated PSAT1 for proteasomal degradation
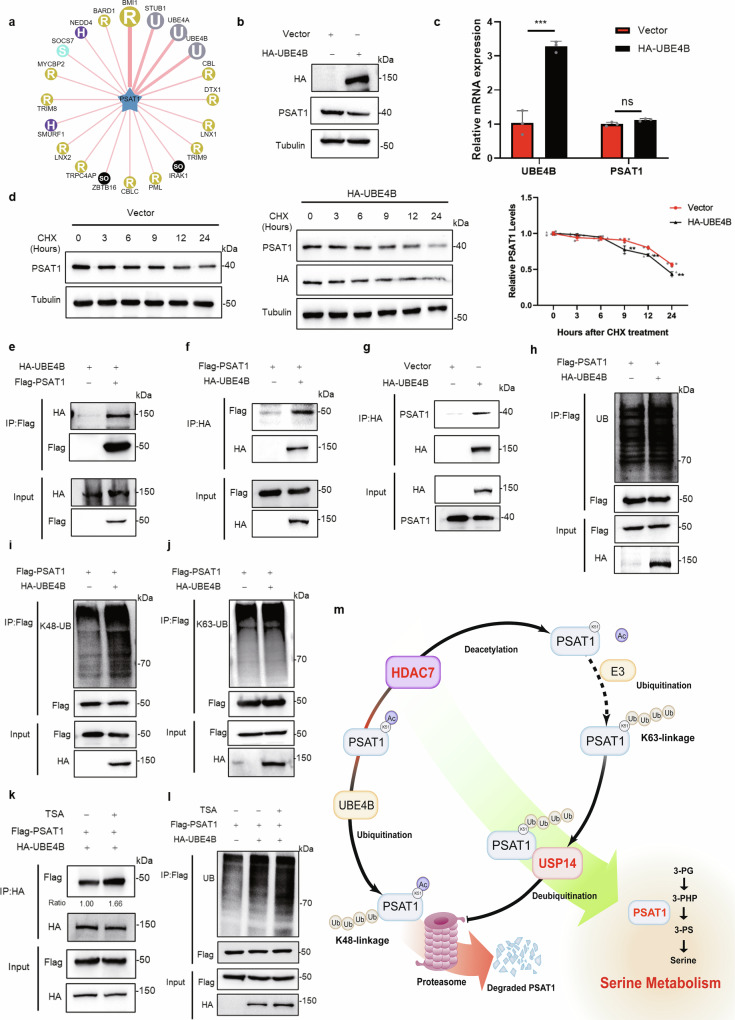
To clarify the E3 ligase that ubiquitinates PSAT1, we used UbiBrowser to identify the potential targets. Figure 7a showed the predicted E3 ligases. We selected the top four proteins (BMI1, STUB1, UBE4A, UBE4B) with the highest score for further validation. Figure 7b and Supplementary Fig. 8a–d showed that overexpressing UBE4B reduced the protein expression of PSAT1 while expressing the others did not show this effect. Overexpressing UBE4B did not affect the transcription of PSAT1 (Fig. 7c), indicating that UBE4B might regulate PSAT1 expression by protein degradation. The protein degradation rates were examined and the result demonstrated that overexpressing UBE4B accelerated the degradation rate of PSAT1 protein (Fig. 7d). Then, we detected the interaction between PSAT1 and UBE4B. Figure 7e–g and Supplementary Fig. 8e showed that PSAT1 interacted with UBE4B. We next examined if UBE4B regulated PSAT1 expression through ubiquitination. Overexpressing UBE4B increased the total and K48-linked ubiquitination of PSAT1, while the K63-linked ubiquitination was not affected (Fig. 7h–j and Supplementary Fig. 8f–h). We further explored if acetylation affected the interaction between UBE4B and PSAT1. TSA treatment was used to increase the acetylation of PSAT1. Figure 7k and Supplementary Fig. 8i showed that TSA treatment increased the interaction between UBE4B and PSAT1. Consistent with this, Supplementary Fig. 8j, k showed that HDAC7 overexpression decreased their interaction. Meanwhile, the ubiquitination levels of PSAT1 were detected with TSA and HDAC7 treatment. Figure 7l and Supplementary Fig. 8l, m revealed that acetylation affected the ubiquitination of PSAT1 by UBE4B, suggesting that acetylation promoted the degradation of PSAT1. These results demonstrated that UBE4B ubiquitinated the acetylated PSAT1 for proteasomal degradation.
Fig. 7. UBE4B ubiquitinates the acetylated PSAT1 for proteasomal degradation.
a UbiBrowser was used to search for E3 ligases of PSAT1. b In H1299 cells, UBE4B was overexpressed to detect PSAT1 protein by Western blot. c H1299 cells were transfected with control Vector or HA-UBE4B plasmid. The mRNA levels of UBE4B and PSAT1 were detected by qPCR. Data represented as the average of three independent experiments (mean ± SD), ns: P > 0.05, ***P < 0.001. d H1299 cells were experimented with CHX (100 μg/mL) for various time points and were transfected with HA-UBE4B. The PSAT1 protein expressions were detected by Western blot. The relative PSAT1 expression compared with that of Tubulin was quantified. Data represented as the average of three independent experiments (mean ± SD), **P < 0.01. e, f H1299 cells were co-transfected with HA-UBE4B and Flag-PSAT1. The immunoprecipitations were blotted with anti-HA or anti-Flag antibodies. g In H1299 cells, endogenous interaction between PSAT1 and UBE4B was tested using anti-HA antibodies for Co-IP. The immunoprecipitations were blotted with anti-PSAT1 and anti-HA antibodies. h–j In H1299 cells, Flag-PSAT1 and HA-UBE4B plasmids were co-transfected. Co-IP was used to detect the total, K48-linked or K63-linked ubiquitination levels of PSAT1. k H1299 cells were co-transfected with Flag-PSAT1 and HA-UBE4B. Then TSA (1 μM) was added 12 h before sample collection. The immunoprecipitations were blotted with anti-Flag or anti-HA antibodies. The relative Flag expression compared with that of HA was quantified. l H1299 cells were transfected with Flag-PSAT1 plasmid alone or co-transfected with HA-UBE4B plasmids and were treated with or without TSA (1 μM). Co-IP was used to detect the ubiquitination level of PSAT1. m Proposed model for the protein stability regulation of PSAT1 by acetylation and ubiquitination.
Taken together, our study demonstrates that the protein stability of PSAT1 is regulated by acetylation and ubiquitination. HDAC7 deacetylates PSAT1 on Lys51, followed by ubiquitination on this site in K63-linkage. Then, USP14 interacts with PSAT1 and deubiquitinates chains with K48-linkage, thus increasing PSAT1 protein stability and enhancing the serine metabolism of lung cancer cells. When PSAT1 is acetylated, UBE4B binds and ubiquitinates PSAT1, promoting its proteasomal degradation (Fig. 7m).
Discussion
Serine is a non-essential amino acid and contributes to the synthesis of proteins, nucleotides, lipids, and antioxidants30. Metabolite profiling in 60 cancer cell lines revealed that cancer cells avidly consume extracellular serine, which is second only to glutamine among amino acids31. Serine can be either obtained from the diet or synthesized de novo via the SSP from an intermediate of glycolysis. During this process, PSAT1 catalyzes the key metabolic step in SSP, converting 3-phosphohydroxylpyruvate to 3-phosphoserine and producing α-ketoglutarate concomitantly32. Thus, PSAT1 is a key transaminase linking metabolic pathways such as serine metabolism and glutamine metabolism. A growing body of evidence indicates that PSAT1 is an oncogene that plays a positive role in lung cancer progression and metastasis33. It has been demonstrated that high expression of PSAT1 inhibits cyclin D1 degradation and subsequently alters Retinoblastoma (RB)- Early 2 factor (E2F) pathway activity, enhancing G1 progression and proliferation of NSCLC cells34. In addition, highly expressed PSAT1 promotes the nuclear translocation of pyruvate kinase M2 (PKM2) in response to epidermal growth factor receptor (EGFR) activation, thus promoting lung cancer progression35. Furthermore, the knockdown of PSAT1 enhances the sensitivity of NSCLC cells to glutamine-limiting conditions and ionizing radiation, indicating that PSAT1 might be a therapeutic target for lung cancer36. These findings suggest that upregulation of PSAT1 maintains lung cancer development and is associated with poor patient outcomes. Therefore, it is particularly important to elucidate the mechanism regulating PSAT1 in lung cancer.
Most researches on the regulatory mechanisms of PSAT1 focus on its transcriptional level. ATF4 was reported to directly bind to the PSAT1 promoter and activate its transcription. NRF2, another transcription factor, regulated PSAT1 expression via ATF4 to support glutathione and nucleotide production8,9. c-Myc was also reported to activate PSAT1 transcription under nutrient deprivation conditions10. However, as a metabolic enzyme, the post-translational modifications of PSAT1 may also play greater roles, because of its advantages in rapid regulation. Until now, little is known about the post-translational modifications on PSAT1. This greatly limits people’s understanding of the regulatory mechanisms of PSAT1. Our study demonstrates that the stability of PSAT1 protein in LUAD is regulated by the interplay between acetylation and ubiquitination. Deacetylation of PSAT1 by HDAC7 facilitates its binding with USP14, leading to the deubiquitination and stabilization of PSAT1.
Acetylated proteins are involved in many metabolic pathways, including fatty acid metabolism, glycolysis, the TCA, and urea cycles37. Lysine acetylation is controlled by two opposing types of enzymes: acetyltransferases and deacetylases. Protein lysine deacetylases consist of two classes, including a total of eighteen proteins (HDAC1–11 and SIRT1–7 in mammals) that deacetylate histones and non-histone proteins38. In our study, we found HDAC7 was the key deacetylase that regulated PSAT1 protein stability. Generally, HDAC7 functions as a transcriptional repressor in the nucleus, and phosphorylation of HDAC7 leads to its nuclear export. HDAC7 shuttles between the nucleus and cytoplasm and is involved in the regulation of cell proliferation, apoptosis, differentiation, and migration39. In lung cancer, HDAC7 promotes tumorigenesis by inhibiting signal transducer and activator of transcription 3 (STAT3) activation via deacetylation40. HDAC7 could also promote NSCLC progression by maintaining the cytoskeletal structure and angiogenesis during the neovascularization of endothelial progenitor cells41. However, the functions of HDAC7 in regulating cancer metabolism, especially in serine metabolism, need to be further clarified. In our study, we demonstrated that HDAC7 could directly interact with and deacetylate PSAT1, resulting in the deubiquitination of K48-linked chains and stabilization of PSAT1 protein. To our knowledge, this is the first study showing that HDAC7 directly regulates metabolic enzymes.
Ubiquitin-mediated degradation is one of the main processes for protein degradation. Proteasome targeting and degradation are primarily accomplished through the formation of K48-linked chains, while K63-linked chains perform more diverse functions including lysosomal degradation, altering target protein structure, localization, or activity42. USP14 can rescue proteins from degradation by disassembling the ubiquitin chain from its substrate’s distal end43. In Fig. 4, we found that USP14 acted as a positive regulator of PSAT1 by removing K48-linked polyubiquitin chains from PSAT1. Our study interestingly found that USP14 did not deubiquitinate K63-linked polyubiquitin chains on PSAT1; instead, K63-linked ubiquitination promoted the interaction between PSAT1 and USP14, thus enhancing its deubiquitination on K48-linked polyubiquitin chains. These results indicate that the deubiquitinase activity of USP14 possesses selectivity according to the substrates and physiological conditions.
Given that PSAT1 is a key enzyme in de novo serine synthesis, which provides important cellular metabolic precursors for biosynthetic metabolism, we examined the effects of PSAT1 acetylation on the serine synthesis pathway. In our study, we found that acetylation of PSAT1 on Lysine 51 regulates serine metabolism and proliferation of LUAD cells. Importantly, the chemotherapy drug DDP could increase PSAT1 expression in LUAD cells by decreasing its acetylation on K51 and enhancing its protein stability. DDP is a DNA-damaging agent. We found PSAT1 could regulate nucleotide synthesis which was required for DNA biosynthesis. Knocking down PSAT1 increased the sensitivity of LUAD cells to DDP. However, treating LUAD cells with PTX, another chemotherapy drug targeting the microtubule cytoskeleton, did not show this effect. These results indicated that inhibiting the function of PSAT1 is an effective strategy to enhance the therapeutic effect of chemotherapy drugs for DNA damage.
Thus, our study discovers a new regulatory mechanism for PSAT1 in LUAD, demonstrating that the interplay between acetylation and ubiquitination regulates PSAT1 protein stability, further affecting serine metabolism and cell proliferation. This further deepens people’s understanding of the regulation mechanism for PSAT1 in cancer and provides a new basis for the development of anticancer drugs targeting serine metabolism.
Methods
Antibodies and reagents
The following primary antibodies were used in this study: anti-PSAT1 (10501-1-AP, Proteintech, 1:1000), anti-USP14 (14517-1-AP, Proteintech, 1:1000), anti-HDAC7 (26207-1-AP, Proteintech, 1:1000), anti-CyclinB1 (55004-1-AP, Proteintech, 1:1000), anti-Tubulin (66362-1-Ig, Proteintech, 1:2000), anti-Acetyllysine (PTM-105RM, PTM BIO, 1:500), anit-Ubiquitin (10201-2-AP, Proteintech, 1:500), anti- SQSTM1 (TA502127, Origene, 1:1000), anti- LC3B (18725-1-AP, Proteintech, 1:1000), anti-K48-linkage Specific Polyubiquitin (8081S, Cell Signaling Technology, 1:500), anti-K63-linkage Specific Polyubiquitin (5621S, Cell Signaling Technology, 1:500), anti-Actin (TA811000, Origene, 1:2000), anti-Flag (66008-4-lg, Proteintech, 1:5000), anti-Myc (16286-1-AP, Proteintech, 1:5000), anti-His (66005-1-lg, Proteintech, 1:5000), anti-HA (51064-2-AP, Proteintech, 1:5000), anti-IgG (B900620, Proteintech, 1:5000). As secondary antibodies for Western blotting, HRP-conjugated goat anti-rabbit (31460, Thermo Scientific, 1:5000) and anti-mouse (31430, Thermo Scientific, 1:5000) antibodies were used. For immunofluorescent staining, CoraLite488-conjugated Goat Anti-Mouse IgG(H+L) (SA00013-1, Proteintech, 1:500) and CoraLite594 – conjugated Goat Anti-Rabbit IgG(H+L) (SA00013-4, Proteintech, 1:500) were used as secondary antibodies. The nucleus was stained with DAPI (0100-20; Southern Biotech).
In this study, the chemotherapy agents cisplatin (DDP, HY-17394, MCE) and paclitaxel (PTX, HY-B0015, MCE) were used. The inhibitors nicotinamide (NAM, 72340, Sigma), trichostatin A (TSA, S1045, Selleck Chemicals), MG132 (1791-5, Biovision), Chloroquine (CQ, C6628, Sigma), Cycloheximide (CHX, S7418, Selleck Chemicals), IU1 (A4002, APExBIO) were used.
Cell culture
BEAS-2B was purchased from the National Collection of Authenticated Cell Cultures and cultured in RPMI 1640 medium (Gibco) supplemented with 10% FBS (ExCell Bio). Lung cancer cell lines A549, H1299, H23, HCC827, H358, PC9, H1975, and breast cancer cell (MCF7), hepatoma cell (HLE) were purchased from the National Collection of Authenticated Cell Cultures and cultured in RPMI 1640 medium supplemented with 10% FBS. All cells were cultured under an atmosphere of 5% CO2 at 37°C.
For serine and glycine deprivation experiments, cells in the control group were cultured in assay medium: MEM (Thermo Scientific) supplemented with 10% FBS (SORFA), 17 mM D-glucose (Sigma), 400 μM serine (Sigma) and 400 μM glycine (Sigma). Cells that were subjected to serine and glycine starvation were cultured in a medium without serine or glycine.
For cell growth assay, with a density of 5000 cells per well in 500 µl of RPMI 1640 medium containing 10% FBS, cells were seeded in 24-well plates. The cell culture medium was replaced every 2 days. At certain times, after fixation with 4% formaldehyde for 30 min the cell was stained using 0.1% crystal violet and extracted with 10% acetic acid. Then, we measured the relative absorbance at 595 nm. Each experiment was conducted in triplicate.
For cell viability assay, with a density of 5000 cells per well in 100 µl of RPMI 1640 medium containing 10% FBS, cells were seeded in 96-well plates. After being treated according to the experimental design, the cells were added with Cell Counting Kit-8 (CCK8) and incubated for 1 h in 5% CO2 at 37 °C. Then, we measured the relative absorbance at 490 nm. Each experiment was conducted in triplicate.
Reverse transcription-polymerase chain reaction (RT-PCR)
Total RNA was extracted using TRIGene reagent (P118-05, Genestar). The RNA was used for reverse transcription to cDNA by HiScript III RT SuperMix for qPCR (+gDNA wiper) (R323-01, Vazyme). For the RT-qPCR assay, we used Real-Time PCR EasyTM-SYBR Green I (QP-01012, Foregene) to conduct experiments. The relative mRNA expression of target genes was calculated using the comparative Ct method. GAPDH was used as a control for the target gene. For all gene primers used for RT-PCR, see Supplementary Table 1.
Gene overexpression and knockdown
For transfection to overexpress certain genes, cells were transiently transfected using the indicated plasmids with the SuperFectin DNA Transfection Reagent kit (2102–100, Pufei). Then the relative transient transfection efficiency in cells was measured by western blot assay with certain antibodies.
For transfection to knock down the gene, cells were transiently transfected using the indicated RNAi nucleotides with the siTran 2.0 siRNA transfection reagent (TT320002, Origene). Then the relative transient transfection efficiency in cells was measured by western blot assay using the relevant antibodies. For all siRNA sequences used for transfection, see Supplementary Table 1.
Immunoprecipitation and western blot
For the immunoprecipitation assay, cells were washed with PBS three times. Then, cells were lysed using NP-40 lysis buffer supplemented with protease inhibitor PMSF (P8340, Solarbio) for 30 min at 4 °C. Cell lysates were centrifuged for 20 min at 4 °C, then the indicated antibodies and protein G agarose beads (11243233001, Roche) were added to the supernatants and incubated overnight at 4 °C. Then the mixed substance was washed by lysis buffer, then added loading buffer, and boiled for 10 min. For detecting the ubiquitination levels, we extracted proteins using the lysis buffer with 1% SDS, heated the lysate for 8 min, and then diluted the cell extract down to 0.1% SDS to perform immunoprecipitation.
For the western blotting assay, we used 10% SDS-PAGE to separate the proteins and transferred these proteins to Polyvinylidene fluoride (PVDF) membranes (IPVH00010, Milipore). The PVDF membranes were blocked with 5% skim milk (36120ES76, Yeasen) for 1 h at room temperature and then incubated with the indicated primary antibodies overnight. The next day, we used TBST to wash the membranes at least three times and used a secondary antibody to incubate the membranes for 1 h at room temperature. Staining was conducted with indicated detection reagents (PA112-01, TIANGEN). Protein levels were visualized and analyzed using a digital gel image analysis system.
Immunofluorescence assay
Cells were seeded on 24-well plate with a cell slide and grew for 18–24 h. The coverslips were washed with PBS for 5 min each time and fixed with 4% paraformaldehyde at room temperature for 20 min. After three washes with PBS, cells were drilled with 0.2% Triton X-100 for 20 min and blocked for 30 min with 5% BSA. Then, coverslips were incubated with indicated primary antibodies overnight at 4 °C. The next day, Coverslips were washed three times with PBS and incubated with a secondary antibody at room temperature for 1 h. After washed three times with PBS, cells were stained with DAPI. Finally, the confocal microscope was used to photograph.
Serine measurement using Serine Assay Kit
The serine concentration in lysates of cells was measured using the DL-Serine Assay Kit (MAK352, Sigma). Briefly, cells were rapidly homogenized with Serine Assay Buffer. After sample cleanup with ultrafiltration tubes and reagents from the kit, we added enzyme mixture and probe solution to the filtrate. 96-well plates were incubated at 37 °C for 60 min, protected from light. Finally, all samples have measured the fluorescence at ex = 535 nm/em = 587 nm in endpoint mode.
Mass spectrometry analysis
H1299 cells were transfected with Vector and PSAT1 plasmids, respectively. When the cell number reached 107, proteins were collected as described in the immunoprecipitation assay. Then the protein samples were sent to APTBIO for mass spectrometry analysis.
Untargeted metabolomics analysis
A549 cells were transfected using the indicated plasmids. Then, the cells were washed and collected with ice-cold PBS. After centrifugation at low speed, we removed the supernatant and collected precipitation from a liquid nitrogen flash freezer. Then the cell samples were sent to Bioprofile for untargeted metabolomics analysis.
Statistics and reproducibility
The statistical data were analyzed using GraphPad Prism software. Each experiment was performed in triplicate. All the data were presented as mean ± SD. Comparisons were performed using two-tailed paired Student’s t-tests or one-way ANOVA. A p-value of lower than 0.05 was considered statistically significant. *, p < 0.05 vs. control; **, p < 0.01 vs. control; ***, p < 0.001 vs. control; ns, på 0.05 vs. control.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Acknowledgements
This work was supported by the National Natural Science Foundation of China (82273258, 82030086, 82260559, 82360474, 82160553), Natural Science Foundation of Jiangxi Province (20212ACB216007), the Science and Technology Innovation High-end Talent Project of Jiangxi Province (jxsq2023201100), the Training Plan for Academic and Technical Leaders of Major Disciplines in Jiangxi Province (20204BCJ23023), Scientific Research Project of Cultivating Outstanding Young People in First Affiliated Hospital of Nanchang University (YQ202104).
Author contributions
T.H. and J.W. designed the study. Y.L., W.X., T.Z., M.H., L.S., G.W., and X.K. performed the experiments. T.H. and Y.L. analyzed the data and wrote the manuscript. J.W. revised the paper. These authors jointly supervised this work.
Peer review
Peer review information
Communications Biology thanks Ashkan Emadi, Chrysa Filippopoulou, and the other, anonymous, reviewer for their contribution to the peer review of this work. Primary Handling Editors: Georgios Giamas and Kaliya Georgieva. A peer review file is available.
Data availability
All data associated with this study are present in this paper or the Supplementary Information. Uncropped western blots are available in Supplementary Fig. 9. Datasets used and analyzed are available in Supplementary Data 1 and 2. All source data behind each graph are shown in Supplementary Data 3. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the identifier PXD056565. Any additional information in this paper will be available from the corresponding author upon reasonable request.
Competing interests
The authors declare no competing interests.
Inclusion and diversity
We support inclusive, diverse, and equitable conduct of research.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Jianbin Wang, Email: jianbinwang@ncu.edu.cn.
Tianyu Han, Email: hantianyu87@163.com.
Supplementary information
The online version contains supplementary material available at 10.1038/s42003-024-07051-2.
References
- 1.Sun, L., Suo, C., Li, S. T., Zhang, H. & Gao, P. Metabolic reprogramming for cancer cells and their microenvironment: beyond the Warburg effect. Biochim. Biophys. Acta Rev. Cancer1870, 51–66 (2018). [DOI] [PubMed] [Google Scholar]
- 2.DeBerardinis, R. J. Serine metabolism: some tumors take the road less traveled. Cell Metab.14, 285–286 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman, A. C. & Maddocks, O. D. K. Serine and functional metabolites in cancer. Trends Cell Biol.27, 645–657 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Yang, M. & Vousden, K. H. Serine and one-carbon metabolism in cancer. Nat. Rev. Cancer16, 650–662 (2016). [DOI] [PubMed] [Google Scholar]
- 5.Amelio, I., Cutruzzola, F., Antonov, A., Agostini, M. & Melino, G. Serine and glycine metabolism in cancer. Trends Biochem. Sci.39, 191–198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang, W. C. et al. Glycine decarboxylase activity drives non-small cell lung cancer tumor-initiating cells and tumorigenesis. Cell148, 259–272 (2012). [DOI] [PubMed] [Google Scholar]
- 7.Possemato, R. et al. Functional genomics reveal that the serine synthesis pathway is essential in breast cancer. Nature476, 346–350 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ye, J. et al. Pyruvate kinase M2 promotes de novo serine synthesis to sustain mTORC1 activity and cell proliferation. Proc. Natl Acad. Sci. USA109, 6904–6909 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.DeNicola, G. M. et al. NRF2 regulates serine biosynthesis in non-small cell lung cancer. Nat. Genet.47, 1475–1481 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun, L. et al. cMyc-mediated activation of serine biosynthesis pathway is critical for cancer progression under nutrient deprivation conditions. Cell Res.25, 429–444 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding, J. et al. The histone H3 methyltransferase G9A epigenetically activates the serine-glycine synthesis pathway to sustain cancer cell survival and proliferation. Cell Metab.18, 896–907 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang, H. et al. Overexpression of PSAT1 regulated by G9A sustains cell proliferation in colorectal cancer. Signal Transduct. Target. Ther.5, 47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lin, H., Su, X. & He, B. Protein lysine acylation and cysteine succination by intermediates of energy metabolism. ACS Chem. Biol.7, 947–960 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhou, F. et al. ARHGEF3 regulates the stability of ACLY to promote the proliferation of lung cancer. Cell Death Dis.13, 870 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhong, Y. et al. The HDAC10 instructs macrophage M2 program via deacetylation of STAT3 and promotes allergic airway inflammation. Theranostics13, 3568–3581 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao, L., Zhao, J., Zhong, K., Tong, A. & Jia, D. Targeted protein degradation: mechanisms, strategies and application. Signal Transduct. Target. Ther.7, 113 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Witt, O., Deubzer, H. E., Milde, T. & Oehme, I. HDAC family: what are the cancer relevant targets? Cancer Lett.277, 8–21 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Swatek, K. N. & Komander, D. Ubiquitin modifications. Cell Res.26, 399–422 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee, B. H. et al. USP14 deubiquitinates proteasome-bound substrates that are ubiquitinated at multiple sites. Nature532, 398–401 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.McClurg, U. L. & Robson, C. N. Deubiquitinating enzymes as oncotargets. Oncotarget6, 9657–9668 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang, F., Ning, S., Yu, B. & Wang, Y. USP14: structure, function, and target inhibition. Front. Pharmacol.12, 801328 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Liu, Y. et al. USP14 regulates cell cycle progression through deubiquitinating CDK1 in breast cancer. Acta Biochim. Biophys. Sin.54, 1610–1618 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu, B. et al. CyclinB1 deubiquitination by USP14 regulates cell cycle progression in breast cancer. Pathol. Res. Pract.215, 152592 (2019). [DOI] [PubMed] [Google Scholar]
- 24.Chau, V. et al. A multiubiquitin chain is confined to specific lysine in a targeted short-lived protein. Science243, 1576–1583 (1989). [DOI] [PubMed] [Google Scholar]
- 25.Zhao, C. et al. A self-amplifying USP14-TAZ loop drives the progression and liver metastasis of pancreatic ductal adenocarcinoma. Cell Death Differ.30, 1–15 (2023). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang, X., Li, C. & Chen, Y. Phosphoserine aminotransferase 1: a metabolic enzyme target of cancers. Curr. Cancer Drug Targets23, 171–186 (2023). [DOI] [PubMed] [Google Scholar]
- 27.Maddocks, O. D. et al. Serine starvation induces stress and p53-dependent metabolic remodelling in cancer cells. Nature493, 542–546 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rose, M. C., Kostyanovskaya, E. & Huang, R. S. Pharmacogenomics of cisplatin sensitivity in non-small cell lung cancer. Genom. Proteom. Bioinform.12, 198–209 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhang, C. C. et al. Chemotherapeutic paclitaxel and cisplatin differentially induce pyroptosis in A549 lung cancer cells via caspase-3/GSDME activation. Apoptosis24, 312–325 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Zhou, X., Tian, C., Cao, Y., Zhao, M. & Wang, K. The role of serine metabolism in lung cancer: From oncogenesis to tumor treatment. Front. Genet.13, 1084609 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jain, M. et al. Metabolite profiling identifies a key role for glycine in rapid cancer cell proliferation. Science336, 1040–1044 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Metcalf, S. et al. Selective loss of phosphoserine aminotransferase 1 (PSAT1) suppresses migration, invasion, and experimental metastasis in triple negative breast cancer. Clin. Exp. Metastasis37, 187–197 (2020). [DOI] [PubMed] [Google Scholar]
- 33.Feng, M. et al. An integrated pan-cancer analysis of PSAT1: A potential biomarker for survival and immunotherapy. Front. Genet.13, 975381 (2022). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yang, Y. et al. PSAT1 regulates cyclin D1 degradation and sustains proliferation of non-small cell lung cancer cells. Int. J. Cancer136, E39–E50 (2015). [DOI] [PubMed] [Google Scholar]
- 35.Biyik-Sit, R. et al. Nuclear pyruvate kinase M2 (PKM2) contributes to phosphoserine aminotransferase 1 (PSAT1)-mediated cell migration in EGFR-activated lung cancer cells. Cancers13, 3938 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jin, H. O. et al. Knock-down of PSAT1 enhances sensitivity of NSCLC cells to glutamine-limiting conditions. Anticancer Res.39, 6723–6730 (2019). [DOI] [PubMed] [Google Scholar]
- 37.Zhao, S. et al. Regulation of cellular metabolism by protein lysine acetylation. Science327, 1000–1004 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shahbazian, M. D. & Grunstein, M. Functions of site-specific histone acetylation and deacetylation. Annu. Rev. Biochem.76, 75–100 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Ho, T. C. S., Chan, A. H. Y. & Ganesan, A. Thirty years of HDAC inhibitors: 2020 insight and hindsight. J. Med. Chem.63, 12460–12484 (2020). [DOI] [PubMed] [Google Scholar]
- 40.Lei, Y. et al. Hdac7 promotes lung tumorigenesis by inhibiting Stat3 activation. Mol. Cancer16, 170 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wei, Y. et al. Endothelial progenitor cells contribute to neovascularization of non-small cell lung cancer via histone deacetylase 7-mediated cytoskeleton regulation and angiogenic genes transcription. Int. J. Cancer143, 657–667 (2018). [DOI] [PubMed] [Google Scholar]
- 42.Hussain, S., Zhang, Y. & Galardy, P. J. DUBs and cancer: the role of deubiquitinating enzymes as oncogenes, non-oncogenes and tumor suppressors. Cell Cycle8, 1688–1697 (2009). [DOI] [PubMed] [Google Scholar]
- 43.Hu, M. et al. Structure and mechanisms of the proteasome-associated deubiquitinating enzyme USP14. EMBO J.24, 3747–3756 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data associated with this study are present in this paper or the Supplementary Information. Uncropped western blots are available in Supplementary Fig. 9. Datasets used and analyzed are available in Supplementary Data 1 and 2. All source data behind each graph are shown in Supplementary Data 3. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium with the identifier PXD056565. Any additional information in this paper will be available from the corresponding author upon reasonable request.