Abstract
Human mitochondrial glutaredoxin 2 (Grx2) is a glutathione-dependent oxidoreductase (active site: Cys-Ser-Tyr-Cys) that facilitates the maintenance of mitochondrial redox homeostasis upon induction of apoptosis by oxidative stress. Here, we have characterized Grx2 as an iron–sulfur center-containing member of the thioredoxin fold protein family. Mössbauer spectroscopy revealed the presence of a four cysteine-coordinated nonoxidizable [2Fe-2S]2+ cluster that bridges two Grx2 molecules via two structural Cys residues to form dimeric holo Grx2. Coimmunoprecipitation of radiolabeled iron with Grx2 from human cell lines indicated the presence of the cluster in vivo. The [2Fe-2S]-bridged dimer was enzymatically inactive, but degradation of the cluster and the resulting monomerization of Grx2 activated the protein. Slow degradation under aerobic conditions was prevented by the presence of glutathione, whereas glutathione disulfide as well as one-electron oxidants or reductants promoted monomerization of Grx2. We propose that the iron–sulfur cluster serves as a redox sensor for the activation of Grx2 during conditions of oxidative stress when free radicals are formed and the glutathione pool becomes oxidized.
Keywords: glutathione, iron–sulfur cluster, mitochondria, thiol disulfide oxidoreductase, oxidative stress
Glutaredoxins (Grxs) are thiol-disulfide oxidoreductases and involved in the maintenance of cellular redox homeostasis. They are glutathione (GSH)-dependent enzymes, first discovered as electron donor for ribonucleotide reductase (1, 2). Grxs belong to the thioredoxin family of proteins, with whom they share a similar structure and overlapping functions (3). Grxs usually contain the active site motif Cys-Pro-Tyr-Cys and reduce disulfides via two distinct mechanisms. The dithiol mechanism requires both active site Cys residues, whereas the monothiol mechanism relies only on the more N-terminal active site cysteine (4). Using these mechanisms, Grxs catalyze the glutathione-dependent reduction of disulfides and glutathione-mixed disulfides, serving as electron donor, e.g., for ribonucleotide or sulfate reduction (2, 5), and controlling the levels of protein GSH-mixed disulfides (6).
In terms of their structure and active site sequences, Grxs can be divided into three categories (7). The first category consists of the classical Grxs of ≈ 9–14 kDa with two cysteines in their active site, exemplified by Escherichia coli Grx1 and the human Grxs. The second category encloses the monothiol Grxs like yeast Grx3, Grx4, and Grx5 (8), lacking the more C-terminal active site cysteine. The third category is defined by the unusual E. coli Grx2 (9). Two Grxs have been identified in mammals so far. Cytosolic Grx1 supports ribonucleotide reductase with electrons and is involved in general disulfide-dithiol exchanges (3), dehydroascorbate reduction (10), cellular differentiation (11), regulation of transcription factors (12–14), and apoptosis (15, 16). The more recently discovered second mammalian Grx (Grx2) is present in two isoforms derived from alternative first exons. Grx2a is targeted to mitochondria, whereas Grx2b was predicted to be localized in the nucleus (17, 18). Both isoforms share the same Grx core, encoded by exons II to IV; the unusual active site (Cys-Ser-Tyr-Cys) is encoded by exon III. Grx2 is a very efficient catalyst of monothiol reactions because of its high affinity for protein glutathione-mixed disulfides and, unlike Grx1, Grx2 is not inhibited by oxidation of structural Cys residues (17). In addition, Grx2 can receive electrons not only from GSH, but also from thioredoxin reductase supporting both monothiol and dithiol reactions (19). Grx2 efficiently catalyzes both glutathionylation and deglutathionylation of mitochondrial complex I (20), which in turn regulates the superoxide production by the complex (21). Small interfering RNA-mediated knock-down of Grx2 dramatically sensitizes HeLa cells to cell death induced by doxorubicin/adriamycin and phenylarsine oxide (22). Overexpression of Grx2a decreased the susceptibility to apoptosis and prevented loss of cardiolipin and cytochrome c release (23). Together, these findings indicate a central role of Grx2 in the cellular response to apoptotic stimuli and oxidative stress at the mitochondrial checkpoint.
In this work, we describe human Grx2 as an iron–sulfur protein. We demonstrate the binding of one [2Fe-2S] cluster to a Grx2 dimer and propose two Cys residues, not conserved in other Grxs, as the place of binding. Holo Grx2 is enzymatically inactive, but loss of the iron–sulfur cluster activates the protein. We present evidence that the cluster is present in vivo, characterizing human Grx2 as an iron–sulfur center-containing member of the thioredoxin family of proteins.
Experimental Procedures
Materials and General Methods. Disposable cell culture materials were purchased from Techno Plastic Products (Trasadingen, Switzerland). Chemicals were purchased from Sigma, unless otherwise stated, and were of analytical grade or better.
Cell Cultures. HeLa cells were cultivated in Dulbecco's modified Eagle's medium (Invitrogen) supplemented with 10% heat-inactivated (30 min, 56°C) FBS (PAA, Pasching, Austria), 2 mM glutamine (PAA), and 100 units·ml-1 penicillin/streptomycin (PAA). The Burkitt's lymphoma cell line BL30 was cultivated by using RPMI medium 1640 (Invitrogen) supplemented in the same way. The cells were incubated at 37°C in a 90% humidified atmosphere containing 5% CO2.
Radiolabeling of cells with 55Fe (PerkinElmer) was done by mixing the desired amount of 55FeCl3 (in HCl) with 5 molar equivalents of citrate followed by titration to pH 8 with 1.5 M NaOH. The final concentration of iron in the medium was adjusted to 10 μM (including the serum-derived iron); specific activity was adjusted to 200 Ci·mol-1 iron (1 Ci = 37 GBq).
Protein Expression and Purification. Grx2 was expressed and purified essentially as described in Lundberg et al. (17). Briefly, 20 liters of LB medium in a fermenter (New Brunswick) was inoculated with E. coli BL21 (DE3) pRIL harboring plasmid pET15bGrx2Δ41–164. After induction with 0.5 mM isopropyl β-d-thiogalactoside at the OD600 of 0.8, cells were propagated at 20°C for 16 h, yielding ≈80 g of bacteria. Seven grams at a time were resuspended in 25 ml of 50 mM sodium phosphate (pH 8) containing 300 mM NaCl and 2 mM GSH and lysed with 0.5 mg·ml-1 lysozyme, followed by sonication. Grx2 was purified from the clarified extract by using nickel nitrilotriacetic acid Superflow beads (Qiagen) equilibrated with 50 mM sodium phosphate, pH 8/300 mM NaCl/2 mM GSH. Grx2 was eluted with 250 mM imidazole. When indicated, buffers were exchanged by using prepacked Sephadex G-25 columns (NAP-5 or PD10, Amersham Pharmacia).
The mutants Cys-28–Ser and Cys-113–Ser were produced by using the QuikChange site-directed mutagenesis kit as described by the manufacturer (Stratagene) using two sets of complementary oligonucleotides harboring the respective mutation (C28S, 5′CAATTTCTGATAATTCTGTGGTGATTTTC-3′; C113S, 5′GCTCCCACTAGTTCATCAGTCTTATTTA-3′) and the pET15b expression plasmid of wild-type Grx2 as template. The mutations were verified by sequencing, and the mutant proteins were expressed and purified as described for the Cys-40–Ser mutant (19).
Mono- and dimeric Grx2 were separated by gel filtration using Sephadex G-50 or Superdex 75 beads (Amersham Pharmacia) equilibrated with 50 mM sodium phosphate buffer, pH 8/300 mM NaCl.
Molecular Mass Determination. The molecular masses of dimeric and monomeric Grx2 were determined by high-performance liquid chromatography (Amersham Pharmacia Smart system) using a Superdex 75 3.2/30 precision column equilibrated with 50 mM sodium phosphate, pH 8/300 mM NaCl. Calibration was done by using the Low Molecular Weight Gel Filtration Calibration kit (Amersham Pharmacia).
Determination of Iron and Sulfide Content. Protein bound iron was quantified as described by Fish (24), and acid-labile sulfide was quantified according to Broderick et al. (25).
Enzymatic Activity. The enzymatic activity of holo and apo Grx2 was determined by using the hydroxyethyl disulfide assay (26) and RNase-SG as substrate as described by Johansson et al. (19).
Western Blotting. SDS/PAGE was run by using Ready-Gels and the Protean 3 Cell (Bio-Rad) according to the manufacturer's instructions. Proteins from SDS gels (8–16% gradients) were transferred to poly(vinylidene difluoride) membranes (Millipore) by wet transfer in a buffer containing 25 mM Tris, 192 mM glycine, 10% methanol, and 0.1% SDS for 2 h, 100 V constant. The membrane was blocked in PBST (137 mM NaCl/3 mM KCl/6.5 mM Na2HPO4/1.5 mM KH2PO4/1% Tween 20) containing 5% nonfat dry milk powder (Semper, Stockholm). Rabbit anti-Grx2 and goat anti-Grx1 antibodies were used at a dilution of 1:2,000. Peroxidase-coupled anti-rabbit and anti-goat antibodies (Dako) were used to identify antigen antibody conjugates using the Western Lightning Chemiluminescence Reagent Plus (PerkinElmer).
Immunoprecipitation. Protein extracts (0.8–2 mg) were adjusted to a volume of 600 μl adding PBS. Preclearing was performed by incubation with 30 μl of protein G Sepharose (Amersham Pharmacia) for 1 h at 4°C. The Sepharose was removed by centrifugation and used as control for unspecifically bound protein. Next, the extract was incubated with 1 μg of affinity-purified anti-Grx1 or anti-Grx2 antibodies (27). After 2 h at 4°C, 30 μl of protein G Sepharose was added and complex formation was allowed at 4°C overnight. The complex was harvested by centrifugation for 5 min at 4,000 rpm (1,500 × g) and washed three times with 1 ml of cold PBS and lysis buffer.
ELISA. Quantification of Grx1 and Grx2 was done by using specific sandwich ELISAs as described by Lundberg et al. (27).
Spectroscopy. UV-visible spectra and kinetic data were recorded with a Shimadzu UV-2100 spectrophotometer.
Mössbauer data were recorded with a spectrometer of the alternating constant acceleration type. The minimum experimental line width was 0.24 mm·s-1 (full width at half height). The sample temperature was maintained constant either in an Oxford Instruments Variox or an Oxford Instruments Mössbauer Spectromag cryostat. Isomer shifts are quoted relative to iron metal at 300 K. 57Fe-labeled Grx2 was expressed and purified as described above by using 57Fe-enriched Vogel–Bonner medium (28).
X-band EPR spectra were recorded with a Bruker Elexsys E500 spectrometer equipped with a helium flow cryostat (Oxford Instruments ESR 910), an NMR gaussmeter, and a Hewlett-Packard frequency counter.
CD spectra were recorded by using an Aviv 202SF spectropolarimeter with a 1-mm path-length sealed cuvette at 25°C and a scan rate of 1 nm·min-1. Reference spectra were subtracted from the average of four spectra. Ellipticity of the chromophore was analyzed from 260 to 650 nm by using 200 μM Grx2 in 50 mM sodium phosphate buffer, pH 8/300 mM NaCl. The secondary structural content of apo and holo Grx2 was analyzed over a range of 190–260 nm by using protein at 7.4–15 μM in a buffer containing 5 mM potassium phosphate, pH 8, and 100 mM KCl. Oxidants or reductants were removed by gel filtration on Sephadex G-25 immediately before recording the spectra.
Results
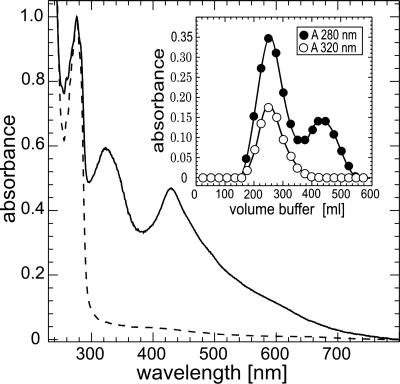
A Chromophore in Human Grx2. Human Grx2 was expressed and purified by using a 25-liter fermenter, yielding ≈4 g of pure protein. Surprisingly, concentrated Grx2 (≈4 mM) was not clear but appeared brownish. This coloration did not completely disappear after 2 days of extensive dialysis. When purified Grx2 (calculated molecular mass, 16.3 kDa) was applied to gel filtration chromatography, it separated into two fractions (Fig. 1 Inset). A larger fraction with an apparent molecular mass of 34.3 ± 0.1 kDa contained the brownish chromophore and a smaller, colorless fraction of 16.9 ± 0.5 kDa. In reducing SDS gels, both fractions yielded bands of ≈16 kDa that were identified as Grx2 by Western blotting (data not shown). The spectral properties of both forms are shown in Fig. 1. Although the monomeric fraction contained no additional features beside of its absorption peak at ≈280 nm, the dimeric form displayed two additional strong bands at ≈320 and 420 nm. The ratio between the absorbance peaks at 280 and 320 nm was 1.67. Using the fourth derivative of the spectrum, we identified absorption peaks at 320, 346, 429, 461, 498, and 622 nm.
Fig. 1.
UV-visible spectra of monomeric apo (dashed line) and dimeric holo (straight line) Grx2. Optical spectra of human apo and holo Grx2 after separation by gel filtration. The spectra were normalized to the respective absorbance at 278 nm. Inset: Elution profile of Grx2 separated by Sephadex G-50. Protein elution was recorded at 280 nm (straight line) and elution of the chromophore at 320 nm (dotted line).
Both fractions of Grx2 were assayed for Grx activity by using 2-mercaptoethanol glutathione disulfide (hydroxyethyl disulfide assay) and glutathionylated RNase as substrates. Whereas the freshly prepared brown dimeric fraction was completely inactive in both assays, the colorless monomeric fraction exhibited essentially the same catalytic rates as published for Grx2 before (Table 1 and ref. 19).
Table 1. Enzymatic activity of apo and holo Grx2.
| Substrate, kcat, s-1
|
||
|---|---|---|
| Enzyme | HED assay | RNase-SG |
| Apo Grx2 | 0.52 ± 0.05 | 0.67 ± 0.07 |
| Holo [2Fe-2S] Grx2 | No activity* | No activity* |
| Holo Grx2 (dithionite-treated) | 0.48 ± 0.04 | 0.59 ± 0.08 |
The reduction of 2-mercaptoethanol GSSG (HED assay) and glutathionylated RNase (RNase-SG) were assayed directly after purification of the proteins by gel filtration at pH 7 in a mixture containing GSH reductase, 200 μM NADPH, 1 mM GSH, and either 0.7 mM HED or 1.5 μM RNase-SG (equivalent to 12 μM GSH moieties). The specific activities were calculated from four experiments using two different enzyme concentrations.
Up to 5 μM Grx2 were assayed
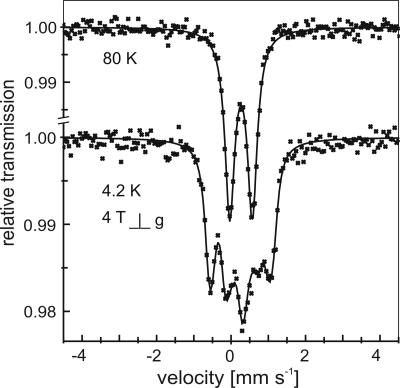
Identification of the Chromophore. The spectral properties of dimeric Grx2 indicated the presence of iron–sulfur complexes in the protein. In fact, we identified 1.4–1.9 iron molecules per Grx2 dimer in freshly prepared fractions and 1.29 ± 0.11 acid labile sulfides per iron by using colorimetric assays. Sulfide was determined in the absence of reductants, thus excluding persulfides as source of the sulfide. Other metals like zinc did not promote dimerization. Most iron–sulfur clusters can exist both in a paramagnetic and a diamagnetic state; however, we did not record any significant iron-specific signal for Grx2 with EPR spectroscopy at 10–60 K, even when the samples were treated with reductants like dithionite or oxidants like ferricyanide. The presence and nature of an EPR-silent iron–sulfur cluster could be well identified by Mössbauer spectroscopy using 57Fe-enriched samples. Therefore, we have expressed the protein in E. coli using 57Fe-enriched mineral medium. The protein was purified as before, and spectra were recorded at 80 K and 4.2 K with a magnetic field of B = 4 T of samples containing 3.4 mM Grx2 (Fig. 2). The zero-field spectrum shows a single symmetric quadrupole splitting with an isomer shift of δ = 0.26 mm·s-1, a quadrupole splitting of ΔEQ = 0.61 mm·s-1, and a Lorentzian line shape of Γfwhm = 0.33 mm·s-1. These Mössbauer parameters are characteristic of FeIII in tetrahedral sulfur coordination, as it is known to occur in oxidized mononuclear [FeIII-(S-Cys)4], tri-nuclear [3Fe-4S]+, and dinuclear [2Fe-2S]2+ clusters. The applied field spectrum shows a simulation for S = 0 with parameters δ = 0.27 mm·s-1, ΔEQ = +0.61 mm·s-1, and asymmetry parameter η = 0.6. The applied-field spectrum proves the total spin state S = 0, seen from the absence of internal fields. Among the known [Fe-S] clusters, only the [2Fe-2S]2+ cluster provides these features. Therefore, we can safely conclude that the chromophores of Grx2 are [2Fe-2S]2+ clusters that are coordinated in the protein by four Cys residues. The presence of other ligands than cysteines is inferred from the typical low Mössbauer isomer shift. A different ligand with a “hard” nitrogen or oxygen donor function would induce a significant higher isomer shift.
Fig. 2.
Identification of a [2Fe-2S] cluster in Grx2 by Mössbauer spectroscopy. Mössbauer spectra of human Grx2 (3.4 mM) recorded at 80 and at 4.2 K with B = 4 T applied perpendicular to the γ-beam, respectively. The solid line in the zero field spectrum is a fit with a symmetric Lorentzian doublet with isomer shift δ = 0.27 mm·s-1 and quadrupole splitting ΔEQ = 0.60 mm·s-1, whereas in the applied field spectrum, a simulation is shown for S = 0 with parameters δ = 0.27 mm·s-1, ΔEQ = +0.61 mm·s-1, asymmetry parameter η = 0.6. These parameters indicate the presence of iron in tetrahedral sulfur coordination with pure valence state (III) and total spin state S = 0 characteristic of a [2Fe-2S]2+ cluster.
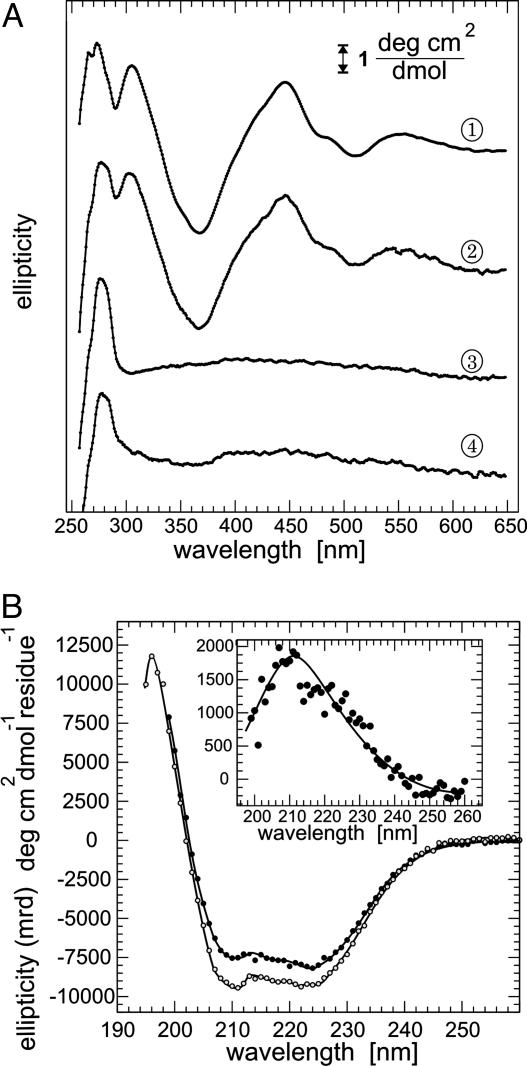
When analyzed by CD spectroscopy, monomeric apo-Grx2 does not contain features above a wavelength of about 290 nm (Fig. 3A, trace 4). The dimer, on the other hand, shows a spectrum rich in features (Fig. 3A, trace 1) with positive ellipticity bands at 305, 445, and 550 nm and a negative band at ≈370 nm. This spectrum does not resemble the features seen in ferredoxin-type [2Fe-2S] proteins. Instead, it shares many features with the spectra obtained from the iron–sulfur cluster synthesis scaffolds Isu/IscU, e.g., human [2Fe-2S] Isu (29).
Fig. 3.
Analysis of the chromophore and secondary structural content of Grx2 by CD spectroscopy. (A) Ellipticity of the Grx2 chromophore in 50 mM sodium phosphate, pH 8/300 mM NaCl. Traces 1, holo Grx2; 2, holo Grx2 treated with 10 mM H2O2; 3, holo Grx2 treated with 10 mM dithionite; 4, apo Grx2. (B) Secondary structural content of apo (open circles) and holo (filled circles) Grx2 in 5 mM potassium phosphate, pH 8/100 mM KCl. (Inset) Difference in ellipticity of apo minus holo Grx2. All spectra represent the average of four spectra recorded at 25°C at a scan rate of 1 nm·min-1 and corrected for the ellipticity of the respective buffer.
The UV CD spectra of both monomeric and dimeric Grx2 are shown in Fig. 3B. Both apo- and holo-Grx2 show similar features indicating a similar secondary structure. The difference spectrum of the dimer minus the monomer exhibits a small peak around 212 nm (Fig. 3B Inset). This may be explained by a small decrease in α-helical content upon dimerization and [Fe-S] cluster binding.
Coordination of the [Fe-S] Cluster. Mössbauer spectroscopy indicated that the iron–sulfur cluster in Grx2 was bound by four Cys residues. Human Grx2 contains four cysteines, from which two form the Cys-Ser-Tyr-Cys active site. The two additional Cys side chains are not conserved in other Grxs. Which one of these coordinate the cluster? The fact that monomeric Grx2 was free of any chromophore strongly suggested that the [2Fe-2S] cluster served to dimerize Grx2, which is in agreement with the determined stoichiometry of two iron and two sulfide molecules per dimer. Molecular modeling indicated that the extra cysteine pair and the active site cysteines are located on opposite sites of the protein. To analyze which cysteine pairs coordinate the cluster, we have prepared mutants of both the active site and the extra cysteines. Grx2 Cys-40–Ser, with the active site changed to Cys-Ser-Tyr-Ser, displayed the same spectral properties as the wild-type protein, whereas both the extra cysteine mutants Cys-28–Ser and Cys-113–Ser lost the absorption bands at 320 and 420 nm (data not shown). Therefore, we propose that the two cysteines outside the active site, Cys-28 and Cys-113, coordinate the cluster.
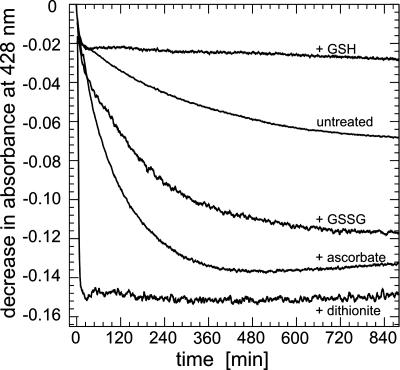
Stability of Holo Grx2. Holo Grx2 gradually lost its chromophore under aerobic conditions. This loss was prevented by the presence of reduced glutathione. Ascorbate or oxidized glutathione, on the other hand, reduced the half life of the chromophore (Fig. 4). Reduction of holo Grx2 with dithionite or oxidation with ferricyanide led to a rapid decrease in absorption between 300 and 600 nm. The products were enzymatically active (Table 1) and spectroscopically identical (e.g., Fig. 3A, trace 3) to the purified monomer indicating the loss of the [2Fe-2S] cluster. In contrast, treatment of holo Grx2 with 10 mM H2O2 for 10–30 min did not effect the [2Fe-2S] chromophore as demonstrated by CD (Fig. 3A, trace 2) and Mössbauer spectroscopy (data not shown).
Fig. 4.
Kinetics of the chromophore loss of Grx2 upon treatment with various compounds. Holo Grx2 (45 μM [Fe-S] center) in 50 mM sodium phosphate, pH 8/300 mM NaCl was incubated with 2 mM of the indicated compounds at 25°C. The loss of the chromophore was recorded after the decrease in absorbance at 428 nm.
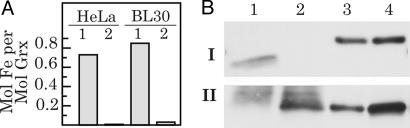
Binding of Iron to Grx2 in Vivo. Is the [Fe-S] cluster bound to Grx2 demonstrated in vitro significant in vivo? We have cultivated the Burkitt's lymphoma cell line BL30 and HeLa cells in the presence of the γ-emitting 55Fe isotope. From these cells, mitochondrial Grx2 and (as control) cytosolic Grx1 were isolated by immunoprecipitation in the presence of reduced glutathione. As depicted in Fig. 5, significantly more 55Fe coprecipitated with Grx2 compared to Grx1. Taking into account the respective protein concentrations determined by ELISA, ≈0.85 molecules Fe per molecule Grx2 coprecipitated with Grx2 from BL30 cells, whereas only 0.03 molecules Fe per molecule of Grx1 were detected. From HeLa cells, ≈0.73 Fe molecules per molecule Grx2 and 0.007 molecules of Fe per Grx1 molecule were isolated. This stoichiometric coimmunoprecipitation of iron with Grx2, but not Grx1, strongly indicated the binding of iron to Grx2 in vivo.
Fig. 5.
Coimmunoprecipitation of iron and Grx2 from two human cell lines. Grx2 was isolated by immunoprecipitation from BL30 and HeLa cells propagated in the presence of the γ-emmitter 55Fe. (A) Iron in immunoprecipitates from BL30 and HeLa was quantified from the specific radioactivity, and protein was determined by ELISA. Lanes: 1, immunoprecipitate of mitochondrial Grx2; 2, immunoprecipitate of cytosolic Grx1. (B) Successful precipitation was confirmed by Western blotting. Lanes: 1, Grx1; 2, Grx2; 3 and 4, pure recombinant Grx1 (7 and 10 ng, I) or Grx2 (5 and 10 ng, II); I, anti-Grx1 Western blot; II, anti-Grx2 Western blot.
Discussion
Human Grx2 is a member of the thioredoxin family of proteins. Thioredoxin (Trxs) and Grxs are ubiquitous proteins that use one or two cysteines to catalyze oxidoreductions such as the reduction of protein disulfides and protein glutathione-mixed disulfides. They have a wide range of functions, which may reflect the general importance of protein thiol groups (3, 30).
Here, we have identified and characterized human Grx2 as an iron–sulfur protein. So far, no other member of the thioredoxin family of proteins has been reported to coordinate an iron–sulfur cluster. Despite of the [2Fe-2S] ferredoxin from Aquifex aeolicus that acquired a thioredoxin-like fold (31), only engineered E. coli Trx1 mutants coordinating [Fe-S] cluster have been described. A mononuclear iron–sulfur center has been rationally designed into Trx by the introduction of two more Cys residues in close proximity to the active site of the protein (32). The introduction of four additional Cys residues enabled Trx to assemble a cuboidal [4Fe-4S] cluster (33). Using a genetic approach, Masip et al. isolated a Cys-Ala-Cys-Cys active site mutant that changed the protein into a [2Fe-2S]-bridged dimer capable of catalyzing O2-dependent disulfide bond formation in vivo (34). Unlike the [2Fe-2S] cluster in human Grx2, all of these engineered iron–sulfur centers could be reversibly oxidized. If not the transfer of electrons, what could be the function of Grx2's iron–sulfur center?
Iron–sulfur centers are multipurpose structures found in all life forms. They can undergo redox reactions, influence protein folding, and act as catalytic centers and as sensors of iron and oxygen (35, 36). In eukaryotic cells, mitochondria are essential for the maturation of cellular iron–sulfur proteins (37). Absence of the mitochondrial monothiol Grx5 in yeast (yGrx5) led to constitutive oxidative damage, iron accumulation in the cell, and inactivation of iron–sulfur containing enzymes. Because these defects could be suppressed by overexpression of proteins involved in [Fe-S] assembly, a function of yGrx5 in iron–sulfur cluster synthesis or repair was suggested (38). Iron–sulfur cluster synthesis is thought to take place on the scaffold protein Isu (IscU or NifU in bacteria) from where the [Fe-S] units are transferred to apo [Fe-S] proteins (overview in refs. 38 and 39). Depletion of yGrx5 from yeast increased the amount of iron bound to Isu1. Therefore, yGrx5 was proposed to be required in a step following [Fe-S] cluster synthesis on Isu1 when the [Fe-S] clusters are inserted into apo proteins (40). Could Grx2 have a similar function in [Fe-S] cluster synthesis in mammals? Yeast contains two mitochondrial Grxs: the monothiol yGrx5 and the dithiol yGrx2, which cannot compensate the loss of yGrx5 (38, 41, 42). In mammals, dithiol Grx2 is the only mitochondrial Grx. Both Grx2 and yGrx5 catalyze the reduction of protein glutathione-mixed disulfides (19, 43), and this could be necessary before the assembly of an [Fe-S] cluster. However, yGrx5 does not bind an iron–sulfur cluster itself. In fact, this feature may be unique to mammalian Grx2 as the cysteines proposed to be responsible for coordination are not conserved in any other Grx. More studies are necessary to investigate whether mammalian Grx2 inherited both the monothiol and dithiol Grx functions of yeast's mitochondrial Grxs.
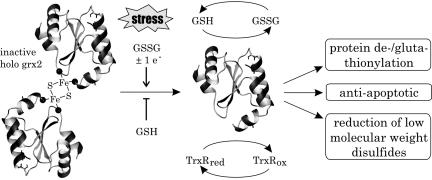
The vulnerability of [Fe-S] centers to oxidative destruction is sometimes used in sensing and regulatory functions. Irreversible cluster rearrangement or total disassembly may facilitate alterations in protein structure or oligomeric state, which may in turn alter the protein's function (44). It is tempting to speculate about a regulative role of Grx2's iron–sulfur cluster in vivo (Fig. 6). Our results indicate that, under nonstress conditions, Grx2 is primary present in its inactive holo form. Loss of the cofactor activated Grx2 in vitro and was induced by glutathione disulfide (GSSG) and one electron redox carriers like ferricyanide and dithionite, but not by hydrogen peroxide. In vivo, activation could therefore take place under conditions of oxidative stress when free radicals are formed and the ratio of GSSG to GSH increases. Unlike its cytosolic counterpart Grx1, apo Grx2 is not inactivated by oxidation (17) and could rely on electrons donated by thioredoxin reductase when the intracellular glutathione pool becomes oxidized or the pH falls (19). Consistent with this model is the fact that silencing or overexpression of Grx2 did not effect the cellular redox state or cell growth per se. Only when oxidative stress was induced, e.g., by doxorubicin, lack of Grx2 rendered the cells hypersensitive (22) and overexpression attenuated apoptosis by preventing cardiolipin oxidation, cytochrome c release, and caspase activation (23). Under conditions of severe oxidative stress, activation of Grx2 could facilitate the regulation of specific protein targets necessary for cell survival, for instance, the regulation of superoxide formation by complex I through glutathionylation/deglutathionylation (20, 21).
Fig. 6.
Model of Grx2 activation in vivo. A high GSH/GSSG ratio stabilizes the holo form of Grx2, whereas the [2Fe-2S] cluster is destroyed by oxidative stress leading to activation of Grx2. Active Grx2 can use electrons from both GSH and Trx reductase for the reduction of low molecular mass as well as protein glutathione-mixed disulfides (19) and protects from apoptosis by preventing cytochrome c release (23).
In conclusion, we have demonstrated that human Grx2 is able to form a [2Fe-2S]2+ cluster-bridged dimer. This dimer is inactive as disulfide reductase, but Grx2 is activated upon loss of the cofactor and monomerization. We propose that the iron–sulfur cluster serves as redox sensor for the activation of Grx2 under conditions of oxidative stress at the crossroad between cell death and survival.
Acknowledgments
We thank Nina Voevodskaya (Stockholm University) for help with EPR spectrometry and critical discussions, Malin Fladvad (Karolinska Institute) for expert help with CD spectrometry, Catrine Johansson (Károlinska Institute) for the oligonucleotides used for mutagenesis, Bernd Mienert for technical assistance, and Lena Ringdén for excellent secretarial work. This investigation was supported by grants from the Karolinska Intitute, the Swedish Cancer Society Grants 961 and 4648-BO2-01VA, and the Wenner-Gren Foundation. C.B. is supported by Fellowship BE3259/1-1 from the Deutsche Forschungsgemeinschaft.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Grx, glutaredoxin; GSH, glutathione; GSSG, glutathione disulfide; Trx, thioredoxin.
References
- 1.Holmgren, A. (1976) Proc. Natl. Acad. Sci. USA 73, 2275-2279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holmgren, A. (1979) J. Biol. Chem. 254, 3672-3678. [PubMed] [Google Scholar]
- 3.Holmgren, A. (1989) J. Biol. Chem. 264, 13963-13966. [PubMed] [Google Scholar]
- 4.Bushweller, J. H., Åslund, F., Wüthrich, K. & Holmgren, A. (1992) Biochemistry 31, 9288-9293. [DOI] [PubMed] [Google Scholar]
- 5.Lillig, C. H., Prior, A., Schwenn, J. D., Åslund, F., Ritz, D., Vlamis-Gardikas, A. & Holmgren, A. (1999) J. Biol. Chem. 274, 7695-7698. [DOI] [PubMed] [Google Scholar]
- 6.Holmgren, A. & Åslund, F. (1995) Methods Enzymol. 252, 283-292. [DOI] [PubMed] [Google Scholar]
- 7.Vlamis-Gardikas, A. & Holmgren, A. (2002) Methods Enzymol. 347, 286-296. [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Manzaneque, M. T., Ros, J., Cabiscol, E., Sorribas, A. & Herrero, E. (1999) Mol. Cell. Biol. 19, 8180-8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vlamis-Gardikas, A., Åslund, F., Spyrou, G., Bergman, T. & Holmgren, A. (1997) J. Biol. Chem. 272, 11236-11242. [DOI] [PubMed] [Google Scholar]
- 10.Wells, W. W., Xu, D. P., Yang, Y. F., Rocque, P. A. (1990) J. Biol. Chem. 265, 15361-15364. [PubMed] [Google Scholar]
- 11.Takashima, Y., Hirota, K., Nakamura, H., Nakamura, T., Akiyama, K., Cheng, F. S., Maeda, M. & Yodoi, J. (1999) Immunol. Lett. 68, 397-401. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura, T., Ohno, T., Hirota, K., Nishiyama, A., Nakamura, H., Wada, H. & Yodoi, J. (1999) Free Radical Res. 31, 357-365. [DOI] [PubMed] [Google Scholar]
- 13.Bandyopadhyay, S., Starke, D. W., Mieyal, J. J. & Gronostajski, R. M. (1998) J. Biol. Chem. 273, 392-397. [DOI] [PubMed] [Google Scholar]
- 14.Hirota, K., Matsui, M., Murata, M., Takashima, Y., Cheng, F. S., Itoh, T., Fukuda, K. & Yodoi, J. (2000) Biochem. Biophys. Res. Commun. 274, 177-182. [DOI] [PubMed] [Google Scholar]
- 15.Chrestensen, C. A., Starke, D. W. & Mieyal, J. J. (2000) J. Biol. Chem. 275, 26556-26565. [DOI] [PubMed] [Google Scholar]
- 16.Daily, D., Vlamis-Gardikas, A., Offen, D., Mittelmann, L., Melamed, E., Holmgren, A. & Barzilai, A. (2001) J. Biol. Chem. 276, 1335-1344. [DOI] [PubMed] [Google Scholar]
- 17.Lundberg, M., Johansson, C., Chandra, J., Enoksson, M., Jacobsson, G., Ljung, J., Johansson, M. & Holmgren, A. (2001) J. Biol. Chem. 276, 26269-26275. [DOI] [PubMed] [Google Scholar]
- 18.Gladyshev, V. N., Liu, A., Novoselov, S. V., Krysan, K., Sun, Q. A., Kryukov, V. M., Kryukov, G. V. & Lou, M. F. (2001) J. Biol. Chem. 276, 30374-30380. [DOI] [PubMed] [Google Scholar]
- 19.Johansson, C., Lillig, C. H. & Holmgren, A. (2004) J. Biol. Chem. 279, 7537-7543. [DOI] [PubMed] [Google Scholar]
- 20.Beer, S. M., Taylor, E. R., Brown, S. E., Dahm, C. C., Costa, N. J., Runswick, M. J. & Murphy, M. P. (2004) J. Biol. Chem. 279, 47939-47951. [DOI] [PubMed] [Google Scholar]
- 21.Taylor, E. R., Hurrell, F., Shannon, R. J., Lin, T. K., Hirst, J. & Murphy, M. P. (2003) J. Biol. Chem. 278, 19603-19611. [DOI] [PubMed] [Google Scholar]
- 22.Lillig, C. H., Lönn, M. E., Enoksson, M., Fernandes, A. P. & Holmgren, A. (2004) Proc. Natl. Acad. Sci. USA 101, 13227-13232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Enoksson, M., Fernandes, A. P., Prast, S., Lillig, C. H., Holmgren, A. & Orrenius, S. (2005) Biochem. Biophys. Res. Commun. 327, 774-779. [DOI] [PubMed] [Google Scholar]
- 24.Fish, W. W. (1988) Methods Enzymol. 158, 357-364. [DOI] [PubMed] [Google Scholar]
- 25.Broderick, J. B., Henshaw, T. F., Cheek, J., Wojtuszewski, K., Smith, S. R., Trojan, M. R., McGhan, R. M., Kopf, M., Kibbey, M. & Broderick, W. E. (2000) Biochem. Biophys. Res. Comm. 269, 451-456. [DOI] [PubMed] [Google Scholar]
- 26.Luthman, M. & Holmgren, A. (1982) J. Biol. Chem. 257, 6686-6689. [PubMed] [Google Scholar]
- 27.Lundberg, M., Fernandes, A. P., Kumar, S. & Holmgren, A. (2004) Biochem. Biophys. Res. Commun. 319, 801-809. [DOI] [PubMed] [Google Scholar]
- 28.Vogel, H. J. & Bonner, D. M. (1956) J. Biol. Chem. 218, 97-106. [PubMed] [Google Scholar]
- 29.Yoon, T. & Cowan, J. A. (2003) J. Am. Chem. Soc. 125, 6078-6084. [DOI] [PubMed] [Google Scholar]
- 30.Holmgren, A. (2000) Antioxid. Redox. Signal 2, 811-820. [DOI] [PubMed] [Google Scholar]
- 31.Yeh, A. P., Chatelet, C., Soltis, S. M., Kuhn, P., Meyer, J. & Rees, D. C. (2000) J. Mol. Biol. 300, 587-595. [DOI] [PubMed] [Google Scholar]
- 32.Benson, D. E., Wisz, M. S., Liu, W. & Hellinga, H. W. (1998) Biochemistry 37, 7070-7076. [DOI] [PubMed] [Google Scholar]
- 33.Coldren, C. D., Hellinga, H. W. & Caradonna, J. P. (1997) Proc. Natl. Acad. Sci. USA 94, 6635-6640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Masip, L., Pan, J. L., Halder, S., Penner-Hahn, J. E., DeLisa, M. P., Georgiou, G., Bardwell, J. C. A. & Collet, J. F. (2004) Science 303, 1185-1189. [DOI] [PubMed] [Google Scholar]
- 35.Beinert, H., Holm, R. H. & Münck, E. (1997) Science 277, 653-659. [DOI] [PubMed] [Google Scholar]
- 36.Beinert, H. (2000) J. Biol. Inorg. Chem. 5, 2-15. [DOI] [PubMed] [Google Scholar]
- 37.Lill, R. & Kispal, G. (2000) Trends Biochem. Sci. 25, 352-356. [DOI] [PubMed] [Google Scholar]
- 38.Rodriguez-Manzaneque, M. T., Tamarit, J., Belli, G., Ros, J. & Herrero, E. (2002) Mol. Biol. Cell 13, 1109-1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mühlenhoff, U. & Lill, R. (2000) Biochim. Biophys. Acta 1459, 370-382. [DOI] [PubMed] [Google Scholar]
- 40.Mühlenhoff, U., Gerber, J., Richhardt, N. & Lill, R. (2003) EMBO J. 22, 4815-4825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pedrajas, J. R., Porras, P., Martinez-Galisteo, E., Padilla, C. S., Miranda-Vizuete, A. & Barcena, J. A. (2002) Biochem. J. 364, 617-623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Molina, M. M., Belli, G., de la Torre, M. A., Rodriguez-Manzaneque, M. T. & Herrero, E. (2004) J. Biol. Chem. 279, 51923-51930. [DOI] [PubMed] [Google Scholar]
- 43.Tamarit, J., Cabiscol, E., Herrero, E. & Ros, J. (2003) J. Biol. Chem. 278, 25745-25751. [DOI] [PubMed] [Google Scholar]
- 44.Beinert, H. & Kiley, P. J. (1999) Curr. Opin. Chem. Biol. 3, 152-157. [DOI] [PubMed] [Google Scholar]