Abstract
Constitutive expression of telomerase prevents senescence and crisis by maintaining telomere homeostasis. However, recent evidence suggests that telomerase is dynamically regulated in normal cells and also contributes to transformation independently of net telomere elongation. Here, we show that suppression of the telomerase catalytic subunit [human telomerase reverse transcriptase (hTERT)] expression abrogates the cellular response to DNA double strand breaks. Loss of hTERT does not alter short-term telomere integrity but instead affects the overall configuration of chromatin. Cells lacking hTERT exhibit increased radiosensitivity, diminished capacity for DNA repair, and fragmented chromosomes, demonstrating that loss of hTERT impairs the DNA damage response.
Keywords: human telomerase reverse transcriptase, histone modification, genomic integrity
Normal human cells exhibit a limited replicative lifespan and eventually enter a growth arrest state, termed replicative senescence, triggered by dysfunctional telomeres (1). However, other stimuli such as oncogene activation (2), increased oxidative potential (3), and genotoxic damage (4) also trigger a cell cycle arrest that shares both morphologic and functional similarities with replicative senescence. Thus, although telomere maintenance plays an important role in regulating the proliferative potential of human cells, the role(s) of telomere biology in replicative senescence induced by successive cycles of cell division and in the senescence-like growth arrest state triggered by other stimuli remains obscure.
Moreover, recent work indicates that senescent human cells show evidence of activation of the DNA damage response pathway (5). Although overexpression of telomerase maintains telomere length and facilitates human cell immortalization (6, 7), accumulating evidence also suggests that telomerase itself plays an additional role in protecting karyotypic stability by “capping” chromosomes (8). Indeed, constitutive overexpression of telomerase reverse transcriptase (TERT) facilitates malignant transformation independently of its effects on overall telomere length (9) and renders cells more resistant to apoptosis (10). Because these observations connect telomerase expression, DNA damage responses, and senescence, we reasoned that human TERT (hTERT) may contribute to the cellular response to genotoxic insults. Here we show the effects of stably suppressing hTERT function in normal human fibroblasts on chromatin architecture and the response to DNA double strand breaks.
Materials and Methods
Stable Expression of Short Hairpin RNA (shRNA). The sequences shown in Table 2, which is published as supporting information on the PNAS web site, were introduced into the pMKO.1-puro vector (11) to create shRNA vectors specific for hTERT and GFP (Fig. 5, which is published as supporting information on the PNAS web site). These vectors were used to make high titer amphotropic retroviruses, which were used to infect human fibroblasts as described (11). Polyclonal cell populations were purified with selection with puromycin (2 μg/ml).
Immunoblotting, Immunofluorescence, FISH, and RT-PCR. For immunofluorescence, cells were fixed in chilled acetone, incubated with the primary antibody, washed, and then incubated with either an Alexa Fluor 568-conjugated or Alexa Fluor 488-conjugated secondary antibody (Pierce) in 1% BSA for 1 h at 37°C. For telomere-specific FISH, we hybridized a peptide nucleic acid (PNA) probe (CCCTAA)3 specific for mammalian telomeres (Applied Biosystems) to acetone-fixed cells at 72°C for 8 min. To remove nonhybridized PNA probes, slides were washed with 0.05% Tween 20 containing PBS at 56°C for 15 min and visualized by using a Nikon Eclipse E800 fluorescence microscope. We did not detect staining of parallel cultures with a single mismatch (CCCTTA)3 PNA probe. The antibodies used in this study were purchased from the following suppliers: rabbit anti-H2AX and mouse anti-hTERT (Novus Biologicals, Littleton, CO); rabbit anti-γ-H2AX, rabbit anti-H2B, mouse anti-H3, rabbit anti-H4, rabbit anti-macro H2A.1, rabbit anti-dimethyl H3 (K9), rabbit anti-acetyl H3 (Lys-9), and rabbit anti-acetyl H4 (K12) (Upstate Biotechnology); goat anti-phospho-specific breast cancer-associated 1 (BRCA1) (Ser-1497) (Santa Cruz Biotechnology); rabbit anti-ATM-pS1981 (Rockland, Gilbertsville, PA); and mouse anti-p53 (Ab6), (Oncogene Science). Cells were lysed in radioimmunoprecipitation assay (RIPA) buffer (12.5 mM sodium phosphate, pH7.2/2 mM EDTA/50 mM NaF/1.25% Nonidet P-40/1.25% SDS/0.1 mM DTT) except when specific conditions are noted. For extraction under low salt conditions, cells were lysed in a buffer comprising 20 mM Tris·HCl, pH 7.4/150 mM NaCl/0.1% Nonidet P-40/0.1 mM DTT. For protein extraction under high salt conditions, 500 mM NaCl was substituted in the low salt buffer. For acid precipitation, cells were homogenized in 0.2 N H2SO4 and centrifuged. Histones were precipitated by adding 0.25× vol of 100% (wt/vol) trichloroacetic acid (TCA). The pellets were suspended in 100% ethanol and centrifuged at 13,000 × g. Ten micrograms of protein was subjected to immunoblotting. The sequences used for the H2AX RT-PCR were as follows: 5′-TCGGGCCGCGGCAAGACTGGCGGCAA-3′ and 5′-GTACTCCTGGGAGGCCTGGGTGGCCTT-3′. Reverse transcription was performed on 500 ng of total RNA for 30 min at 42°C, followed by PCR (25 cycles: 94°C for 45 s, 60°C for 45 s, and 72°C for 90 s).
Analysis of Telomere Structure. Telomere length was measured by hybridizing a 32P-labeled telomeric (CCCTAA)3 probe to HinfI- and RsaI-digested genomic DNA. Quantitative FISH (Q-FISH) analysis was performed as described (12). Results of Q-FISH analysis are expressed in kilobases as determined by comparison with plasmid DNA containing telomere inserts. The telomeric 3′ single-stranded overhang was analyzed by a telomeric 3′-overhang ligation assay (T-OLA) (13).
Micrococcal Nuclease Assay. Cells (1 × 106) were suspended in 1 ml of nuclei buffer [25 mM Hepes, pH 7.8/1.5 mM MgCl2/10 mM KCl/0.1% Nonidet P-40/1 mM DTT/protease inhibitor mixture (Roche Applied Science, Indianapolis)]. Nuclei were obtained by Dounce homogenization (20 strokes, pestle A) and sedimented by centrifugation at 1400 × g at 4°C for 20 min through 1 ml of a solution containing 10 mM Tris·HCl, pH 7.4/15 mM NaCl/60 mM KCl/0.15 mM spermine/0.5 mM spermidine/10% sucrose. The nuclear pellet was then resuspended in 350 μl of digestion buffer (50 mM Tris·HCl, pH 7.5/15 mM NaCl/5 mM KCl/3 mM MgCl2/1 mM CaCl2/10 mM NaHS04/0.25 M sucrose/0.15 mM spermine/0.5 mM spermidine/0.15 mM mercaptoethanol) containing micrococcal nuclease (9 units/ml, Roche). Fifty microliters from this reaction mixture was mixed with 50 μl of stop solution (200 mM EDTA/200 mM EGTA, pH 7.5) to stop the reaction. Digested DNA was recovered by QIAquick columns (Qiagen, Valencia, CA), subjected to agarose gel electrophoresis, and visualized by staining with ethidium bromide. Direct application of digested chromatin without further purification to agarose gels gave similar results.
Clonogenic Assay. Clonogenic assays were performed by using two different seeding protocols. In some experiments, 200 cells were seeded into 9.6-cm2 plates in triplicate and exposed to ionizing radiation after 24–48 h. Cells were allowed to proliferate for 10–12 days, trypsinized, and replated into plates to eliminate cell debris. Cells were counted after an additional 5–7 days by using a Coulter particle counter. In other experiments, 1,000 cells were seeded into 9.6-cm2 plates in triplicate, irradiated after 24–48 h, incubated 21 days, and stained with crystal violet (0.2%) to identify colonies. Colonies containing >20 cells were counted manually. Identical results were obtained by using these two methods, and the first method was used for the experiment shown in Fig. 4A.
Fig. 4.
Functional effects of suppressing hTERT expression. (A) Effects of hTERT suppression on clonogenic growth after ionizing radiation (IR). BJ cells expressing a control vector (filled circles), an hTERT-specific shRNA (triangles), a3′ UTR hTERT-specific shRNA (filled squares), a 3′ UTR hTERT-specific shRNA together with WT-hTERT (diamonds), and WT-hTERT (open circles), respectively, were exposed to γ-irradiation. Relative cell survival was calculated as the percentage of viable cells after irradiation relative to unexposed cells. Mean ± SD is shown. In some cases, the error bars are covered by the symbol. (B) Effects of hTERT suppression on DNA repair. BJ cells expressing the indicated shRNA or WT hTERT were irradiated (2 Gy). The fraction of DNA breaks induced by ionizing radiation that was repaired at 4 h was measured by pulse-field gel electrophoresis and normalized to the control shRNA samples. Each bar represents the mean ± SD for three independent experiments.
DNA Repair Assay. The DNA repair assay was performed as described (14). Briefly, cells were mock irradiated or irradiated (2 Gy), allowed to recover at 37°C for 0, 2, and 4 h, trypsinized, and cast into 0.75% SeaPlaque agarose (FMC). These agarose cell plugs were placed in lysis buffer and incubated at 50°C for 38 h, washed with TE buffer, and equilibrated. The plugs were then subjected to pulse field gel electrophoresis in 0.7% agarose gels, dried, and stained with SYBR Green (Molecular Probes), and the fluorescence signal was measured by imagequant software. The fraction of DNA entering the gel was determined by the following equation: [(signal in lane)/(signal in lane + signal in plug)] × 100. The relative fraction of DNA breaks repaired at 4 h was determined by calculating the ratio of DNA entering the gel at 4 h to that present immediately after irradiation (0 h). The measured value of signal present in unirradiated cells was subtracted for each sample. The data were normalized to the control shRNA sample and presented as bars representing the mean ± standard deviation.
Cytogenetic Analyses. Before or after exposure to 5 Gy of γ-radiation, cells were incubated at 37°C for 24 h and subjected to a standard cytogenetic protocol (15). Cytogenetic abnormalities were scored by a blinded observer.
Results
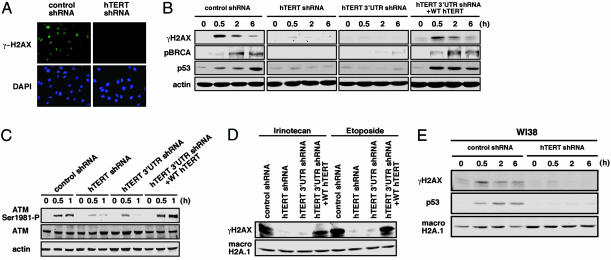
To assess the effects of suppressing hTERT function on the response to ionizing radiation, we examined well characterized changes in several proteins implicated in the response to DNA damage in diploid human fibroblasts, which only transiently express low levels of hTERT in S-phase (11). As expected, irradiation of human BJ or WI38 fibroblasts expressing a control, GFP-specific shRNA vector led to the phosphorylation of H2AX (γ-H2AX) (Fig. 1 A, B, and E), to phosphorylation of the ataxia–telangiectasiamutated (ATM) (Fig. 1C) and BRCA1 tumor suppressor proteins (Fig. 1B), and to the up-regulation of the p53 protein from basal levels (Fig. 1 B and E). Treatment of these fibroblasts with irinotecan or etoposide, chemotherapeutic agents that induce DNA double-strand breaks, also induced phosphorylation of H2AX (Fig. 1D).
Fig. 1.
Impaired DNA damage response in cells lacking hTERT. (A) Effects of hTERT suppression on H2AX phosphorylation. BJ fibroblasts expressing the indicated shRNA vectors were irradiated (10 Gy), incubated for 1 h, fixed, and stained with an anti-γ-H2AX Ab. (Magnification: ×400.) (B) Expression of DNA damage proteins. BJ cells stably expressing the indicated shRNA vectors were irradiated (10 Gy), incubated, and lysed. Whole cell lysates (100 μg) were resolved by SDS/PAGE and immunoblotted with the indicated antibodies. pBRCA1, phosphorylated BRCA1 (Ser-1497). (C) Autophosphorylation of ATM. BJ cells expressing or lacking hTERT were irradiated as in B, and the amounts of phosphorylated (ATM Ser-1981-P) and total ATM protein were determined by immunoblotting. (D) BJ cells stably expressing the indicated shRNA vectors were treated with the chemotherapeutic drugs (10 μM) for 4 h, and immunoblotting on whole cell lysates (100 μg) was performed. (E) DNA damage response in WI38 fibroblasts. WI38 cells expressing the indicated shRNA vectors were irradiated as in B.
Surprisingly, exposure of parallel cultures of fibroblasts stably expressing either an hTERT-coding sequence-specific shRNA (hTERT shRNA) or an hTERT 3′untranslated region-specific shRNA (hTERT 3′ UTR shRNA) (Fig. 5) to ionizing radiation, irinotecan, or etoposide failed to induce a similar degree of H2AX phosphorylation (Fig. 1 A, B, D, and E) or accumulation of Nijmegan Breakage Syndrome (NBS-1) in nuclear foci (data not shown). In addition, the autophosphorylation of ATM was diminished (Fig. 1C), and we failed to observe the phosphorylation of BRCA1 or the stabilization of p53 protein levels in cells lacking hTERT expression (Fig. 1 B and E). These findings indicate that the DNA damage response in cells lacking hTERT is impaired. Expression of WT hTERT, which is resistant to the effects of the hTERT 3′ UTR-specific shRNA (Fig. 5), in cells expressing this shRNA rescued telomerase activity (Fig. 2G) and permitted cells to respond to DNA damage (Fig. 1 B–D). We also found that fibroblasts expressing a catalytically inactive hTERT mutant (DN hTERT), which inhibits the catalytic activity of telomerase (16), also showed an impaired DNA damage response (Fig. 6, which is published as supporting information on the PNAS web site). Thus, chronic loss of hTERT function either by RNA interference or catalytic inhibition abrogates the cellular response to DNA damage, implicating hTERT as a critical regulator of the DNA damage response pathway.
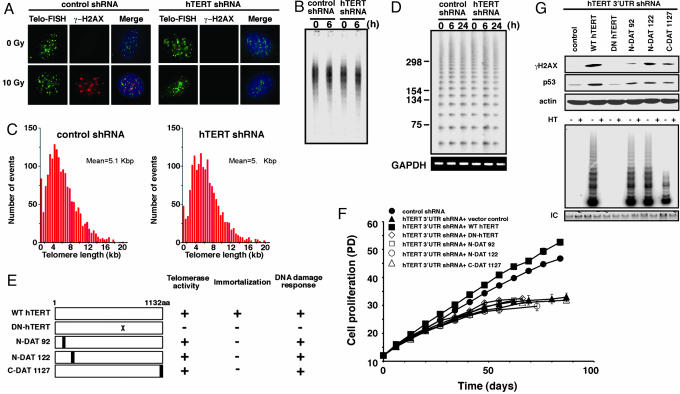
Fig. 2.
DNA damage, telomeres, and suppression of hTERT. (A) Colocalization with γ-H2AX and telomeres. BJ fibroblasts expressing the indicated shRNA were exposed to ionizing radiation (10 Gy) and incubated for 1 h. Fixed cells were hybridized with an FITC-conjugated, telomere-specific peptide nucleic acid probe. After FISH, cells were stained with an anti-γ-H2AX Ab (red) and DAPI (blue). We note that 7% of γ-H2AX foci colocalized with telomeres. The average number of telomere signals and γ-H2AX foci per cell was 23 ± 4.1 and 7.6 ± 2.1, respectively. The average number of colocalized telomeres and γ-H2AX foci per cell was 0.5 ± 0.6; however, in half of the cells analyzed, no colocalization of telomeres and γ-H2AX foci was observed. (Magnification: ×1,000.) (B) Effects of ionizing radiation on telomere length. BJ cells expressing either a control shRNA or an hTERT-specific shRNA were irradiated (5 Gy); genomic DNA was isolated immediately (0 h) or 6 h later; and telomere length was determined by Southern blotting for telomere restriction fragments (TRF). (C) Q-FISH telomere length analysis. For each cell line, at least 400 chromosomes were analyzed; shown is the mean fluorescence intensity correlated to telomere length. Because the cells used in this study arrest after irradiation, Q-FISH could not be performed on irradiated cells. No significant difference in the number of telomere ends lacking a fluorescence signal was observed in either of these cell populations. (D) Effects of irradiation on telomeric single-stranded overhangs. BJ cells expressing the indicated shRNA were irradiated (5 Gy); genomic DNA was isolated immediately (0 h) or at the indicated time points; and the telomeric 3′-overhang ligation assay (T-OLA) was performed. Molecular weight markers are noted in nucleotides. PCR for GAPDH confirmed that equivalent amounts of DNA were analyzed in each lane. (E) Schematic summary of hTERT mutants. X represents substitution of Asp and Val residues at positions 731 and 732 with Ala and Ile. The black bars represent sites where the endogenous hTERT sequence was substituted with the peptide sequence NAAIRS. (F) Effects of hTERT mutant expression on the replicative lifespan in cells that lack endogenous hTERT expression by suppression with a 3′ UTR hTERT-specific shRNA. The symbols representing the proliferation of BJ cells expressing each combination of shRNA and hTERT expression constructs are shown in the panel. The means ± SD for three determinations are shown. In some cases, the symbol covers the error bars. PD, population doubling. (G) Effects of hTERT mutant expression on the DNA damage response in cells that lack endogenous hTERT. BJ cells expressing a 3′ UTR hTERT-specific shRNA together with a control vector (control), WT hTERT, DN hTERT, N-DAT92, N-DAT122, or C-DAT1127 were irradiated (10 Gy), incubated for 1 h, and lysed. Immunoblotting was performed on whole cell lysates (100 μg). Telomerase activity was measured by using the telomere-repeat amplification protocol (TRAP) assay. Although N-DAT92 shows telomerase activity in the TRAP assay, this mutant exhibits diminished activity in non-PCR based assays (21). HT refers to heat-treated samples. IC refers to the internal PCR control for the TRAP assay.
Although overexpression of hTERT stabilizes telomere length in human cells (6), over the short time periods encompassed by these experiments, we did not detect alterations in overall telomere length (Fig. 2 A–C) or changes in the length of the 3′ telomeric single-stranded overhang (13) (Fig. 2D) either before or after irradiation of cells expressing an hTERT-specific shRNA as compared with cells expressing a control shRNA. Moreover, we noted that only 7% of nuclear foci containing γ-H2AX colocalized with telomeres after treatment with ionizing radiation (Fig. 2A), confirming recent observations (17). Although we failed to detect changes in overall telomere length, it remains possible that loss of hTERT exacerbates t-loop deletions produced by homologous recombination (18). Moreover, although suppression of hTERT expression induces premature entry into senescence in human fibroblasts (at population doubling 50–60) (11), we performed these studies in parallel, exponentially dividing cultures at early passage (population doubling 12) to ensure that any differences in the DNA damage response observed in senescent cells did not contribute to these experiments. Indeed, in unirradiated cells, we failed to identify evidence of karyotypic abnormalities in cells expressing either the control shRNA or an hTERT-specific shRNA before irradiation (See legend to Table 1), confirming that the suppression of hTERT in early passage fibroblasts does not, by itself, result in immediate telomere dysfunction.
Table 1. Statistical analysis of cytogenetic abnormalities.
| Comparison group | Comparison values | P value |
|---|---|---|
| No. of fragments per metaphase | ||
| WT hTERT vs. vector control | 0.524 vs. 0.718 | 0.41 |
| Vector control vs. hTERT shRNA | 0.718 vs. 1.31 | 0.02 |
| WT hTERT vs. hTERT shRNA | 0.524 vs. 1.31 | 0.008 |
| Proportion of normal metaphases | ||
| WT hTERT vs. vector control | 0.619 vs. 0.462 | 0.25 |
| Vector control vs. hTERT shRNA | 0.462 vs. 0.241 | 0.058 |
| WT hTERT vs. hTERT shRNA | 0.619 vs. 0.241 | 0.008 |
Metaphase chromosomes were examined at 24 h after exposure to ionizing radiation (5 Gy) from BJ cells expressing a control vector, an hTERT-specific shRNA, or WT hTERT. P values were obtained by applying the Wilcoxon rank-sum test. Comparison values were calculated by dividing the number of the indicated findings by the number of metaphases examined. We also observed that total number of cytogenetic abnormalities found in hTERT shRNA-expressing cells was slightly increased when compared with vector control cells (P = 0.09). The number of metaphase cells examined was as follows: cells expressing WT hTERT (21 cells), cells expressing the hTERT-specific shRNA (29 cells), and cells expressing a control vector (40 cells). Examination of metaphases from unirradiated cells expressing the control shRNA (13 metaphases) or the hTERT-specific shRNA (15 metaphases) revealed no fragments.
To determine further whether the telomere elongation function of hTERT was required for the DNA damage response, we introduced several hTERT mutants into cells in which the endogenous hTERT was suppressed by the expression of the hTERT 3′ UTR-specific shRNA. Specifically, we expressed hTERT mutants that harbor mutations in the amino (N)- and carboxyl (C)-terminal DAT (dissociates activities of telomerase) domains (N-DAT92, N-DAT122, and C-DAT1127) as well as the DN hTERT mutant (Fig. 2E) (16, 19–21). These DAT mutants have previously been shown to reconstitute telomerase biochemical activity yet fail to elongate telomeres or to confer an immortal phenotype when expressed in human cells (19–21). We confirmed that these hTERT mutants exhibited telomerase activity (Fig. 2G) and failed to rescue the premature senescence phenotype found in human fibroblasts that lack endogenous hTERT expression (Fig. 2F) (11). Despite this defect in telomere maintenance, these hTERT mutants restored the ability of human fibroblasts to phosphorylate H2AX and stabilize p53 after exposure to ionizing radiation (Fig. 2G). We note that the N-DAT92 and C-DAT1127 only partially rescue the DNA damage response (Fig. 2G). The C-DAT1127 mutant shows less telomerase activity in the telomere-repeat amplification protocol (TRAP) assay, whereas the N-DAT92 mutant also exhibits catalytic defects when assessed in telomerase assays that are not based on PCR amplification (21). Consistent with this finding, the catalytically inactive DN hTERT mutant failed to rescue DNA damage responses in cells expressing the hTERT 3′ UTR-specific shRNA (Fig. 2G). Therefore, the effect of hTERT suppression on the DNA damage response does not seem to be related primarily to effects on overall telomere length.
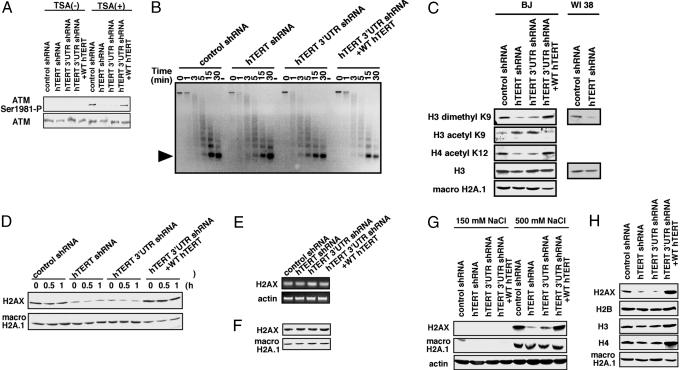
Autophosphorylation of ATM occurs rapidly in response to changes in chromatin structure induced by exposure to ionizing radiation, as well as to agents such as trichostatin A (TSA), even in the absence of DNA double-strand breaks (22). To determine whether chronic hTERT suppression also affected the activation of ATM after treatment with TSA independently of the DNA damage induced by ionizing radiation, we treated cells that transiently express or constitutively lack hTERT and found that TSA-induced ATM phosphorylation was also significantly impaired in cells that constitutively lack hTERT expression (Fig. 3A). These findings suggest that suppression of hTERT expression modulates overall chromatin architecture. To investigate this possibility further, we treated nuclear preparations from cells expressing or lacking hTERT with micrococcal nuclease (MN). We found that chromatin derived from cells lacking hTERT was slightly more susceptible to MN digestion, particularly at earlier time points compared with control cell lines (Fig. 3B). Although others have reported that shorter telomeres exhibit an unusual telomeric chromatin pattern (23), these experiments were performed with cells harboring similar telomere lengths (Fig. 2 B and C), making it unlikely that these effects are limited to telomeric heterochromatin.
Fig. 3.
Suppressing hTERT expression alters chromatin state. (A) Effects of hTERT suppression on chromatin alterations induced by TSA. Cells were treated with TSA (10 μM) for 8 h. Phosphorylated ATM and total ATM protein levels were determined by immunoblotting. (B) MN digestion of nuclei derived from cells expressing the indicated shRNA vectors. Nuclei isolated from 1 × 106 cells were treated with MN for the indicated time, subjected to gel electrophoresis, and stained with ethidium bromide. The arrowhead indicates the migration of mononucleosomes. (C) Histone tail modifications. BJ cells expressing the indicated shRNA were lysed in radioimmunoprecipitation assay (RIPA), and immunoblotting was performed. (D) Extraction of H2AX from chromatin. BJ cells expressing the indicated shRNA were irradiated (10 Gy), incubated for the indicated time, and lysed with RIPA buffer, and immunoblotting was performed on whole cell lysates (100 μg). (E) H2AX mRNA expression. Total RNA (500 ng) was used for RT-PCR with primers specific for H2AX and β-actin. (F) Precipitation of H2AX from chromatin under acidic conditions from BJ cells expressing the indicated shRNA vectors. (G) Extraction of histones under low and high ionic strength. Cells were lysed with low salt buffer and high salt buffer and immunoblotted as indicated. (H) Extraction of core histones from chromatin. BJ cells expressing the indicated shRNA were lysed in RIPA buffer, and immunoblotting was performed on whole cell lysates (100 μg).
Because MN treatment provides a nonspecific measure of chromatin structure and the differences we observed were subtle, we also investigated whether particular posttranslational modifications of histone tails were also affected by hTERT suppression. We found decreased levels of histone H3-lysine (K) 9 dimethylation and increased amounts of H3-K9 acetylation in cells lacking hTERT (Fig. 3C). The heterochromatic proteins 1 (HP1) associate with di- and tri-methylated but not acetylated forms of H3-K9 to form heterochromatin (24). In assessing other histone modifications that may be important for heterochromatin organization, we noted that the degree of H4-K12 acetylation was also decreased in cells lacking hTERT expression (Fig. 3C). The combination of decreased H3-K9 dimethylation and H4-K12 acetylation is reminiscent of that seen in Suv39h histone methyltransferase-deficient cells, which also exhibit impaired genomic stability (25).
Phosphorylation of H2AX plays an important role in the response to DNA damage and is involved in both homologous recombination and nonhomologous end joining (26, 27). Because we noted that suppression of hTERT expression alters overall chromatin structure and impaired DNA damage responses including H2AX phosphorylation, we examined total H2AX levels in fibroblasts expressing the control or either of the two hTERT-specific shRNAs. When we lysed cells in detergent-based buffers over a wide range of salt concentrations, we detected 75% less H2AX protein in whole cell lysates derived from cells lacking hTERT (Fig. 3 D and G). This decrease in soluble H2AX was not the result of altered H2AX expression (Fig. 3E) but instead correlated with enhanced association of H2AX with the insoluble cell fraction. Indeed, when we precipitated whole cell proteins under acidic conditions, we were able to recover equal amounts of H2AX in cells that expressed or lacked hTERT (Fig. 3F). In contrast, we failed to detect differences in the amounts of soluble macro H2A.1 H2B, H3, and H4 in cells that expressed or lacked hTERT expression (Fig. 3 G and H), although we note that we consistently found slightly increased levels of H3 and H4 in cells overexpressing hTERT (Fig. 3H). These observations indicate that chronic suppression of hTERT expression not only alters the overall chromatin architecture but also disturbs H2AX solubility. Taken together, these findings suggest that sustained loss of hTERT expression through several cycles of cell division alters the overall state of chromatin into a configuration that inhibits the activation of the DNA damage response.
Because loss of even one copy of H2AX (26, 27) dramatically impairs the DNA damage response and affects genome stability, we ascertained the functional consequences of treating human fibroblasts unable to express hTERT with ionizing radiation. We noted that cells expressing either of the two hTERT-specific shRNAs showed a substantial increase in their sensitivity to ionizing radiation, as assessed in clonogenic growth assays (Fig. 4A). Coexpression of WT hTERT in cells expressing the hTERT 3′ UTR-specific shRNA rescued this increased sensitivity to ionizing irradiation (Fig. 4A). In consonance with these findings, we found that stable suppression of hTERT expression also altered the capacity of these cells to repair DNA after treatment with ionizing radiation, as assessed by the electrophoretic migration rates of genomic DNA into pulse-field agarose gels (Fig. 4B). Finally, irradiated human cells lacking hTERT expression rapidly accumulated statistically significant increased numbers of chromosomal fragments compared with cells expressing hTERT either transiently (vector control) or constitutively (WT hTERT) (Table 1). These findings demonstrate that hTERT plays a functionally important role in allowing cells to repair genotoxic damage.
Discussion
Several lines of evidence now indicate that telomerase, together with other telomere-specific binding proteins, maintains telomere heterochromatin, thereby preventing telomere degradation and the activation of the DNA damage response pathway (8). Prior work in both budding yeast (28–30) and mammalian cells (31, 32) indicates that chromosome breaks at locations distinct from telomeres are occasionally repaired by telomere addition; however, this mechanism for chromosome healing occurs much less frequently than other forms of DNA repair. The observations presented herein implicate hTERT as a regulator of the DNA damage response pathway through its actions on chromatin structure. Surprisingly, hTERT seems to participate in chromatin maintenance in a manner different from its known role in telomere length maintenance. Although we failed to find evidence of significant telomere loss in cells lacking hTERT, we cannot eliminate the possibility that chronic hTERT suppression renders cells incapable of responding to DNA damage such as has been observed in yeast after elimination of a single telomere (33). However, because hTERT is transiently up-regulated in S-phase and fails to maintain overall telomere length in normal human cells (11), the observations presented here suggest that this regulated expression of hTERT plays an important role in resetting chromatin during DNA replication.
Moreover, these findings establish a direct connection between the mechanisms that maintain telomeres and those that sense and repair breaks in intrachromosomal DNA. Indeed, because the telomere binding protein TRF2 seems to localize transiently to DNA breaks (34) and many proteins implicated in the response to DNA damage associate with telomeres (35), these observations corroborate accumulating evidence that the mechanisms that maintain telomeres and repair DNA breaks are intimately related. Although the experiments presented here were performed in exponentially proliferating, early passage cells, recent work also indicates that senescent cells accumulate heterochromatic foci (36) and activate the DNA damage response (5). Because loss of hTERT function in normal fibroblasts hastens entry into a senescent-like state (11), telomerase may thus facilitate immortalization through effects on both telomeric and nontelomeric chromatin.
Genetically manipulated mice lacking the telomerase RNA component mTerc also show impaired responses to agents that damage DNA; however, the effects of mTerc deletion are apparent only in late generation mTerc-null mice that show significant telomere shortening and dysfunction (14, 37). Although mice lacking the telomerase catalytic subunit mTert are viable, such mice are unable to elongate (38) or maintain telomeres (39). The effects of mTert deficiency on chromatin structure and DNA damage responses have not yet been reported; however, because regulation of chromatin plays a critical role in mammalian development (40), developmental compensation, as has been observed in mice lacking the retinoblastoma gene (41), may also occur in mice lacking mTert, masking the effects of germ-line mTert loss on chromatin structure and DNA damage. Interestingly, some investigators have reported that mice lacking one mTert allele maintain shorter telomeres and exhibit increased genomic instability than control mice (42), suggesting that haploinsufficiency at the mTert locus impairs telomerase function.
In addition, despite harboring long telomeres and basal telomerase activity, murine tumors show evidence of increased telomerase activity (43). Because recent work in both transgenic mice and human cells suggests that increased TERT expression contributes to malignant transformation even in cells harboring long telomeres (9), the effects of hTERT on chromatin may provide a plausible mechanism for such additional functions of hTERT in maintaining chromosomal stability and suggest how TERT may contribute to cell transformation independently of its effects on telomere maintenance.
Supplementary Material
Acknowledgments
We thank E. Chavez for assistance with the Q-FISH analysis and R. A. DePinho, D. M. Livingston, R. Maser, S. Murakami, Y. Nakatani, R. A. Weinberg, K. Wong, and members of the W.C.H. laboratory for comments and discussion. We thank T. Pandita (Washington University, St. Louis) for the gift of the polyclonal anti-hTERT antibody. This work was supported in part by National Institutes of Health Grants K01 CA94223 and R01 AG023145 (to W.C.H.), a Doris Duke Clinical Scientist Development Award (to W.C.H.), a Claudia Adams Barr Research Award (to W.C.H.), a Gillette Fellowship (to S.G.), a Uehara Memorial Foundation Research Fellowship (to K.M.), a Charles A. King Trust Postdoctoral Fellowship (to K.M.), a Howard Hughes Medical Institute Predoctoral Fellowship (to R.P.), a National Cancer Institute of Canada Fellowship (to J.M.Y.W.), and a Research Scholar Grant from the American Cancer Society (to K.C.).
Author contributions: K.M., R.P., K.C., and W.C.H. designed research; K.M., R.P., J.M.Y.W., J.L.C., Z.T., and S.G. performed research; K.M., R.P., Z.T., J.B.M., S.G., P.M.L., K.C., and W.C.H. analyzed data; J.M.Y.W., Z.T., J.B.M., S.G., P.M.L., K.C., and W.C.H. contributed new reagents/analytic tools; and K.M., R.P., and W.C.H. wrote the paper.
Abbreviations: Q-FISH, quantitative FISH; MN, micrococcal nuclease; TERT, telomerase reverse transcriptase; hTERT, human TERT; DN hTERT, catalytically inactive hTERT mutant; shRNA, short hairpin RNA; ATM, ataxia–telangiectasia-mutated; BRCA1, breast cancer-associated 1; DAT, dissociates activities of telomerase; TSA, trichostatin A.
References
- 1.Wright, W. E. & Shay, J. W. (2001) Curr. Opin. Genet. Dev. 11, 98-103. [DOI] [PubMed] [Google Scholar]
- 2.Serrano, M., Lin, A. W., McCurrach, M. E., Beach, D. & Lowe, S. W. (1997) Cell 88, 593-602. [DOI] [PubMed] [Google Scholar]
- 3.Parrinello, S., Samper, E., Krtolica, A., Goldstein, J., Melov, S. & Campisi, J. (2003) Nat. Cell Biol. 5, 741-747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Schmitt, C. A., Fridman, J. S., Yang, M., Lee, S., Baranov, E., Hoffman, R. M. & Lowe, S. W. (2002) Cell 109, 335-346. [DOI] [PubMed] [Google Scholar]
- 5.d'Adda di Fagagna, F., Reaper, P. M., Clay-Farrace, L., Fiegler, H., Carr, P., Von Zglinicki, T., Saretzki, G., Carter, N. P. & Jackson, S. P. (2003) Nature 426, 194-198. [DOI] [PubMed] [Google Scholar]
- 6.Bodnar, A. G., Ouellette, M., Frolkis, M., Holt, S. E., Chiu, C. P., Morin, G. B., Harley, C. B., Shay, J. W., Lichtsteiner, S. & Wright, W. E. (1998) Science 279, 349-352. [DOI] [PubMed] [Google Scholar]
- 7.Vaziri, H. & Benchimol, S. (1998) Curr. Biol. 8, 279-282. [DOI] [PubMed] [Google Scholar]
- 8.Chan, S. W. & Blackburn, E. H. (2002) Oncogene 21, 553-563. [DOI] [PubMed] [Google Scholar]
- 9.Blasco, M. A. & Hahn, W. C. (2003) Trends Cell Biol. 13, 289-294. [DOI] [PubMed] [Google Scholar]
- 10.Kang, H. J., Choi, Y. S., Hong, S. B., Kim, K. W., Woo, R. S., Won, S. J., Kim, E. J., Jeon, H. K., Jo, S. Y., Kim, T. K., et al. (2004) J. Neurosci. 24, 1280-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Masutomi, K., Yu, E. Y., Khurts, S., Ben-Porath, I., Currier, J. L., Metz, G. B., Brooks, M. W., Kaneko, S., Murakami, S., DeCaprio, J. A., et al. (2003) Cell 114, 241-253. [DOI] [PubMed] [Google Scholar]
- 12.Martens, U. M., Zijlmans, J. M., Poon, S. S., Dragowska, W., Yui, J., Chavez, E. A., Ward, R. K. & Lansdorp, P. M. (1998) Nat. Genet. 18, 76-80. [DOI] [PubMed] [Google Scholar]
- 13.Stewart, S. A., Ben-Porath, I., Carey, V. J., O'Connor, B. F., Hahn, W. C. & Weinberg, R. A. (2003) Nat. Genet. 33, 492-496. [DOI] [PubMed] [Google Scholar]
- 14.Wong, K. K., Chang, S., Weiler, S. R., Ganesan, S., Chaudhuri, J., Zhu, C., Artandi, S. E., Rudolph, K. L., Gottlieb, G. J., Chin, L., et al. (2000) Nat. Genet. 26, 85-88. [DOI] [PubMed] [Google Scholar]
- 15.Barch, M. J., Knutsen, T. & Spurbeck, J. L. The AGT Cytogenetics Laboratory Manual (1997) (Lippincott–Raven, Philadelphia).
- 16.Hahn, W. C., Stewart, S. A., Brooks, M. W., York, S. G., Eaton, E., Kurachi, A., Beijersbergen, R. L., Knoll, J. H., Meyerson, M. & Weinberg, R. A. (1999) Nat. Med. 5, 1164-1170. [DOI] [PubMed] [Google Scholar]
- 17.Sedelnikova, O. A., Horikawa, I., Zimonjic, D. B., Popescu, N. C., Bonner, W. M. & Barrett, J. C. (2004) Nat. Cell Biol. 6, 168-170. [DOI] [PubMed] [Google Scholar]
- 18.Wang, R. C., Smogorzewska, A. & de Lange, T. (2004) Cell 119, 355-368. [DOI] [PubMed] [Google Scholar]
- 19.Armbruster, B. N., Banik, S. S., Guo, C., Smith, A. C. & Counter, C. M. (2001) Mol. Cell. Biol. 21, 7775-7786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Banik, S. S., Guo, C., Smith, A. C., Margolis, S. S., Richardson, D. A., Tirado, C. A. & Counter, C. M. (2002) Mol. Cell. Biol. 22, 6234-6246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lee, S. R., Wong, J. M. & Collins, K. (2003) J. Biol. Chem. 278, 52531-52536. [DOI] [PubMed] [Google Scholar]
- 22.Bakkenist, C. J. & Kastan, M. B. (2003) Nature 421, 499-506. [DOI] [PubMed] [Google Scholar]
- 23.Tommerup, H., Dousmanis, A. & de Lange, T. (1994) Mol. Cell. Biol. 14, 5777-5785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Grewal, S. I. & Rice, J. C. (2004) Curr. Opin. Cell Biol. 16, 230-238. [DOI] [PubMed] [Google Scholar]
- 25.Peters, A. H., O'Carroll, D., Scherthan, H., Mechtler, K., Sauer, S., Schofer, C., Weipoltshammer, K., Pagani, M., Lachner, M., Kohlmaier, A., et al. (2001) Cell 107, 323-337. [DOI] [PubMed] [Google Scholar]
- 26.Celeste, A., Difilippantonio, S., Difilippantonio, M. J., Fernandez-Capetillo, O., Pilch, D. R., Sedelnikova, O. A., Eckhaus, M., Ried, T., Bonner, W. M. & Nussenzweig, A. (2003) Cell 114, 371-383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bassing, C. H., Suh, H., Ferguson, D. O., Chua, K. F., Manis, J., Eckersdorff, M., Gleason, M., Bronson, R., Lee, C. & Alt, F. W. (2003) Cell 114, 359-370. [DOI] [PubMed] [Google Scholar]
- 28.Kramer, K. M. & Haber, J. E. (1993) Genes Dev. 7, 2345-2356. [DOI] [PubMed] [Google Scholar]
- 29.Myung, K., Datta, A. & Kolodner, R. D. (2001) Cell 104, 397-408. [DOI] [PubMed] [Google Scholar]
- 30.Stellwagen, A. E., Haimberger, Z. W., Veatch, J. R. & Gottschling, D. E. (2003) Genes Dev. 17, 2384-2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Flint, J., Craddock, C. F., Villegas, A., Bentley, D. P., Williams, H. J., Galanello, R., Cao, A., Wood, W. G., Ayyub, H. & Higgs, D. R. (1994) Am. J. Hum. Genet. 55, 505-512. [PMC free article] [PubMed] [Google Scholar]
- 32.Sprung, C. N., Reynolds, G. E., Jasin, M. & Murnane, J. P. (1999) Proc. Natl. Acad. Sci. USA 96, 6781-6786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sandell, L. L. & Zakian, V. A. (1993) Cell 75, 729-739. [DOI] [PubMed] [Google Scholar]
- 34.Bradshaw, P. S., Stavropoulos, D. J. & Meyn, M. S. (2005) Nat. Genet. 37, 193-197. [DOI] [PubMed] [Google Scholar]
- 35.D'Adda Di Fagagna, F., Teo, S. H. & Jackson, S. P. (2004) Genes Dev. 18, 1781-1799. [DOI] [PubMed] [Google Scholar]
- 36.Narita, M., Nunez, S., Heard, E., Lin, A. W., Hearn, S. A., Spector, D. L., Hannon, G. J. & Lowe, S. W. (2003) Cell 113, 703-716. [DOI] [PubMed] [Google Scholar]
- 37.Goytisolo, F. A., Samper, E., Martin-Caballero, J., Finnon, P., Herrera, E., Flores, J. M., Bouffler, S. D. & Blasco, M. A. (2000) J. Exp. Med. 192, 1625-1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chiang, Y. J., Hemann, M. T., Hathcock, K. S., Tessarollo, L., Feigenbaum, L., Hahn, W. C. & Hodes, R. J. (2004) Mol. Cell. Biol. 24, 7024-7031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuan, X., Ishibashi, S., Hatakeyama, S., Saito, M., Nakayama, J., Nikaido, R., Haruyama, T., Watanabe, Y., Iwata, H., Iida, M., et al. (1999) Genes Cells 4, 563-572. [DOI] [PubMed] [Google Scholar]
- 40.Meehan, R. R. (2003) Semin. Cell Dev. Biol. 14, 53-65. [DOI] [PubMed] [Google Scholar]
- 41.Sage, J., Miller, A. L., Perez-Mancera, P. A., Wysocki, J. M. & Jacks, T. (2003) Nature 424, 223-228. [DOI] [PubMed] [Google Scholar]
- 42.Erdmann, N., Liu, Y. & Harrington, L. (2004) Proc. Natl. Acad. Sci. USA 101, 6080-6085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blasco, M. A., Rizen, M., Greider, C. W. & Hanahan, D. (1996) Nat. Genet. 12, 200-204. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.