Abstract
Telomerase is essential for maintaining telomere length and chromosome stability in stem cells, germline cells, and cancer cells. The telomerase ribonucleoprotein complex consists of two essential components, a catalytic protein component and an RNA molecule that provides the template for telomeric repeat synthesis. A pseudoknot structure in the human telomerase RNA is conserved in all vertebrates and is essential for telomerase activity. It has been proposed that this highly conserved structure functions as a dynamic structure with conformational interchange between the pseudoknot and a hairpin with intraloop base pairings. To examine the structural and functional requirements of the pseudoknot structure, we made mutations in the proposed base-paired regions in the pseudoknot. Although mutations that disrupted the pseudoknot P3 helix abolished activity as predicted, mutations that disrupted the intraloop hairpin base pairings did not reduce telomerase activity, indicating that the intraloop hairpin is not required for telomerase function. This functional study thus provides evidence against the previous proposed molecular-switch model of telomerase pseudoknot function and supports a static pseudoknot structure. The mutational analysis further suggests that telomerase RNA can function independent of the proposed intermolecular pairings between pseudoknot regions on two RNA molecules.
Keywords: reverse transcriptase, ribonucleoprotein, telomere
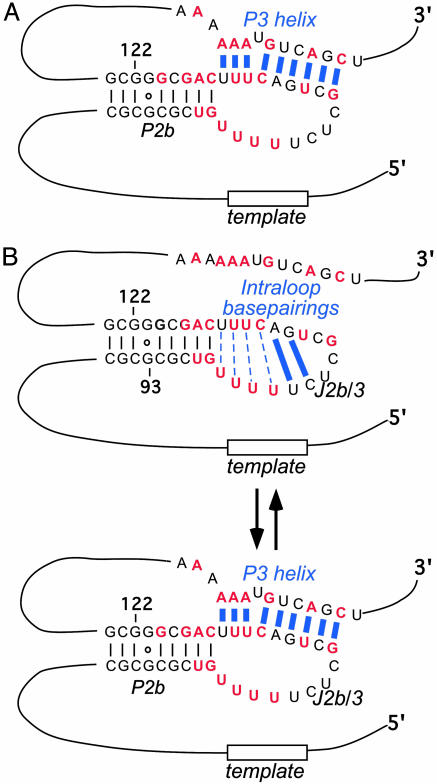
Telomerase is a unique ribonucleoprotein complex that is essential for telomere maintenance. Vertebrate telomeres are required for the stable chromosome maintenance; they consist of simple tandemly repeated TTAGGG sequences and telomere-associated proteins. Telomerase maintains telomeres by adding telomeric repeats to chromosome ends to counter-balance the natural shortening that occurs during DNA replication. Telomerase consists of two essential core components, the catalytic protein component telomerase reverse transcriptase (TERT), and the telomerase RNA (TR) that specifies the repeat sequence added. Vertebrate TR comprises three highly conserved structural domains: the template/pseudoknot domain, CR4–CR5 domain, and the small Cajal-body RNA domain (1–4). The template/pseudoknot domain contains the template region for telomeric DNA synthesis and a conserved pseudoknot structure essential for telomerase activity (Fig. 1A). The template region and the pseudoknot structure are brought into proximity by long-range base pairing interactions, i.e., the P1 helix. The integrity of the P1 helix plays an important role in template boundary definition (5). The presence of the pseudoknot structure formed by helices P2b and P3 was predicted based on phylogenetic analysis (1) and confirmed by mutational analysis (6). Although the pseudoknot structures are conserved in ciliate and vertebrate TRs, the molecular mechanism of their function in telomerase activity is unclear. A “molecular switch” hypothesis for pseudoknot function was proposed based on a NMR structure of the P2b stem–loop (Fig. 1B) (7, 8). This model predicted a conformational conversion between the pseudoknot inferred from the phylogenetic study and the intraloop base pairings seen in the NMR structure of the P2b–stem–loop RNA (nucleotides 93–122).
Fig. 1.
Two models for the functional role of telomerase pseudoknot structure. (A) The stable pseudoknot model suggests that the P3 helix is stable and does not undergo conformational switching. The Watson–Crick base pairings are shown as solid lines, and the noncanonical base pairings are shown as dashed lines. (B) The molecular switch model suggests that the RNA conformation switches between the intraloop base pairings and the P3 helix pseudoknot structure.
In addition, human TR (hTR) can dimerize in vitro through a P3 helix formed in trans between two RNAs (9). However, this experiment involved two mutant RNAs, and it is not known whether this intermolecular pseudoknot formation occurs in the WT TR. To further understand the functional role of the essential pseudoknot structure, we examined the effect of specific mutation in the pseudoknot on telomerase function in vitro and in vivo.
Materials and Methods
Mutagenesis of TR. Point mutations in the TR gene were generated essentially by using an overlap extension PCR strategy as described in ref. 10. Briefly, segments of the RNA gene were PCR-amplified by using combinations of an upstream hTR(-79) forward primer (5′-GAGCCGCGAGAGTCAGCTTG-3′), a downstream hTR(326) reverse primer (5′-CCCGCGGCTGACAGAGCCCAACTC-3′), and the internal primer pairs that contained desired mutant sequence in the middle of the primer. The amplified fragments were PCR-assembled, and the full-length PCR products covering sequence from -79 to 326 were gel-purified. The purified PCR DNAs were digested with NotI at position -40 and BanII at position 310 and cloned into the NotI–BanII-digested phTR-AflII–HindIII plasmid DNA. The phTR–AflII–HindIII plasmid contains a 1.5-kb AflII–HindIII-digested genomic DNA fragment that encodes the hTR gene. All mutant RNA genes were sequenced to confirm the presence of only the intended mutations.
In Vitro Reconstitution of Telomerase. Human and mouse telomerase complexes were reconstituted in vitro or in vivo as described in refs. 11 and 12. Briefly, for in vitro reconstitution of telomerase, the RNA gene was PCR-amplified from plasmids containing either hTR or mouse TR (mTR) mutant genes by using oligonucleotides that contain a T7 promoter sequence in the forward primer. TRs were generated by T7 in vitro transcription with PCR DNA templates. Epitope-tagged human and mouse TERT proteins were expressed from plasmid DNAs phTERT-HA2 and pmTERT-HA2 encoding hemagglutinin (HA)-tagged human TERT (hTERT) and mouse TERT (5) by using the TnT transcription/translation system (Promega). In vitro reconstituted telomerase complexes were immunoaffinity-purified by using anti-HA F7 agarose beads (Santa Cruz Biotechnology) in the presence of 1× immunoprecipitation buffer (10 mM Hepes, pH 7.5/100 mM potassium glutamate/1 mM MgCl2/1 mM DTT/10% glycerol) and assayed for telomerase activity by using the direct telomerase assay protocol.
Direct Telomerase Assay. Activity of immunopurified telomerase was assayed directly without amplification (11). The reaction mixture (20 μl) contained 1× telomerase assay buffer (50 mM Tris·HCl, pH 8.0/50 mM KCl/1 mM MgCl2/5 mM 2-mercaptoethanol/1 mM spermidine), 1.0 μM telomere primer (TTAGGG3), 0.5 mM dATP, 0.5 mM dTTP, 2 μM dGTP, and 1.25 μM α-32P-dGTP [800 Ci/mmol (1 Ci = 37 GBq)] with 6 μl of immunopurified telomerase complex. The reaction was incubated at 30°C for 1 h. The products were subjected to ethanol precipitation, followed by polyacrylamide electrophoresis. The gels were dried and exposed to a Fuji BAS1500 phosphorimager screen for an appropriate period.
In Vivo Reconstitution of Telomerase. For in vivo reconstitution of telomerase, we established a VA13 cell line that stably expressed the native hTERT gene but not the hTR gene. The hTERT gene was cloned into pIRESpuro3 (Clontech) at a site after the puromycin resistant gene. Both genes are expressed under control of the same CMV promoter. After transfection of the VA13 cells with the pIRESpuro3-hTERT DNA, a stable hTERT-expressing VA13 cell line was selected in the presence of 2 μg/ml puromycin. After transient transfection and expression of the hTR mutant genes in the hTERT-expressing VA13 cell line, cells were trypsinized and centrifuged for 5 min at 500 × g. Cell pellets were washed twice in PBS and resuspended in 1× CHAPS buffer (10 mM Tris·HCl, pH 8.0/1 mM MgCl2/1 mM EGTA/0.5% CHAPS/10% glycerol/5 mM 2-mercaptoethanol/1 mM DTT). One tablet of Mini Complete protease inhibitor mixture (Roche) was added to 10 ml of CHAPS buffer. After incubation on ice for 30 min, extracts were centrifuged for 15 min at 4°C at 14,000 × g. Supernatants were then collected and assayed for telomerase activity by the telomere repeat amplification protocol (TRAP) assay (13). The TRAP assay was carried out by using the TRAPeze telomerase detection kit according to the vendor's instruction (Intergen, Purchase, NY).
Results and Discussion
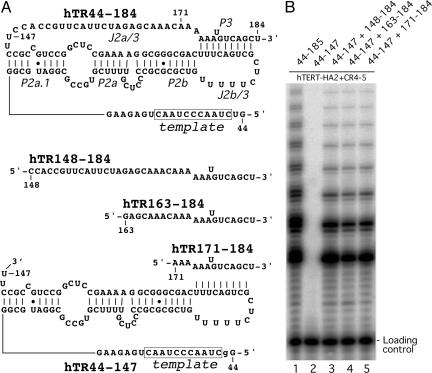
The P3 Helix Can Be Functionally Reconstituted in Trans. Telomerase activity can be reconstituted in vitro by using T7-transcribed RNA fragments containing the template/pseudoknot domain hTR44–184 and the CR4–CR5 domain (14, 15). The base pairings in the P3 helix of the pseudoknot are essential for activity (6). To further dissect the essential structural role of the pseudoknot, we first tested whether the P3 helix can be functionally reconstituted in trans from two independent RNA fragments. The hTR44–184 RNA fragment was divided into two separate RNA fragments, hTR44–147 and hTR148–184 (Fig. 2). Although reconstitution of telomerase with hTR44–147 and CR4–CR5 RNA alone generated no activity (Fig. 2, lane 2), telomerase reconstituted with three RNA fragments, CR4–CR5, hTR44–147, and hTR148–184 had similar activity as a mixture with an uninterrupted 44–185 fragment (Fig. 2, compare lane 3 with lane 1). This result indicates that the essential base-paired structure of P3 helix can form from separate fragments in trans and that the specific topological configuration of a pseudoknot is not needed for telomerase activity.
Fig. 2.
Trans reconstitution of P3 helix in vitro. (A) The RNA fragments used to generate the pseudoknot domain hTR44–184, hTR44–147, hTR148–184, hTR163–184, and hTR171–184 in the in vitro reconstitution are shown. The nomenclature of the helical and single-stranded regions is from ref. 1. (B) Direct telomerase activity assay. The RNA fragments used to reconstitute telomerase activity are indicated above each lane. The reconstitution was carried out in the presence of HA-tagged TERT (hTERT-HA2) protein and the CR4–CR5 RNA fragment hTR200–278, as described in Materials and Methods.
We next tested whether the J2a/3 region, the unpaired bases in the 5′ portion of the hTR148–184 fragment, was required for telomerase activity. Two shorter RNA fragments with truncations in the J2a/3 region, hTR163–184 and hTR171–184, together with hTR44–147 also fully reconstituted telomerase activity (Fig. 2, lanes 4 and 5). Thus, as predicted from its low sequence conservation, the single-stranded region J2a/3 between P2a and P3 is dispensable for activity. Further truncations that removed conserved residues 171A, 172A, and 173A from the 5′ end of hTR171–184 significantly reduced telomerase activity (data not shown).
Mutational Analysis of the P3 Helix and the J2b/3 Loop Support a Static Pseudoknot Structure. An NMR solution structure of a short RNA oligonucleotide representing the P2b stem–loop (nucleotides 93–122) indicated that the loop region of this RNA can form four noncanonical pyrimidine:pyrimidine base pairings and two Watson–Crick base pairings when the complementary strand of the P3 helix is not present (7). The intraloop base pairing was thus suggested to be in thermodynamic equilibrium with the P3 helix (Fig. 1B). A “molecular switch” model was therefore proposed in which these intraloop base pairings represent an alternative conformation of the P3 pseudoknot structure (Fig. 1B). In this model, the RNA structure switches between the P3 pseudoknot and the intraloop base pairing conformations during telomerase elongation (7, 8). However, in our minimal RNA reconstitution system, telomerase activity was fully restored by using a short 14-mer RNA oligonucleotide (nucleotides 171–184) that forms the P3 helix in trans with the P2 stem–loop RNA fragment (nucleotides 44–147) (Fig. 2). This enzyme with a trans P3 helix was immunoprecipitated by anti-H A antibody against the hTERT-HA protein and washed extensively to remove unbound RNA oligonucleotides. Thus, dissociation of the P3 helix base pairings would result in a loss of the 14-mer RNA fragment from the complex because of the very low concentration of the reconstituted enzyme used in the telomerase assay. The fact that activity is still seen after these washes indicates that the P3 helix is stable once it is formed (Fig. 1 A).
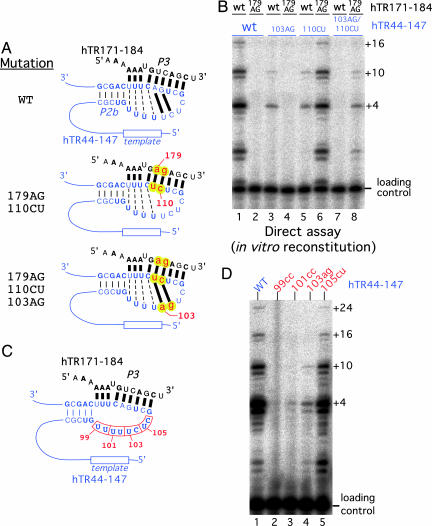
To more directly examine the functional role of the P3 and the intraloop base pairings, we designed a mutation, 110CU, that disrupted the two Watson–Crick base pairings (103U:111A and 104C:110G) in the intraloop configuration (Fig. 3A). This mutant RNA was then reconstituted with a 14-mer RNA oligonucleotide hTR171–184 that had a compensatory mutation, 179AG, that allowed the P3 pairing in trans. Reconstitution of telomerase by using the 179AG mutated hTR171–184 RNA and the 110CU mutated hTR44–147 RNA restored the P3 helix and telomerase activity (Fig. 3B, compare lane 1 with lane 6). In contrast, when the same 179AG mutated hTR171–184 RNA was reconstituted with a WT hTR44–147 RNA, no activity was seen, because of the disruption of P3 helix (Fig. 3B, lane 2). This result suggests that the P3 helix, but not the intraloop base pairings, is essential for telomerase activity.
Fig. 3.
Functional analysis of P3 helix and residues in the J2b/3 loop region. (A) Potential base pairing within the P3 helix and the intraloop base pairing is shown for the WT and the mutant RNAs with double mutations 179AG/110CU or triple mutations 179AG/110CU/103AG. (B) Activity analysis of in vitro reconstituted telomerase by using hTR171–184 RNA fragments (WT or 179AG mutant) and hTR44–147 RNA fragments (WT, 103AG, 110CU, or 103AG/110CU). The RNAs added to the reaction are indicated above each lane. The reconstitution was carried out in the presence of hTERT-HA2 protein and the CR4–CR5 RNA fragment, hTR200–278. (C) The position of dinucleotide mutations introduced in the J2b/3 single-stranded loop region are marked by boxes, and the nucleotide numbers are indicated. (D) The activity of in vitro reconstituted TR mutants. The mutations in the hTR44–147 fragment are indicated above each lane. The 99cc mutant contains mutations U99C and U100C. The 101cc mutant contains mutations U101C and U102C. The 103ag mutant contains mutations U103A and C104G. The 105cu mutant contains mutations U105C and C106U.
To further test for a role of the intraloop pairings, we made a third mutation (103AG) within the loop region to restore the potential intraloop base pairings of the hTR44–147 110CU mutant RNA (Fig. 3A). Reconstitution of this RNA with the 179AG mutant P3 RNA oligonucleotide, which generated a triple compensatory mutant, resulted in a even lower activity than the simple compensatory mutant which restored only the P3 base pairings (Fig. 3B, compare lane 6 with lane 8). This finding suggests that this sequence of the J2b/3 loop, rather than the base pairing ability in the loop region, plays a critical role for telomerase activity.
The Sequence in the J2b/3 Loop Is Important for Telomerase Function. To investigate the functional importance of the sequence in the J2b/3 loop region, we made dinucleotide substitutions of all of the residues in the loop region and tested the reconstituted mutant TR for telomerase activity (Fig. 3 C and D). The dinucleotide mutations, 99UU→99cc and 101UU→101cc, severely impaired telomerase activity (Fig. 3D, lanes 2 and 3). Mutation of nucleotides 103–104 from UC to AG also significantly reduced telomerase activity (Fig. 3 B, lane 3, and D, lane 4). In contrast, mutations from UC to CU at the less conserved residues, 105 and 106, did not affect telomerase activity (Fig. 3D, lane 5). These data indicate that the identity of the residues in the single-stranded region from nucleotide 99 to nucleotide 104 is critical for telomerase activity. We hypothesize that these nucleotides may be involved in RNA–RNA (20) or RNA–protein interactions that are important for telomerase function.
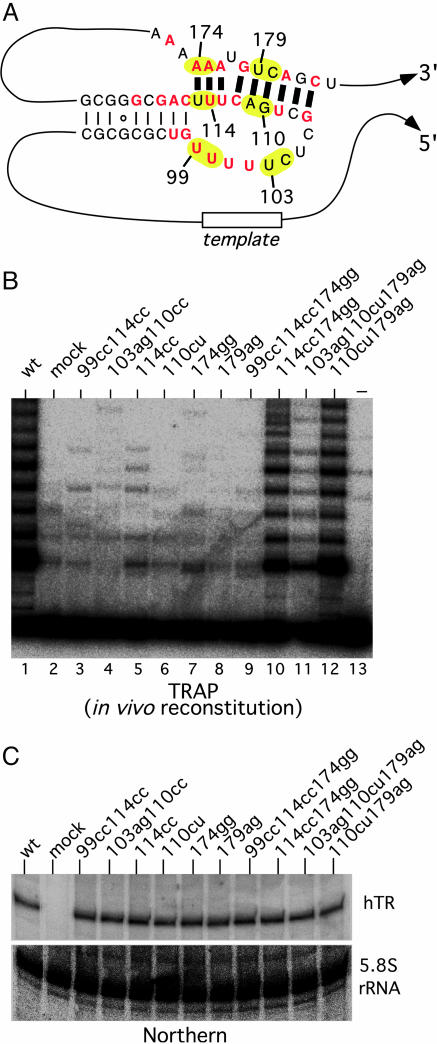
In Vivo Confirmation of the Pseudoknot Structure and Function. Although our in vitro reconstitution experiments argue against a functional role of the intraloop base pairings in telomerase activity in vitro, it is not clear whether they play any role in telomerase function in vivo. In Tetrahymena, mutations in the pseudoknot region have a much greater effect on in vivo activity than in vitro activity (16). Thus, we generated TR genes with mutations in the P3 helix and the J2b/3 loop and expressed them in a telomerase-negative, VA13-derived cell line that expresses the hTERT gene but not the endogenous hTR gene. The cell lysate of transfected cells was analyzed for telomerase activity by using the TRAP, a PCR-based assay for telomerase activity (Fig. 4B). The expression levels of all mutant RNAs were similar as assayed by Northern blotting (Fig. 4C). All of the mutations that disrupt base pairing of the helix P3 or change the sequence of the P2b/3 loop severely reduced the in vivo reconstituted telomerase activity to background level (Fig. 4B, lanes 3–9). In contrast, the compensatory double point mutations, 114cc174gg and 110cu179ag, which disrupt the potential interloop base pairing but maintain P3 helix pairing, reconstituted telomerase activity in vivo at the WT level (Fig. 4B, lanes 10 and 12). These results confirm that the P3 pairing in the pseudoknot is important for telomerase activity both in vitro and in vivo, whereas the intraloop base pairings in the P2b stem–loop are not. Furthermore, the WT level of activity from the compensatory changes in P3 indicates that it is the helical structure, not the sequence, that is important for telomerase activity.
Fig. 4.
In vivo reconstituted telomerase with pseudoknot mutations. (A) The position of mutations introduced in the P3 helical and the J2b/3 single-stranded loop region are indicated with nucleotide position numbers and highlighted in yellow. (B) Telomerase activity of in vivo reconstituted TR with mutations. Telomerase activity of the cell lysates from in vivo reconstituted mutant telomerase were analyzed by the TRAP. (C) Northern blot of in vivo expression levels of mutant TR. 5.8S rRNA was quantitated as loading control.
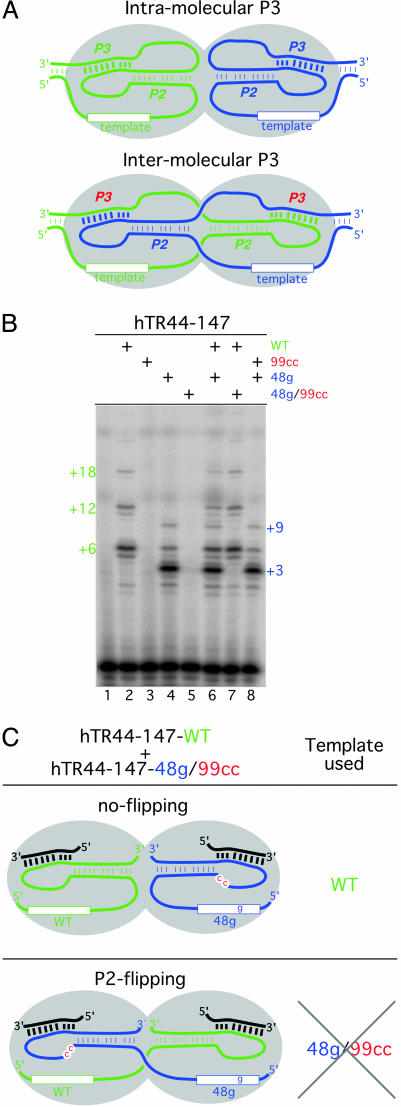
Intramolecular Versus Intermolecular P3 Pseudoknot Structure. Human telomerase enzyme functions as a dimer (17–19). However, the RNA conformation within the telomerase dimer complex is not well studied. Recent experiments showed that, in the absence of TERT protein, the pseudoknot domain of TR can form intermolecular P3 base pairings that result in dimerization of the RNA in vitro (9). When two nonfunctional RNAs each with P3 mutations that allowed only compensatory pairing in trans between two molecules were reconstituted together, a low level of telomerase activity was detected (9). It was thus proposed that the pseudoknot can form intermolecular RNA dimers through the P3 pairing region (Fig. 5A). However, in our minimal reconstitution system in which the P3 helix was formed from two separate RNA fragments, such intermolecular P3-dependent dimerization of TR is not possible; yet high levels of activity are seen (Fig. 2B, lane 5). These data argue against a functional role of the proposed P3-dependent RNA dimerization as depicted in Fig. 5A.
Fig. 5.
A template rescue analysis to test for possible P2 helix flipping. (A) Two possible configurations of the pseudoknot structure: Intramolecular or intermolecular P3 helix might form between the two TRs in a dimer. The TERT dimer is shown as the two gray ovals. (B) The activity of telomerase reconstituted in vitro with one or a mixture of two different RNA mutants with WT template or 48g mutant template. Telomerase reconstituted with WT RNA generated telomere products at positions +6, +12, and +18. Telomerase with 48g mutant template generated unique telomerase product at positions +3 and +9. (C) Model for P2 helix swapping in an in vitro reaction. Two possible models for pseudoknot conformation generated by using two independent RNA fragments. In the no-flipping model, the template of the inactive RNA fragment hTR44–147–48g99cc is not used. In the helix P2 flipping model, the functional pseudoknot domain of the RNA with WT template will flip into the other catalytic site and allow the utilization of 48g template of the inactive RNA, hTR44–147–48g/99cc. Only the WT pattern was seen when the hTR44–147–WT and hTR44–147–48g/99cc RNAs were mixed, indicating that helix P2 flipping does not occur.
Although the experiments above indicate that dimerization through P3 is not required for activity, it is still possible that, even in the absence of a trans pseudoknot formation, the P2 helix can flip from one dimer subunit to another (Fig. 5C). To test whether such P2 helix flipping occurs, we made a TR mutant with a mutation at the residue 48 in the template region that generates a distinct elongation pattern in the telomerase activity assay. This template mutation allows us to monitor the template utilization during telomerase reaction (Fig. 5B, lanes 2 and 4). Reconstitution with WT template RNA alone generated the expected telomere elongation pattern with pausing at position +6, +12, and + 18 (Fig. 5B, lane 2), whereas the RNA with a 48g template mutation showed a similar level of activity but with a +3 and + 9 pattern (Fig. 5B, lane 4). This pausing pattern presumably results from the premature dissociation at the mutated 48g residue and reassociation at the 3′ end of the template for a second round of elongation. WT and 48g templates showed no activity when a 99cc mutation was present (Fig. 5B, lanes 3 and 5). When the WT and 48g template RNAs were added to the same reaction, both elongation patterns were seen (Fig. 5B, lane 6).
To test whether P2 helix flipping between RNAs occurs, we added the WT RNA to a reaction with the inactive double mutant 48g/99cc. If a P2 helix can flip between two catalytic sites in a telomerase dimer, the RNA with WT template should be able to rescue the inactive 99cc mutant RNA with 48g template and allow the 48g template to be used (Fig. 5 B, lane 7, and C). Interestingly, only the WT pattern with pausing at +6, +12, and +18 was seen (Fig. 5B, lane 7). Conversely, when the 48g template was mixed with the inactive 99cc mutant that contains a WT template, only the mutant +3 and +9 pattern was seen (Fig. 5B, lane 8). Thus, intermolecular formation of pseudoknot is not required for telomerase enzyme activity.
The pseudoknot region of the TR is essential for enzyme activity. We tested the role of base pairings and proposed structural elements within the pseudoknot and found that it is the base pairing not the topology of the helical region P3 that is essential for activity. We also found no evidence for the functional role of the proposed J2b/3 loop intraloop pairings; thus, we propose that the essential P3 helix of the pseudoknot structure is stable during telomere elongation. Recently the solution structure of the pseudoknot was reported, and the solution structure revealed a unique triple helix surrounding the helical junction in the pseudoknot (20). It was also shown that the stable tertiary structure of the pseudoknot is strongly correlated with telomerase activity (20). The highly conserved nucleotides in the J2b/3 region were shown to be involved in tertiary RNA–RNA interactions that are critical for the formation of a stable pseudoknot structure and, thus, essential for telomerase activity as shown in this study. With the availability of a solution structure of the pseudoknot RNA, it would be interesting to know whether there is a conformational change in the pseudoknot structure upon the binding of TERT protein or during telomerase reaction. Further elucidation of the pseudoknot structure and function in the context of a telomerase complex will provide a detailed mechanistic understanding of this unique enzyme and may help in the rational design of specific telomerase inhibitors for cancer therapy.
Acknowledgments
This research was supported by National Institutes of Health Grant AG09383 (to C.W.G.). J.-L.C. was supported by a fellowship from the Leukemia and Lymphoma Society.
Author contributions: J.-L.C. designed research; J.-L.C. performed research; J.-L.C. contributed new reagents/analytic tools; J.-L.C. and C.W.G. analyzed data; J.-L.C. and C.W.G. wrote the paper; and C.W.G. was the principal investigator.
This contribution is part of the special series of Inaugural Articles by members of the National Academy of Sciences elected on April 29, 2003.
Abbreviations: TERT, telomerase reverse transcriptase; hTERT, human TERT; TR, telomerase RNA; hTR, human TR; mTR, mouse TR; HA, hemagglutinin; TRAP, telomere repeat amplification protocol.
See accompanying Biography on page 8077.
References
- 1.Chen, J.-L., Blasco, M. A. & Greider, C. W. (2000) Cell 100, 503-514. [DOI] [PubMed] [Google Scholar]
- 2.Chen, J.-L. & Greider, C. W. (2004) Trends Biochem. Sci. 29, 183-192. [DOI] [PubMed] [Google Scholar]
- 3.Mitchell, J. R., Cheng, J. & Collins, K. (1999) Mol. Cell. Biol. 19, 567-576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jady, B. E., Bertrand, E. & Kiss, T. (2004) J. Cell Biol. 164, 647-652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chen, J.-L. & Greider, C. W. (2003) Genes Dev. 17, 2747-2752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ly, H., Blackburn, E. H. & Parslow, T. G. (2003) Mol. Cell. Biol. 23, 6849-6856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Theimer, C. A., Finger, L. D., Trantirek, L. & Feigon, J. (2003) Proc. Natl. Acad. Sci. USA 100, 449-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Comolli, L. R., Smirnov, I., Xu, L., Blackburn, E. H. & James, T. L. (2002) Proc. Natl. Acad. Sci. USA 99, 16998-17003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ly, H., Xu, L., Rivera, M. A., Parslow, T. G. & Blackburn, E. H. (2003) Genes Dev. 17, 1078-1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ge, L. & Rudolph, P. (1997) BioTechniques 22, 28-30. [DOI] [PubMed] [Google Scholar]
- 11.Chen, J.-L. & Greider, C. W. (2003) EMBO J. 22, 304-314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen, J.-L., Opperman, K. K. & Greider, C. W. (2002) Nucleic Acids Res. 30, 592-597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, N. W. & Wu, F. (1997) Nucleic Acids Res. 25, 2595-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mitchell, J. R. & Collins, K. (2000) Mol. Cell 6, 361-371. [DOI] [PubMed] [Google Scholar]
- 15.Tesmer, V. M., Ford, L. P., Holt, S. E., Frank, B. C., Yi, X., Aisner, D. L., Ouellette, M., Shay, J. W. & Wright, W. E. (1999) Mol. Cell. Biol. 19, 6207-6216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gilley, D. & Blackburn, E. H. (1999) Proc. Natl. Acad. Sci. USA 96, 6621-6625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Beattie, T. L., Zhou, W., Robinson, M. O. & Harrington, L. (2001) Mol. Cell. Biol. 21, 6151-6160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moriarty, T. J., Huard, S., Dupuis, S. & Autexier, C. (2002) Mol. Cell. Biol. 22, 1253-1265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wenz, C., Enenkel, B., Amacker, M., Kelleher, C., Damm, K. & Lingner, J. (2001) EMBO J. 20, 3526-3534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Theimer, C. A., Blois, C. A. & Feigon, J. (2005) Mol. Cell 17, 671-682. [DOI] [PubMed] [Google Scholar]