Abstract
Signal transducer and activator of transcription (STAT)3 is a member of a family of DNA-binding factors that function to induce expression of responsive genes. STAT3 can act as an oncogene, and its function has been shown to be critical for cellular transformation by a number of oncogenic tyrosine kinases. The role of STAT3 as a DNA-binding transcription factor naturally depends on its ability to gain entrance to the nucleus. In this study, we provide evidence that STAT3 is distinct from previously characterized STAT molecules in that it dynamically shuttles between cytoplasmic and nuclear compartments and maintains prominent nuclear presence. Although tyrosine phosphorylation is required for STAT3 to bind to specific DNA target sites, nuclear import takes place constitutively and independently of tyrosine phosphorylation. We identify a region within the coiled-coil domain of the STAT3 molecule that is necessary for nuclear import and demonstrate that this region is critical for its recognition by specific import carrier importin-α3. RNA interference studies were used to verify the role and specificity of importin-α3 in STAT3 nuclear translocation. These results distinguish STAT3 cellular localization from other STAT molecules and identify a feature that may be targeted in the clinical intervention of STAT3-dependent neoplasia.
Keywords: nuclear translocation, signal transduction, transcription factor
Proper cellular localization is critical for the effective function of most signaling molecules, and nuclear translocation is central to the function of transcription factors. Signal transducers and activators of transcription (STATs) comprise a family of transcription factors that play critical roles in immunity, development, and proliferation (1–4). STATs can be activated by tyrosine phosphorylation after cytokine binding or growth factor binding to cell surface receptors or by intracellular oncogenic tyrosine kinases. The mammalian STAT family includes seven members that share a similar structural arrangement of functional domains, including an amino-terminal coiled-coil domain, a central DNA-binding domain, a Src homology 2 (SH2) domain, and a carboxyl transactivation domain. Phosphorylation of a conserved tyrosine downstream of the SH2 domain leads to dimerization of two STAT molecules by reciprocal SH2–phosphotyrosine interactions. This dimerization alters the conformation of the STAT proteins to allow them to recognize target DNA and induce gene expression.
Because protein synthesis occurs in the cytoplasm, transcription factors must transit nuclear pore complexes (NPC) in the nuclear membrane to enter the nucleus (5, 6). Proteins larger than ≈50 kDa require specific signaling motifs to move into or out of the nucleus that are recognized by soluble carriers known as karyopherins or importins/exportins (7, 8). Most nuclear localization signals (NLSs) identified thus far are recognized directly by a family of mammalian importin-α carriers (9). The importin-α proteins associate with importin-β1 which mediates transit of the complex through the NPC. Export of proteins from the nucleus to the cytoplasm is similarly regulated by signal sequences known as nuclear export signals (NESs). NESs that are rich in leucine or hydrophobic residues are recognized by a common exportin carrier known as CRM1 that mediates exit through the NPC (10).
Our understanding of STAT nuclear trafficking is mostly derived from studies of STAT1 activated in response to interferons (11–14). The nuclear accumulation of STAT1 depends on tyrosine phosphorylation and subsequent dimer conformation, which is recognized by a specific import carrier, importin-α5. Much attention has focused on another STAT family member, STAT3, because accumulating evidence demonstrates its role as a potential oncogene (15–17). A gain-of-function mutation in STAT3 transforms cells (18), constitutive STAT3 activation is associated with several human tumors, and STAT3 function is essential for transformation by several oncogenic tyrosine kinases (17, 19). Furthermore, it was recently reported that unphosphorylated STAT3 may play a role in oncogenesis (20). Consequently, understanding nuclear trafficking of STAT3 may provide a means by which to intervene its transforming activity in cancer development. In this report, we provide evidence that, in contrast to STAT1, STAT3 has prominent nuclear presence before and after tyrosine phosphorylation. In addition, we identify a distinct region of STAT3 critical for its nuclear import and necessary for its recognition by specific importin-α carriers.
Materials and Methods
Cell Culture and Reagents. HeLa, COS1, NIH-3T3, Hep3B, and HUVEC cells were cultured according to American Type Culture Collection guidelines or as described in ref. 21. Recombinant murine or human IL-6 (BioSource International, Camarillo, CA), IL-6 soluble receptor (R & D Systems) and human EGF (Sigma) were used at 10 ng/ml. Leptomycin B (LMB) was a gift from Barbara Wolff-Winiski (Novartis Research Institute, Vienna, Austria) and was used at 10 nM. DNA transfections were performed with FuGENE 6 (Roche). Anti-GFP (Roche), anti-phosphotyrosine STAT3 (B7, Santa Cruz Biotechnology), anti-STAT3 (F2, H190, K15 and C20; Santa Cruz Biotechnology and Cell Signaling Technology, Beverly, MA) were used at a 1:1,000 dilution for Western Blots and 1:200 for immunofluorescence.
Plasmid Construction and Recombinant Protein Purification. cDNA encoding murine STAT3 and mutants were generated by PCR and cloned into the pEGFP-N1 vector to generate GFP fusions. STAT3 was cloned into vector pEF1/V5-His to generate V5 epitope-tagged fusion and pFastBacHTb (Invitrogen) for baculoviral expression in Sf9 insect cells. Oligonucleotides encoding simian virus 40 NLS and STAT1 NES were cloned into the STAT3–GFP plasmid to generate STAT3–NLS–GFP and STAT3–NES–GFP fusions (14). Plasmids encoding importins are described in ref. 22 and were a kind gift from Matthias Köhler (The Franz–Volhard Clinic, Berlin). Importin-β1 binding (IBB) domain-deleted importin-αs and fragments of importin-α3 were generated by PCR and cloned into vector pGEX-KG for bacterial expression. Recombinant proteins tagged with GST were purified by standard binding to glutathione agarose beads (Sigma).
Fluorescence Microscopy. Cells were seeded on glass coverslips for 24 h and serum-starved for 18–24 h before treatment and fixation in 4% paraformaldehyde for 15 min at 4°C. Cells were permeabilized in 0.2% Triton X-100 for 5 min and blocked with 10% goat serum. STAT3 was detected with anti-STAT3 (Cell Signaling Technology) or anti-V5 (Invitrogen) antibody and rhodamine-conjugated secondary antibody. Images were captured with a Zeiss Axioskop fluorescence microscope and presented with photoshop (Adobe Systems, San Jose, CA).
Heterokaryon Assay. HeLa cells were seeded on coverslips and transfected with the STAT3–NLS–GFP construct. Murine NIH-3T3 cells were plated onto the HeLa cultures for 3 h in the presence of 50 μg/ml cycloheximide, washed with saline, and fused with Sigma HybriMax for 2 min. Fused cultures were placed in medium with 100 μg/ml cycloheximide for 2 h and processed for fluorescence microscopy. Nuclei of the two cell types were distinguished by staining with 20 ng/ml Hoechst 33258 (Sigma).
Importin-α Protein Binding Assay. Purified recombinant GST-importin-α(ΔIBB) protein or GST protein (25 μg each) was used in each binding reaction. Mammalian cell lysates containing STAT3 were prepared by combining cytosol from a hypotonic lysis with a high-salt extraction of nuclei (23). Cellular protein (500 μg) was incubated with importin-α proteins immobilized on GST beads, washed, and eluted. Binding reactions with STAT3 expressed in insect cells was performed similarly.
RNA Interference (RNAi). siGENOME SMARTpool short interfering RNA (siRNA) duplexes specific for human importin-α1, -α3, and -α5 were prepared (Dharmacon Research, Layfayette, CO) and transfected with Lipofectamine 2000 (Invitrogen). Forty-eight hours after siRNA transfection, cells were transfected with STAT3-RY–GFP and processed 14–16 h later. For statistical analyses, >1,000 STAT3-RY–GFP-expressing cells were counted per sample.
Real-Time Quantitative RT-PCR. RNA extraction was performed with TRIzol (Invitrogen) and cDNA was synthesized with SuperScript II reverse transcriptase (Invitrogen). Real-time PCRs were performed by using the following primers at their optimal condition as described for the LightCycler-FastStart DNA Master SYBR Green I kit (Roche Molecular Biochemicals): importin-α1, forward 5′-CGCAGAATAGAGGTCAATGTG and reverse 5′-CGGAGAAGTAGCATCATCAGG; importin-α3, forward 5′-ATGCTTCAAGTGATAACCAGGG and reverse 5′-CAAGACAATGGACTAAAATGG; importin-α5, forward 5′-TCGCCTGAAAAGTTACAAGAA and reverse 5′-AGAAGTGATGACACCACCTGG; and β-actin, forward 5′-GAGCTACGAGCTGCCTGACG and reverse 5′-GTAGTTTCGTGGATGCCACAG.
Results
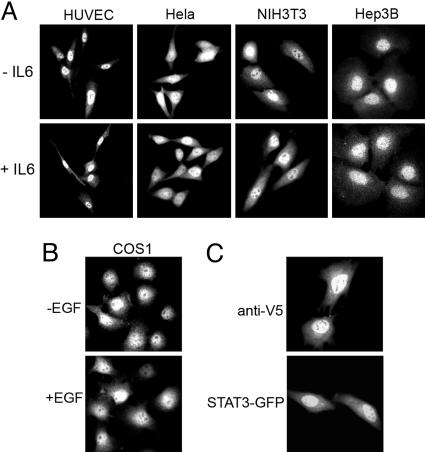
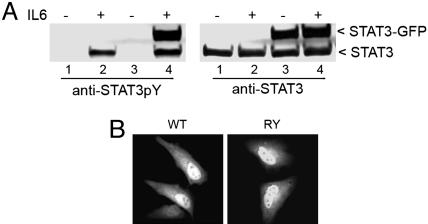
STAT3 Predominates in the Nucleus Independent of Tyrosine Phosphorylation. The cellular distribution of endogenous STAT3 was evaluated in cells of different origins either in the absence or presence of activating stimuli. Unexpectedly, unphosphorylated STAT3 was found to reside prominently in the nucleus of primary cells or immortalized cells, even after serum deprivation. The nuclear localization was similar to that after tyrosine phosphorylation of STAT3 by treatment with IL-6 (Fig. 1A) or EGF (Fig. 1B). A variety of cell types and species were tested and showed similar STAT3 nuclear presence when evaluated with various sources of antibodies that specifically detected endogenous STAT3 by immunofluorescence (Fig. 7, which is published as supporting information on the PNAS web site). To verify the constitutive nuclear presence of STAT3, we generated a V5-epitope-tagged STAT3 and a STAT3–GFP fusion for detection by immunofluorescence or direct fluorescence. These STAT3 proteins were expressed in HeLa cells and were also found to accumulate in the nucleus similar to endogenous STAT3 (Fig. 1C). To eliminate the possibility that nuclear presence of STAT3 was due to elevated tyrosine phosphorylation, the phosphorylation state of STAT3 was evaluated. Cells were untransfected or transfected with STAT3–GFP and either serum deprived or stimulated with IL-6. Western blot analysis with specific STAT3 phosphotyrosine antibodies detected phosphorylation only after IL-6, indicating that the STAT3 constitutive nuclear presence was independent of tyrosine phosphorylation (Fig. 2A). In addition to microscopy, cellular fractionation analyses were used to demonstrate the nuclear presence of endogenous unphosphorylated STAT3 (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 1.
Constitutive nuclear presence of STAT3 in various cell lines. Images represent >95% of the cell population. (A) Immunofluorescence of endogenous STAT3 in human umbilical cord vein endothelial cells (HUVEC), HeLa, NIH-3T3 and Hep3B cells that were serum-starved (-IL6) or treated with IL-6 for 30 min (+IL6). Cells were stained with polyclonal anti-STAT3 (Cell Signaling Technology) followed by rhodamine-conjugated secondary antibodies. (B) Immunofluorescence of endogenous STAT3 in COS1 cells serum-starved (-EGF) or treated with EGF for 30 min (+EGF) and stained as in A. (C) Cells expressing V5-STAT3 were stained with monoclonal anti-V5 antibody followed by rhodamine-conjugated secondary antibodies. Cells expressing STAT3–GFP were processed for direct fluorescence microscopy.
Fig. 2.
STAT3 nuclear translocation occurs independently of tyrosine phosphorylation. (A) HeLa cells (lanes 1 and 2) and HeLa cells transfected with STAT3–GFP (lanes 3 and 4) were untreated (-) or treated (+) with human IL-6 for 30 min. Whole-cell lysates were subjected to SDS/PAGE and Western blot with specific anti-STAT3 phosphotyrosine antibody (Left) or anti-STAT3 antibody (Right). (B) HeLa cells were transfected with wild-type STAT3–GFP or STAT3-RY–GFP, and localization of the proteins was examined by fluorescence microscopy. More than 95% of the transfected cells exhibited localization of the STAT3–GFP fusion as shown in the images.
To confirm the premise that tyrosine phosphorylation is not required for STAT3 nuclear translocation and to eliminate possible dimerization with endogenous STATs, we generated a STAT3 double mutation to eliminate reciprocal SH2–phosphotyrosine interactions. The tyrosine that is a target of phosphorylation was substituted with phenylalanine (Y705F), and the critical arginine within the SH2 domain was substituted with alanine (R609A) to create STAT3-RY–GFP. This mutant protein was expressed in cells and found to localize in the nucleus to the same extent as wild-type STAT3 (Fig. 2B). Because the DNA-binding region of the STAT1 protein plays a critical role in nuclear trafficking, the localization of characterized STAT3 DNA-binding mutants was evaluated (14, 24). Unlike STAT1, the STAT3 DNA-binding mutants were found prominently in the nucleus (data not shown).
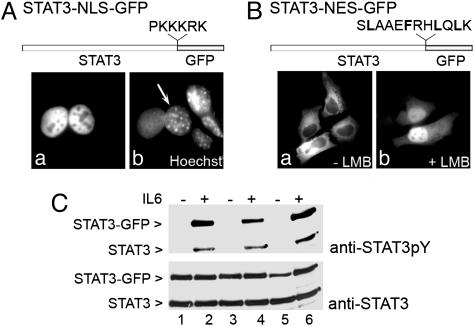
STAT3 Constitutively Shuttles Between Cytoplasm and Nucleus. Although STAT3 is prominently nuclear, it is known to be a substrate of Janus kinases resident in the cytoplasm of the cell. For this reason, it seemed likely that STAT3 actively shuttled between the cytoplasm and nucleus. To determine whether STAT3 constitutively exits the nucleus, we performed a heterokaryon assay in which human and murine cells were fused and a redistribution of STAT3–GFP from one nucleus to the other provides evidence of active export (25, 26). Murine nuclei can be distinguished from human nuclei by a punctate staining pattern with Hoechst dye. The assay was initiated with completely nuclear STAT3 by expressing STAT3 with the NLS of the simian virus 40 T antigen (Fig. 3A). This STAT3–NLS–GFP was expressed in human HeLa cells, and these cells were fused with murine NIH-3T3 cells. Within 2 h after cell fusion in the presence of cycloheximide to inhibit new protein synthesis, the STAT3–NLS–GFP was present in the murine nuclei of heterokaryons, indicating that STAT3 was actively exported from the HeLa nuclei and not anchored in the nucleus.
Fig. 3.
Nuclear–cytoplasmic shuttling of STAT3. (A) HeLa cells expressing STAT3–NLS–GFP were fused with NIH-3T3 cells. (a) Localization of the STAT3–NLS–GFP in heterokaryons was examined by fluorescent microscopy. (b) Nuclei were distinguished with Hoechst stain. NIH-3T3 nucleus within a heterokaryon containing STAT3–NLS–GFP is noted with a characteristic punctate staining (arrow). (B) HeLa cells transfected with STAT3–NES–GFP were untreated (a) or treated (b) with 10 nM LMB for 15 min. Localization of the GFP fusion was examined by fluorescent microscopy. STAT3–NES–GFP accumulated in 100% of transfected cells in response to LMB treatment. (C) HeLa cells transfected with STAT3–GFP (lanes 1 and 2), STAT3–NES–GFP (lanes 3 and 4), or STAT3–NLS–GFP (lanes 5 and 6) were untreated (-) or treated (+) with human IL-6 for 30 min. Cell lysates were subjected to SDS/PAGE and Western blot with specific anti-STAT3 phosphotyrosine (Upper) and anti-STAT3 (Lower) antibodies.
To demonstrate that STAT3 undergoes dynamic and rapid nuclear import, we evaluated the localization of a protein containing a CRM1-dependent NES inserted between STAT3 and GFP (Fig. 3B). This protein was expressed in HeLa cells and found to accumulate in the cytoplasm because of dominant nuclear export. However, when the cells were treated with the CRM1 inhibitor LMB, the STAT3–NES–GFP protein accumulated in the nucleus within minutes (Fig. 3B) (27, 28). The STAT3–NES–GFP is too large to passively diffuse through the nuclear pore complex; therefore, an intrinsic NLS in STAT3 must have mediated import of the fusion protein into the nucleus when CRM1 was inhibited.
Although STAT3–NLS–GFP was prominently nuclear and STAT3–NES–GFP was prominently cytoplasmic, these proteins could both be tyrosine phosphorylated within 30 min of IL-6 treatment (Fig. 3C). This observation provides additional evidence that STAT3 actively shuttles between cytoplasm and nucleus.
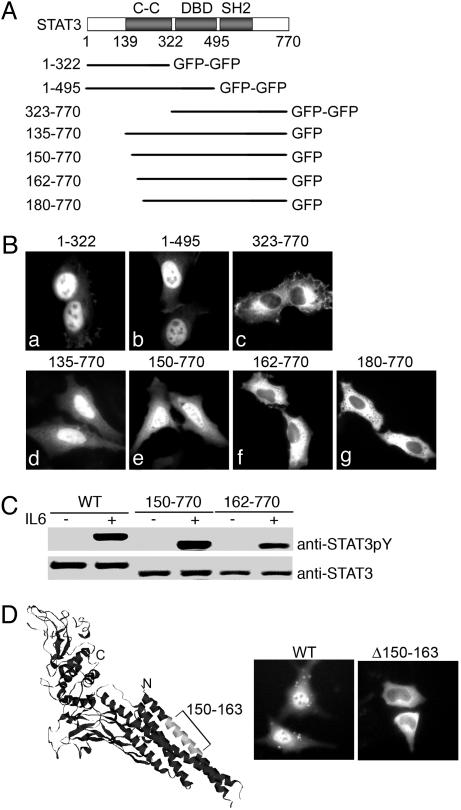
Amino Acid Sequence 150–162 Is Indispensable for STAT3 Nuclear Import. To identify a region in STAT3 critical for nuclear translocation, the localization of STAT3 truncations was evaluated. Small fragments of STAT3 were tagged with two tandem GFP molecules to ensure that they did not passively diffuse into the nucleus (Fig. 4A). STAT3 fragments encoding the amino terminus and coiled-coil domain (amino acids 1–322 and 1–495) were found to accumulate in the nucleus (Fig. 4B). However, the carboxyl portion of STAT3, including the DNA-binding domain and SH2 domain (amino acids 323–770), accumulated in the cytoplasm. A region required for nuclear import therefore resided within amino acids 1–322. To further delineate the sequence necessary for nuclear import, additional deletion mutations in this region were tested. Proteins encoding amino acids 135–770 or 150–770 accumulated in the nucleus similar to wild-type STAT3. Conversely, deletions of the coiled-coil domain encoding proteins of amino acids 162–770 or 180–770 resided in the cytoplasm. The cytoplasmic STAT3(162–770) protein maintained its integrity as a substrate for Janus kinases because it was tyrosine phosphorylated in response to IL-6 comparably to the STAT3 wild type or STAT3(150–770) protein (Fig. 4C). Therefore the lack of nuclear import was not due to an overall disruption of protein structure.
Fig. 4.
Cellular localization of STAT3 deletion mutations. (A) Linear diagram of STAT3 deletion constructs. (B) Localization of STAT3 domains fused with two tandem GFPs and STAT3 amino-terminal deletions fused with GFP. Images are representative of >90% of the cell population. (C) HeLa cells transfected with the STAT3–GFP, STAT3(150–770)–GFP, or STAT3(162–770)–GFP were untreated (-) or treated (+) with human IL-6 for 30 min. Cell lysates were subjected to SDS/PAGE and Western blot with specific anti-STAT3 phosphotyrosine (Upper) and anti-STAT3 (Lower) antibodies. (D) STAT3–GFP with an internal deletion of the amino acids 150–163 region (Δ150–163) is defective in nuclear import. (Left) Position of the amino acids 150–163 region in the crystal structure of STAT3β (Protein Data Bank ID code 1BG5) in a ribbon representation. (Center and Right) Cellular localization of the wild type (Center) and STAT3(Δ150–163)–GFP mutant (Right).
Based on this study with STAT3 deletions, the sequence between amino acids 150–162 (DVRKRQDLEQKM) appears necessary for nuclear import. In the STAT3 crystal structure, this region is located within the first helix of the coiled-coil domain (Fig. 4D). To determine the effect of a specific deletion of this sequence from otherwise full-length STAT3, we evaluated the localization of STAT3 lacking amino acids 150–162 [STAT3(Δ150–163)]. The results shown in Fig. 4D demonstrate cytoplasmic localization of STAT3(Δ150–163) and thereby a deficiency in nuclear import. This finding further indicates that the amino acid sequence 150–162 is required for STAT3 nuclear import. In addition, the STAT3(Δ150–163) protein can be tyrosine phosphorylated in response to IFN or EGF, indicating that the deletion does not disrupt STAT3 response to cytokine or growth factor signals (data not shown). To determine the contribution of the basic amino acids of this sequence to nuclear import, they were replaced in the context of full-length STAT3; however, this substitution did not reduce STAT3 nuclear accumulation (data not shown). For this reason, amino acids 150–162 may play a role in a conformational structure that is required for nuclear import.
Accumulating evidence suggests that STAT proteins can associate in an unphosphorylated state in a distinct conformation relative to their tyrosine phosphorylated state (29–31). To investigate whether the defect in nuclear import of STAT3(162–770) reflects a lack of STAT3–STAT3 association, we evaluated its ability to bind unphosphorylated STAT3. Coimmunoprecipitation analyses demonstrated its ability to bind full-length STAT3 equivalent to STAT3 or STAT3(150–770) (Fig. 9A, which is published as supporting information on the PNAS web site). In addition, a triple mutation in STAT3 of sequences homologous to those required for self-association of unphosphorylated STAT1 did not effect STAT3 nuclear import (V77, L78, F174) (31) (Fig. 9B). Together these data indicate that unphosphorylated dimer formation is not correlated with STAT3 nuclear translocation.
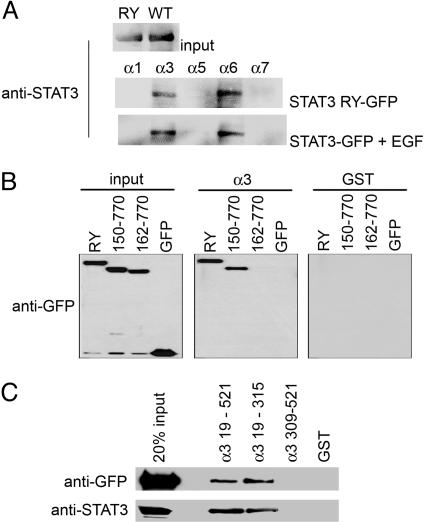
Nuclear Import of STAT3 Is Mediated by the Importin-α/β Pathway. There are six characterized mammalian importin-α proteins that bind to classical NLS motifs (7, 8, 32). To investigate whether any of these importin-α proteins recognized unphosphorylated STAT3, we performed in vitro binding assays. The importin-α proteins were produced as GST fusions. The six mammalian importin-α proteins have an IBB domain at their amino terminus, central Armadillo repeats, and a less-conserved carboxyl-terminal region (33–36). Solving the crystal structure of importin-α revealed that the IBB domain contains an NLS-like sequence that has an autoinhibitory effect by interacting with the Armadillo repeats (37). To eliminate this autoinhibition, the IBB domain was deleted from the importin-α(ΔIBB) proteins. Equal amounts of importin-α(ΔIBB) proteins were bound to glutathione agarose beads as estimated from Coomassie blue staining (data not shown). As a source of unphosphorylated STAT3 protein, mammalian cells were transfected with the double mutation STAT3-RY–GFP to ensure that STAT3 was not phosphorylated and did not form tyrosine phosphorylated dimers. As a source of tyrosine phosphorylated protein, cells were transfected with wild-type STAT3–GFP and stimulated with EGF. Cell lysates were incubated with the GST-importin beads, bound proteins were eluted, and the presence of STAT3 was detected by Western blot. Unphosphorylated STAT3 and tyrosine-phosphorylated STAT3 dimers were found to interact with the same importin-α isoforms, importin-α3 and importin-α6 (Fig. 5A). Because binding was independent of phosphorylation, this result is in accord with a constitutive NLS in STAT3.
Fig. 5.
STAT3 binds to importin-α3 and importin-α6. (A) GST–importin-α(ΔIBB) proteins bound to glutathione agarose beads were incubated with lysate from COS cells transfected with STAT3-RY–GFP or cells transfected with wild-type STAT3–GFP and treated with EGF for 30 min. Interacting proteins were analyzed by Western blot using monoclonal STAT3 antibody. (B) GST–importin-α3(ΔIBB) or GST proteins bound to glutathione agarose beads were incubated with lysates from cells expressing GFP, STAT3-RY–GFP, STAT3(150–770)–GFP or STAT3(162–770)–GFP. Interacting proteins were analyzed by Western blot using anti-GFP antibody. (C) GST–importin-α3ΔIBB (amino acids 19–521), GST–importin-α3 central Armadillo repeats (amino acids 19–315), GST–importin-α3 carboxyl terminus (amino acids 309–521), or GST proteins were immobilized on glutathione agarose beads and incubated with lysates from COS1 cells expressing STAT3-RY–GFP (Upper) or from Sf9 insect cells (Lower) expressing His-6–STAT3-RY. Bound STAT3 proteins were detected by Western blot with anti-GFP antibody (Upper) or anti-STAT3 antibody (Lower).
Importin-α3 has been described to be ubiquitously expressed; however, significant levels of importin-α6 mRNA are restricted to testes (38, 39). Importin-α3 could therefore be the primary carrier for STAT3 nuclear import in most cells. If the interaction of STAT3 with importin-α3 was critical for nuclear import, a deletion that abolished nuclear import would be expected to lack this interaction. To test this prediction we evaluated binding to the STAT3-RY mutation, the STAT3(150–770) that accumulates in the nucleus, and the STAT3(162–770) that is defective for nuclear translocation (Fig. 4). STAT3–GFP fusions expressed in cells and lysates were incubated with GST-importin-α3(ΔIBB) or GST. Bound STAT3 proteins were eluted and detected by Western blot with anti-GFP antibody. The data clearly indicated that the STAT3-RY mutation and STAT3(150–770) bound to importin-α3. However, STAT3(162–770), which is defective for nuclear import, did not bind importin-α3 (Fig. 5B). Because importin-α3 is expressed ubiquitously, it appears to be a primary mediator of STAT3 nuclear translocation by recognition of a critical element in the coiled-coil domain of STAT3. Because our results indicate that the amino acids 150–163 region of STAT3 is essential for nuclear import, we evaluated the ability of this peptide to bind importin-α3. The results indicated a weak but detectable in vitro association (Fig. 10, which is published as supporting information on the PNAS web site).
Proteins with conventional basic NLS sequences have been shown to interact with the shallow grooves formed by Armadillo repeats 2–4 and 7 and 8 of the importin-α molecules (36) The STAT1 tyrosine-phosphorylated dimer is somewhat unconventional in its specific interaction with repeats 9 and 10 and the carboxyl terminus of importin-α5 (11–13). To determine whether the STAT3 protein binds to the central region or carboxyl region of the importin-α3 protein, interactions were evaluated between STAT3 and GST-importin-α3 Armadillo repeats 1–8 (amino acids 19–315) or the GST–importin-α3 carboxyl terminus (amino acids 309–521) (Fig. 5C). The source of STAT3 was either from mammalian cells transfected with STAT3-RY–GFP or from Sf9 insect cells expressing His-6-STAT3-RY. Both sources of STAT3 interacted with the more conventional central Armadillo repeat region of importin-α3.
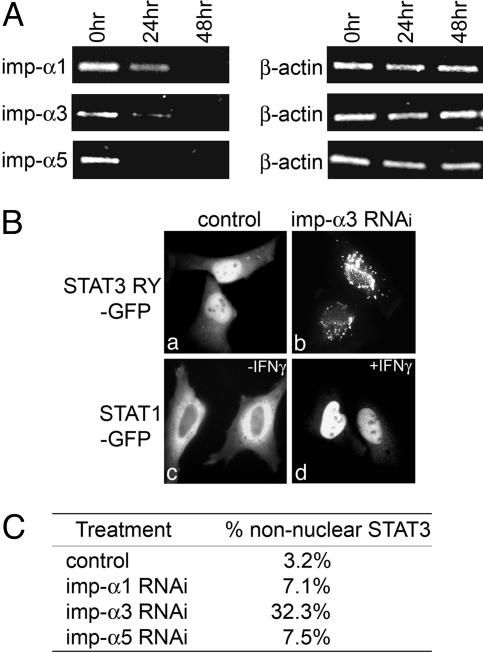
To test the hypothesis that importin-α3 is a primary carrier of STAT3 into the nucleus, we used RNAi to inhibit the expression of importin-α3 and evaluated the effect on the localization of STAT3-RY–GFP. siRNA duplexes were transfected into HeLa cells specific to importin-α3 or to controls importin-α1 or importin-α5 to evaluate any nonspecific effects caused by small dsRNA transfection (40–42). The effect of RNAi on the corresponding RNA levels was examined by quantitative real-time PCR (Fig. 6A). In addition, the effect of RNAi on importin-α3 protein expression was evaluated (Fig. 11, which is published as supporting information on the PNAS web site). After cultures were treated with siRNA, the STAT3-RY–GFP expression plasmid was transfected and cellular localization was evaluated. It was readily apparent that cultures transfected with importin-α3 siRNA showed a distinct perinuclear localization of STAT3-RY-GFP with some punctate cytoplasmic staining (Fig. 6B a and b). In contrast, cultures transfected with siRNA to importin-α1 or importin-α5 showed minimal effects on the localization of STAT3-RY-GFP (Fig. 6C). The slight effect of importin-α1 and importin-α5 siRNA may indicate that they play a minor role in STAT3 import. More significantly, transfection of importin-α3 siRNA had no effect on the cellular localization of control STAT1-GFP. After stimulation with IFN-γ, STAT1-GFP accumulated in the nucleus of all cells in the importin-α3 siRNA treated cultures (Fig. 6B c and d). These results demonstrate the functional and specific interaction of a particular importin-α with STAT3.
Fig. 6.
Effect of importin-α3 siRNA on STAT3 nuclear accumulation. (A) Total RNA was prepared from cells 0, 24, or 48 h after specific siRNA transfection. Reduction of mRNAs encoding importin-α1, importin-α3 or importin-α5 proteins was detected by quantitative real-time RT-PCR using specific primers. β-actin mRNA was amplified as internal control. (B) Images of cells expressing STAT3-RY–GFP in control cultures or after importin-α3 siRNA transfection (imp-α3 RNAi). Images of cells expressing STAT1–GFP in control cultures (-IFN-γ) or importin-α3 siRNA transfected cells after stimulation by IFN-γ (+IFN-γ). (C) Percentage of cells that exhibited aberrant STAT3-RY–GFP localization for each siRNA in this experiment. Results are representative of three independent experiments.
Discussion
STAT3 was first identified as a transcription factor activated by IL-6 able to mediate induction of genes involved in acute inflammatory responses (43, 44). It is now known to be activated by a wide variety of cytokines, growth factors, G-protein coupled receptors, as well as oncogenic kinases such as v-Src and bcr-Abl (16, 45). The fact that STAT3 has been found to be activated in a large number of tumors, and that it is essential for the transforming activity of various oncoproteins has placed it on center stage of cancer biology.
Because STAT1 is the founder member of the STAT family, it was presumed that cellular localization of STAT1 would serve as the paradigm for all STATs. That is, tyrosine phosphorylation would be requisite for nuclear accumulation (11–13, 16, 45, 46). This basic premise does not apply to STAT3. In this report we demonstrate that the cellular localization of STAT3 is prominently nuclear at all times (Figs. 1 and 2). Treatment with IL-6 at a concentration that efficiently stimulates tyrosine phosphorylation did not significantly alter the localization pattern of STAT3 assayed by fluorescence microscopy (Fig. 1). Further characterization of STAT3 demonstrated that it actively shuttles between the cytoplasm and nucleus, consistent with its role as a signal transducer of cytokine stimulation. Active shuttling between nuclear and cytoplasmic compartments requires both nuclear import and export. Nuclear export of STAT3 may result from both leucine-rich NES sequences recognized by CRM1 (47), and CRM1-independent NES signals, because LMB was not able to completely block export in the heterokaryon assay (data not shown). In this report we show that nuclear import of STAT3 is mediated by a constitutively active NLS that requires the STAT3 coiled-coil domain. By generating a series of deletions in STAT3, we determined amino acids 150–162 in the coiled-coil domain to be indispensable for its nuclear translocation. It was reported recently that arginine residues 214 and 215 in the coiled-coil domain were necessary for STAT3 nuclear accumulation in response to cytokine stimulation (48). However, in our studies the RR214/215AA mutation had no effect on STAT3 constitutive nuclear localization (data not shown).
We have demonstrated that specific importin-α carriers recognize STAT3 independent of its phosphorylation state. Both importin-α3 and importin-α6 can effectively bind STAT3 produced in mammalian cells or insect cells. The same profile of importin-αs binds unphosphorylated or tyrosine phosphorylated STAT3, in accordance with the use of the same constitutive NLS before or after phosphorylation. In addition, the importin-αs that bind STAT3 are distinct from the importin-α5 that binds STAT1, supporting the distinct nature of the constitutive STAT3 NLS in contrast to the dimerization-dependent STAT1 NLS. It has been suggested that tyrosine phosphorylation is not a mandatory event for nuclear translocation of STATs (49). Our report demonstrates unphosphorylated STAT3 interacts with importin-α carriers and constitutively translocates to the nucleus. Although the six characterized mammalian importin-α carriers share significant homology and consist in major part of helical Armadillo repeats that bind basic NLSs, it is becoming more evident that they have distinct substrate specificities (33, 39). The 150–162 region of STAT3 is critical for its NLS function and interaction with importin-α3. Although this sequence is indispensable for nuclear translocation of full-length STAT3, a peptide corresponding to amino acids 150–163 has only weak in vitro binding to importin-α3, suggesting it may be part of a structural nuclear localization motif (Fig. 10).
RNAi was also used to corroborate the function of importin-α3 in STAT3 nuclear import. Transient transfections of importin-α3 siRNA produced dramatic effects on the localization of STAT3-RY–GFP. A considerable population of cells showed STAT3-GFP fluorescence restricted to the rim of the nucleus with punctate staining in the cytoplasm. The punctate staining pattern remains to be characterized but may indicate that importin-α3 plays a more general chaperone role for STAT3 (50). Importin-α3 siRNA had a minimal effect on endogenous nuclear STAT3, which may reflect the ability of importin-α6 to mediate import in spite of its low expression level. The newly synthesized STAT3–GFP appears more sensitive to the absence of importin-α3. Future structural information of the STAT3–importin-α protein complex will be critical in understanding the exact nature of the STAT3 NLS–importin-α interaction.
The nuclear accumulation of unphosphorylated STAT3 is distinct from the primarily cytoplasmic localization of unphosphorylated STAT1. Although there is partial sequence conservation between these proteins in the amino acids 150–163 region, there are distinct charge differences in the nonconserved residues (Fig. 12A, which is published as supporting information on the PNAS web site). In addition, a comparison of the residues accessible on the surface of the crystal structures is distinct, and this may contribute to recognition of STAT3 by importin-α3 (Fig. 12B).
Previous reports have described the cytoplasmic-to-nuclear localization of STAT3 after tyrosine phosphorylation. It should be noted that certain of these studies used IFN treatment for STAT3 activation and did not eliminate the gain of NLS function for STAT1 in STAT1:STAT3 tyrosine-phosphorylated heterodimers. Other reports used commercial antibodies to STAT3 that appear to crossreact with nonspecific proteins in immunofluorescence studies or used amino-terminal GFP-tagged STAT3 that, in our experience does not induce gene expression (Fig. 7 and data not shown). All of our experimental analyses, including cellular fractionation, demonstrate significant levels of unphosphorylated STAT3 in the nucleus. These findings do not oppose the role of STAT3 as a cytokine-stimulated signal transducer because STAT3 constitutively shuttles between nuclear and cytoplasmic compartments.
The localization of unphosphorylated STAT3 in the nucleus and cytoplasm may serve to allow its rapid response to activating stimuli in the nucleus or its response in the cytoplasm to extracellular stimuli. STAT3 may be a target of tyrosine kinases in the nucleus, and this could play a role in STAT3-dependent proliferation. Alternatively, STAT3 may influence gene expression in an unphosphorylated state (20). Although unphosphorylated STAT3 cannot bind the characterized STAT DNA target site, it may bind distinct DNA targets in cooperation with other transcription factors. For example, cooperative influences on gene transcription have been described for complexes between STAT3 and c-Jun (51, 52).
Targeting nuclear transport of transcription factors has arisen as a new strategy in recent years to develop cancer drugs (53, 54). The prominent nuclear presence of STAT3 distinguishes it from the other STATs and may offer a mechanism for functional intervention. Understanding the specific regulation of STAT3 nuclear import should support approaches to interfere with its function in aberrant proliferation while maintaining normal translocation of other STATs and critical nuclear proteins.
Supplementary Material
Acknowledgments
We thank the members of the N.C.R. laboratory, especially for the assistance of Sarah Van Scoy and the cooperative efforts of Gregg Banninger, and our colleagues Dafna Bar-Sagi, Michael Hayman, Patrick Hearing, and W. Todd Miller for advice along the way. This study was supported by National Institutes of Health Grants RO1CA50773 and PPGCA28146 and in part by a Carol Baldwin Breast Cancer Research Program grant (to N.C.R.).
Author contributions: L.L., K.M.M., and N.C.R. designed research; L.L. and K.M.M. performed research; L.L. and N.C.R. analyzed data; and L.L. and N.C.R. wrote the paper.
Abbreviations: STAT, signal transducer and activator of transcription; SH2, Src homology 2; NLS, nuclear localization signal; NES, nuclear export signal; siRNA, short interfering RNA; RNAi, RNA interference; IBB, importin-β1 binding; LMB, leptomycin B.
References
- 1.Akira, S. (1999) Stem Cells 17, 138-146. [DOI] [PubMed] [Google Scholar]
- 2.Bromberg, J. & Darnell, J. E., Jr. (2000) Oncogene 19, 2468-2473. [DOI] [PubMed] [Google Scholar]
- 3.Darnell, J. E., Jr. (1997) Science 277, 1630-1635. [DOI] [PubMed] [Google Scholar]
- 4.Ihle, J. N. (2001) Curr. Opin. Cell Biol. 13, 211-217. [DOI] [PubMed] [Google Scholar]
- 5.Davis, L. I. (1995) Annu. Rev. Biochem. 64, 865-896. [DOI] [PubMed] [Google Scholar]
- 6.Rout, M. P. & Aitchison, J. D. (2001) J. Biol. Chem. 276, 16593-16596. [DOI] [PubMed] [Google Scholar]
- 7.Gorlich, D. & Kutay, U. (1999) Annu. Rev. Cell Dev. Biol. 15, 607-660. [DOI] [PubMed] [Google Scholar]
- 8.Macara, I. G. (2001) Microbiol. Mol. Biol. Rev. 65, 570-594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dingwall, C. & Laskey, R. A. (1991) Trends Biochem. Sci. 16, 478-481. [DOI] [PubMed] [Google Scholar]
- 10.Fornerod, M., Ohno, M., Yoshida, M. & Mattaj, I. W. (1997) Cell 90, 1051-1060. [DOI] [PubMed] [Google Scholar]
- 11.McBride, K. M., Banninger, G., McDonald, C. & Reich, N. C. (2002) EMBO J. 21, 1754-1763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melen, K., Fagerlund, R., Franke, J., Kohler, M., Kinnunen, L. & Julkunen, I. (2003) J. Biol. Chem. 278, 28193-28200. [DOI] [PubMed] [Google Scholar]
- 13.Sekimoto, T., Imamoto, N., Nakajima, K., Hirano, T. & Yoneda, Y. (1997) EMBO J. 16, 7067-7077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride, K. M., McDonald, C. & Reich, N. C. (2000) EMBO J. 19, 6196-6206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bowman, T., Garcia, R., Turkson, J. & Jove, R. (2000) Oncogene 19, 2474-2488. [DOI] [PubMed] [Google Scholar]
- 16.Bromberg, J. (2002) J. Clin. Invest. 109, 1139-1142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bowman, T., Broome, M. A., Sinibaldi, D., Wharton, W., Pledger, W. J., Sedivy, J. M., Irby, R., Yeatman, T., Courtneidge, S. A. & Jove, R. (2001) Proc. Natl. Acad. Sci. USA 98, 7319-7324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bromberg, J. F., Wrzeszczynska, M. H., Devgan, G., Zhao, Y., Pestell, R. G., Albanese, C. & Darnell, J. E., Jr. (1999) Cell 98, 295-303. [DOI] [PubMed] [Google Scholar]
- 19.Zamo, A., Chiarle, R., Piva, R., Howes, J., Fan, Y., Chilosi, M., Levy, D. E. & Inghirami, G. (2002) Oncogene 21, 1038-1047. [DOI] [PubMed] [Google Scholar]
- 20.Yang, J., Chatterjee-Kishore, M., Staugaitis, S. M., Nguyen, H., Schlessinger, K., Levy, D. E. & Stark, G. R. (2005) Cancer Res. 65, 939-947. [PubMed] [Google Scholar]
- 21.Gomez, D. & Reich, N. C. (2003) J. Immunol. 170, 5373-5381. [DOI] [PubMed] [Google Scholar]
- 22.Kumar, K. P., McBride, K. M., Weaver, B. K., Dingwall, C. & Reich, N. C. (2000) Mol. Cell. Biol. 20, 4159-4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Weaver, B. K., Kumar, K. P. & Reich, N. C. (1998) Mol. Cell. Biol. 18, 1359-1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Horvath, C. M., Wen, Z. & Darnell, J. E., Jr. (1995) Genes Dev. 9, 984-994. [DOI] [PubMed] [Google Scholar]
- 25.Nakielny, S. & Dreyfuss, G. (1996) J. Cell Biol. 134, 1365-1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pinol-Roma, S. & Dreyfuss, G. (1992) Nature 355, 730-732. [DOI] [PubMed] [Google Scholar]
- 27.Wolff, B., Sanglier, J. J. & Wang, Y. (1997) Chem. Biol. 4, 139-147. [DOI] [PubMed] [Google Scholar]
- 28.Kudo, N., Wolff, B., Sekimoto, T., Schreiner, E. P., Yoneda, Y., Yanagida, M., Horinouchi, S. & Yoshida, M. (1998) Exp. Cell Res. 242, 540-547. [DOI] [PubMed] [Google Scholar]
- 29.Braunstein, J., Brutsaert, S., Olson, R. & Schindler, C. (2003) J. Biol. Chem. 278, 34133-34140. [DOI] [PubMed] [Google Scholar]
- 30.Ota, N., Brett, T. J., Murphy, T. L., Fremont, D. H. & Murphy, K. M. (2004) Nat. Immunol. 5, 208-215. [DOI] [PubMed] [Google Scholar]
- 31.Mao, X., Ren, Z., Parker, G. N., Sondermann, H., Pastorello, M. A., Wang, W., McMurray, J. S., Demeler, B., Darnell, J. E., Jr., & Chen, X. (2005) Mol. Cell 17, 761-771. [DOI] [PubMed] [Google Scholar]
- 32.Mattaj, I. W. & Englmeier, L. (1998) Annu. Rev. Biochem. 67, 265-306. [DOI] [PubMed] [Google Scholar]
- 33.Fontes, M. R., Teh, T. & Kobe, B. (2000) J. Mol. Biol. 297, 1183-1194. [DOI] [PubMed] [Google Scholar]
- 34.Chook, Y. M. & Blobel, G. (2001) Curr. Opin. Struct. Biol. 11, 703-715. [DOI] [PubMed] [Google Scholar]
- 35.Cingolani, G., Petosa, C., Weis, K. & Muller, C. W. (1999) Nature 399, 221-229. [DOI] [PubMed] [Google Scholar]
- 36.Conti, E., Uy, M., Leighton, L., Blobel, G. & Kuriyan, J. (1998) Cell 94, 193-204. [DOI] [PubMed] [Google Scholar]
- 37.Kobe, B. (1999) Nat. Struct. Biol. 6, 388-397. [DOI] [PubMed] [Google Scholar]
- 38.Kohler, M., Ansieau, S., Prehn, S., Leutz, A., Haller, H. & Hartmann, E. (1997) FEBS Lett. 417, 104-108. [DOI] [PubMed] [Google Scholar]
- 39.Kohler, M., Speck, C., Christiansen, M., Bischoff, F. R., Prehn, S., Haller, H., Gorlich, D. & Hartmann, E. (1999) Mol. Cell. Biol. 19, 7782-7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Moss, E. G. & Taylor, J. M. (2003) Nat. Cell Biol. 5, 771-772. [DOI] [PubMed] [Google Scholar]
- 41.Sledz, C. A., Holko, M., de Veer, M. J., Silverman, R. H. & Williams, B. R. (2003) Nat. Cell Biol. 5, 834-839. [DOI] [PubMed] [Google Scholar]
- 42.Gitlin, L., Karelsky, S. & Andino, R. (2002) Nature 418, 430-434. [DOI] [PubMed] [Google Scholar]
- 43.Akira, S., Nishio, Y., Inoue, M., Wang, X. J., Wei, S., Matsusaka, T., Yoshida, K., Sudo, T., Naruto, M. & Kishimoto, T. (1994) Cell 77, 63-71. [DOI] [PubMed] [Google Scholar]
- 44.Zhong, Z., Wen, Z. & Darnell, J. E., Jr. (1994) Science 264, 95-98. [DOI] [PubMed] [Google Scholar]
- 45.Levy, D. E. & Darnell, J. E. J. (2002) Nat. Rev. Mol. Cell. Biol. 3, 651-662. [DOI] [PubMed] [Google Scholar]
- 46.Begitt, A., Meyer, T., van Rossum, M. & Vinkemeier, U. (2000) Proc. Natl. Acad. Sci. USA 97, 10418-10423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bhattacharya, S. & Schindler, C. (2003) J. Clin. Invest. 111, 553-559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Benekli, M., Baer, M. R., Baumann, H. & Wetzler, M. (2003) Blood 101, 2940-2954. [DOI] [PubMed] [Google Scholar]
- 49.Meyer, T., Gavenis, K. & Vinkemeier, U. (2002) Exp. Cell Res. 272, 45-55. [DOI] [PubMed] [Google Scholar]
- 50.Jakel, S., Mingot, J. M., Schwarzmaier, P., Hartmann, E. & Gorlich, D. (2002) EMBO J. 21, 377-386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ivanov, V. N., Bhoumik, A., Krasilnikov, M., Raz, R., Owen-Schaub, L. B., Levy, D., Horvath, C. M. & Ronai, Z. (2001) Mol. Cell 7, 517-528. [DOI] [PubMed] [Google Scholar]
- 52.Zhang, X., Wrzeszczynska, M. H., Horvath, C. M. & Darnell, J. E., Jr. (1999) Mol. Cell. Biol. 19, 7138-7146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kau, T. R. & Silver, P. A. (2003) Drug Discovery Today 8, 78-85. [DOI] [PubMed] [Google Scholar]
- 54.Henderson, B. (2003) Drug Discovery Today 8, 249. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.