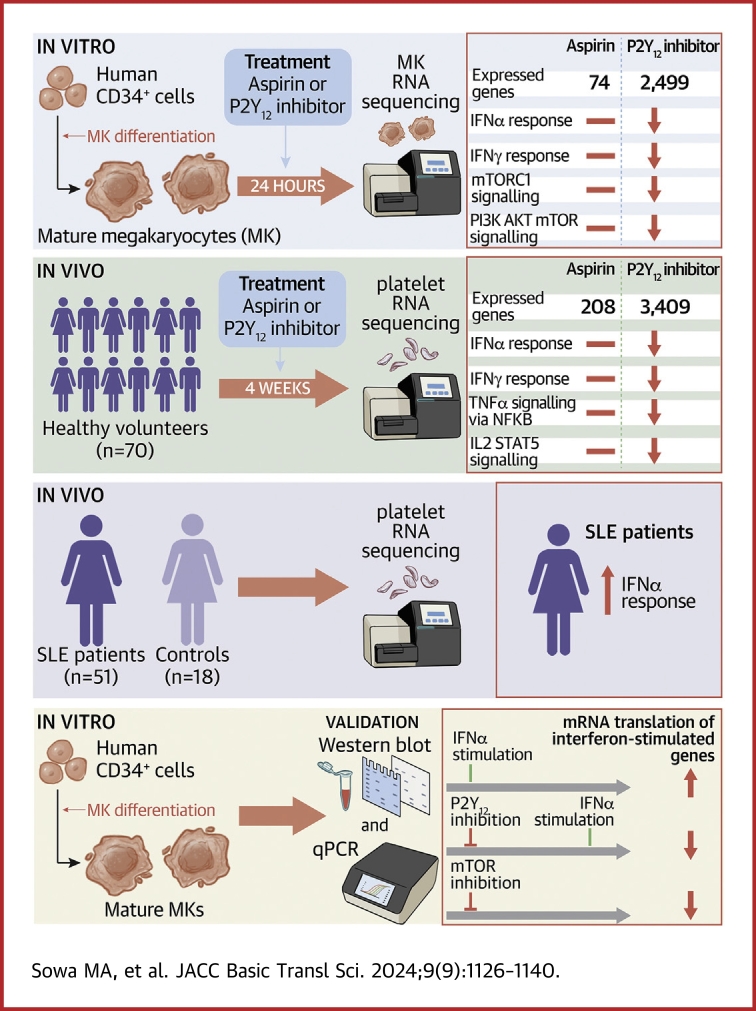
Visual Abstract
Key Words: antiplatelet therapy, megakaryocytes, platelet inhibitor, transcriptomics
Highlights
-
•
Platelets and MKs are implicated in inflammation and immunity, prompting the nonhemostatic investigation of platelet-directed therapies.
-
•
P2Y12 inhibitors, distinct from ASA, attenuate the IFNα response pathway in both MKs and platelets. Notably, the IFNα pathway is increased in platelets isolated from patients with SLE. P2Y12 inhibitors could hold therapeutic potential for inhibiting platelet-mediated inflammation in proinflammatory conditions like SLE.
-
•
Using MKs as an in vitro model to study platelet transcriptome dynamics offers a powerful approach to understand the complex biology of platelets.
-
•
Exploring the clinical applicability of P2Y12 inhibition in targeting inflammatory and autoimmune diseases and investigating combination therapies for enhanced efficacy provides new insights into reducing platelet-mediated thromboinflammation and improving patient outcomes.
Summary
The authors investigated the impact of antiplatelet therapy on the megakaryocyte (MK) and platelet transcriptome. RNA-sequencing was performed on MKs treated with aspirin or P2Y12 inhibitor, platelets from healthy volunteers receiving aspirin or P2Y12 inhibition, and platelets from patients with systemic lupus erythematosus (SLE). P2Y12 inhibition reduced gene expression and inflammatory pathways in MKs and platelets. In SLE, the interferon (IFN) pathway was elevated. In vitro experiments demonstrated the role of P2Y12 inhibition in reducing IFNα-induced platelet-leukocyte interactions and IFN signaling pathways. These results suggest that P2Y12 inhibition may have therapeutic potential for proinflammatory and autoimmune conditions like SLE.
Platelets are the smallest cells circulating in the blood, which play an important role in wound healing and blood clot formation. During vessel injury, platelets are involved in the first wave of hemostasis, forming a hemostatic plug to attenuate bleeding. Despite its essential role in maintaining hemostasis, platelet hyper-reactivity is associated with thrombotic disorders, including myocardial infarction and stroke.1 Commonly prescribed antiplatelet therapies (APT) for the prevention of atherothrombosis are aspirin (ASA) and P2Y12 receptor antagonists (eg, clopidogrel, prasugrel, ticagrelor).2
Megakaryocytes (MKs) are large polyploidic blood cells produced in the bone marrow and lungs.3 In their mature state, MKs give rise to anucleate platelets, which retain significant amounts of MK-derived transcripts and proteins that regulate platelet function.4 Primary MKs can be generated in vitro by differentiating CD34+ hematopoietic stem cells in the presence of thrombopoietin (TPO).4,5 Culture-derived MKs are routinely used as nucleate platelet surrogates6, 7, 8 and have the potential to provide regulatory insight into the pleiotropic effects of platelet-directed therapies.
The interferon (IFN) pathway is a crucial component of the innate immune response that triggers various antiviral mechanisms, activates immune cells, and helps regulate the balance between pro- and anti-inflammatory processes.9,10 Patients with systemic lupus erythematosus (SLE) are characterized by an activation of type I IFN signaling, which leads to the increased expression of IFN-stimulated genes (ISGs).11,12 Previous findings from our group demonstrated up-regulation of ISGs in the platelet transcriptome from patients with SLE, with the IFNα- and IFNγ-response pathways showing robust enrichment.13,14 In lupus-prone mice, administration of P2Y12 inhibitor reduced lupus-like disease severity and improved overall survival.15 Despite these intriguing studies, the relation between the IFN pathway and P2Y12 inhibition is unknown.
This study was designed to investigate the effect of APT on the MK and platelet transcriptome and its impact on proinflammatory pathways. We demonstrated a significant effect on the MK and platelet transcriptome following P2Y12 receptor inhibition. Specifically, we found a significant down-regulation of IFN pathways with P2Y12 receptor inhibition. In addition, we noted a negative correlation in ISGs between the platelet transcriptome from patients with SLE (vs control patients) and transcriptomes of MKs and platelets treated with P2Y12 inhibitors. Finally, these results were validated in vitro, demonstrating the suppressive role of P2Y12 inhibition on the expression of ISG in IFNα-stimulated MKs and whole blood.
Methods
MK culturing and antiplatelet drug treatment
CD34-positive cells collected from 4 healthy donors (Fred Hutch Cancer Center) were plated in 6-well plates at 2 × 105 cells/mL in StemSpan Serum-Free Expansion Medium II (#09655, STEMCELL Technologies) supplemented with 50 ng/mL recombinant human TPO (#288-TPE-050, R&D Systems), 25 ng/mL recombinant human stem cell factor (#255-SC-050, R&D Systems), 100 U/mL penicillin and 100 μg/mL streptomycin (#15140122, Gibco) and cultured at 37 °C and 5% CO2. Cultures were transferred into fresh TPO-containing media on days 3, 6, and 9.6 On day 10; cultures were labelled with PE Mouse Anti-Human CD42a antibody (#558819, BD Biosciences), and MKs isolated with the EasySep Release Human PE Positive Selection Kit (#17654, STEMCELL Technologies) and then plated onto 12-well plates at a concentration of 2 × 105 cells/mL. On day 11, cells were treated with 100 μmol/L ASA (reconstituted in phosphate-buffered saline; #70260, Cayman Chemical), 5 μmol/L P2Y12 inhibitor AZD1283 (reconstituted in 0.025% dimethyl sulfoxide [DMSO], #27649, Cayman Chemicals), or dual antiplatelet therapy (DAPT) for 24 hours. Phosphate-buffered saline or 0.025% DMSO were vehicle control solutions for ASA and AZD1283/DAPT, respectively. ASA and AZD1283 concentrations chosen effectively inhibit arachidonic acid- or adenosine diphosphate (ADP)-induced platelet aggregation, respectively, as determined by light-transmission aggregometry (Supplemental Figure 1) and as previously reported.16,17 For clinical validation, MKs were additionally treated with 1 μmol/L ticagrelor (#15425, Cayman) or 10 μmol/L (S)-(+)-clopidogrel sulfate (#21002, Cayman) for 24 hours. Following incubation, MKs were lysed in QIAzol Lysis Reagent (#79306, QIAGEN) or radio immunoprecipitation assay (RIPA) buffer and stored at −80 °C until use.
MK IFN stimulation
MKs at day 11 were incubated with 5 μmol/L P2Y12 inhibitor AZD1283 or 0.025% DMSO for 20 hours at 37 °C in a 5% CO2 and then 4 hours with 1,000 U/mL recombinant IFNα (#IF007, Sigma) or phosphate-buffered saline (control solution). After 24 hours, samples were lysed in QIAzolLysis Reagent or RIPA buffer and stored at −80 °C until use.
MK and platelet RNA-seq
Total RNA was isolated using Direct-zol RNA MicroPrep columns (#R2062, Zymo Research). RNA quality and quantity were determined with a Bioanalyzer 2100 (Agilent Technologies). Sequencing libraries were barcoded and prepared using the Clontech SMART-Seq HT with Nxt HT kit (Takara Bio USA), and libraries were sequenced single end (51 bp) on an Illumina NovaSeq 6000 at the Genome Technology Center at New York University Langone Health.
Data processing and quality control were performed using the RNAseq PE snakemake pipeline.18 In short: quality control of sequencing reads was assessed using FastQC (version 0.11.9)19 and MultiQC (version 1.10.1).20 Reads were trimmed using fastp (version 0.20.1)21 and mapped to human reference genome hg38 using STAR (version 2.7.7a).22 Genomic features were then assigned using Subread featureCounts (version 1.6.3).23 The data are available through the Gene Expression Omnibus: GSE242369.
RNA-sequencing (RNA-seq) data from MKs were analyzed in R (version 4.0.2, R Foundation) with the package DESeq2 (version 1.24.0)24 using a linear model that accounts for donor source (∼donor + treatment). Statistical significance was calculated using the Wald test on the treatment factor. Genes with low counts (base mean <30) were excluded from the analysis. Gene set enrichment analysis25,26 of Molecular Signatures Database hallmark gene set27 was performed on genes with P < 0.10 (nominal) using the ClusterProfiler package from BioConductor (version 3.18.1).28,29
Raw and processed RNA-seq data of clinical studies used to compare with MK transcriptome were previously described13,14,30, 31, 32 and are available through the Gene Expression Omnibus (APT—GSE158765; SLE—GSE226147; COVID-19—GSE176480).
Differentially expressed genes were identified independently in each data set. The log2-fold changes (Log2FC) and P values were compared between the data sets.
Clinical cohorts
The crossover study to assess the effects of APT on platelet gene expression was performed at the Duke Clinical Research Unit and has been previously described.30,31 This study was approved by the Duke Institutional Review Board. The cohorts of 51 patients with SLE and 18 matched control subjects as well as 8 patients with COVID-19 and 10 matched control subjects were recruited at New York University Langone Health and have been previously described.13,14,32 SLE and COVID-19 studies were approved by the New York University Grossman School of Medicine Institutional Review Board and performed with a waiver of informed consent. Demographics and clinical characteristics of the cohorts are provided in Supplemental Tables 1 to 3.
Light transmission aggregometry
Whole blood was collected into tubes containing 3.2% sodium citrate. Platelet-rich plasma was obtained by centrifugation of the tubes at 200g for 20 minutes at room temperature. Light transmission aggregometry was performed using an aggregometer AggRAM (Helena Biosciences) in a final volume of 300 μL. Then 260 μL of platelet-rich plasma were added into an AggRAM Cuvette with a stirring bar inside and incubated for 30 minutes with 10 μL of inhibitor (1, 10, 100, or 500 μmol/L ASA; 0.5, 1, 5, 25 μmol/L AZD1283; or 100 μmol/L ASA with 5 μmol/L AZD1283). Under stirring conditions (1,200 rev/min at 37 °C), platelets were then stimulated with 30 μL of agonist: arachidonic acid (0.8 mmol/L) or ADP (10 μmol/L). Light transmission was monitored for 6 minutes. Platelet-poor plasma was used as a blank (100% of aggregation). The same method was used to investigate the effect of other P2Y12 inhibitors, ticagrelor and (S)-(+)-clopidogrel, on ADP-induced platelet aggregation.
Quantitative real-time polymerase chain reaction
To generate cDNA, iScript Reverse Transcription Supermix for RT-qPCR kit (#1708841, Bio-Rad) was used per the manufacturer’s instructions. Quantitative real-time polymerase chain reactions were conducted using fast SYBR Green master mix (#4385614, Applied Biosystems) on a QuantStudio 3 (Applied Biosystems). Primers: IFITM3 5′-TGTCCAAACCTTCTTCTCTCC-3′ and 5′-CGTCGCCAACCATCTTCC-3′; IRF7 5′-GCTGGACGTGACCATCATGTA-3′ and 5′–GGGCCGTATAGGAACGTGC-3′; ISG15 5′-CGCAGATCACCCAGAAGATCG-3′ and 5′-TTCGTCGCATTTGTCCACCA-3′; DHX34 5′-TCCGTGAAGAGGATTACATCCG-3′ and 5′-GGGTCGTAAGTGCGAGGTAG-3′; GAPDH 5′-GCGAGATCCCTCCAAAATCA-3′ and 5′-GACTGTGGTCATGAGTCCTTC-3′. Relative gene expression was calculated using the 2−ΔΔCt method and GAPDH was used as a housekeeping gene to normalize messenger RNA.
MEG-01 cell culturing and treatment
MEG-01 cells were cultured in RPMI-1640 medium (#30-2001, ATTC) supplemented with 10% heat inactivated fetal bovine serum (#16-140-071, Gibco), 100 U/mL penicillin and 100 μg/mL streptomycin at 37 °C and 5% CO2. The cells were plated in 24-well plates at a concentration of 2 × 105 cells/mL and next day treated with 2.5 μmol/L Torin-1 (#10997, Cayman), or 0.1% DMSO (control solution) for 24 hours. After 24 hours of treatment, cells were lysed with cold RIPA buffer and stored at −80° C until use.
Western blot
CD34+-derived MKs or MEG-01 cells were harvested by centrifugation for 5 minutes at 800g and lysed in cold RIPA buffer (#89900, Thermo Fisher Scientific) containing protease inhibitor cocktail (cOmplete Mini, #118361530001, Roche Diagnostics) for 30 minutes at 4 °C. Lysates were subjected to centrifugation for 20 minutes at 15,000g at 4 °C to obtain the supernatant. Total protein concentration was determined using Pierce BCA Protein Assay Kit (#23225, Thermo Fisher Scientific) as per manufacturers’ instructions. Equal protein amounts of the lysates were mixed with LDS Sample buffer (#NP0007, Invitrogen), denatured, and loaded onto 4%-12% Bis-Tris Protein Gels (#NP0341BOX, Invitrogen) along with SeeBlue Plus2 Pre-stained Standard (#LC5925, Invitrogen). Proteins were transferred onto the polyvinylidene difluoride membrane using iBlot2 Transfer stacks (#IB24002, Invitrogen) and iBlot2 Gel Transfer Device (#IB21001, Invitrogen). Membranes were then blocked with 5% (weight/volume) bovine serum albumin in Tris-buffered saline (200 mmol/L Tris base, 1.37 mol/L NaCl, pH 7.6) containing 0.1% (volume/volume) Tween-20 (TBS-T) for 1 hour. The blocked membranes were incubated overnight at 4 °C with primary antibodies. Afterward, the membrane was washed 3 times for 10 minutes with TBS-T, incubated with horseradish peroxidase–conjugated secondary antibodies for 1 hour, and again washed 3 times for 10 minutes with TBS-T. The membranes were developed with Pierce ECL Western blotting substrate (#32109, Thermo Fisher Scientific) and bands were visualized using the ChemiDoc MP imaging system (Bio-Rad). The following primary antibodies were used: anti-IFITM3 (#MA5-32798, Invitrogen), anti-Phospho-S6 Ribosomal Protein (#2211S, Cell Signaling Technology), anti-ISG15 (sc-166755, Santa Cruz). As a loading control, anti-α-tubulin (#sc-53030, Santa Cruz) or anti-GAPDH (P/N 926-42216, LI-COR) were used. As a secondary antibodies, the following horseradish peroxidase–conjugated antibodies were used: goat anti-rat (#31470, Invitrogen), goat anti-mouse (#62-6520, Invitrogen), and goat anti-rabbit (#31460, Invitrogen).
Leukocyte-platelet aggregate formation
A total of 500 μL of whole blood was incubated in the presence of AZD1283 (5 μmol/L) or DMSO (as a control solution) for 30 minutes at room temperature, stimulated for another 30 minutes with recombinant IFNα (250 U/mL) and then fixed with 600 μL of 1% formaldehyde. Then 50 μL of fixed blood was stained with 5 μL CD61-FITC (#130-098-682, Miltenyi Biotec) to identify platelets and 1 μL CD45-VioGreen (#130-110-638, Miltenyi Biotec) and 1 μL CD14-PE-Vio 770 (#130-110-521 Miltenyi Biotec) for 15 minutes. Red blood cells were lysed by adding 200 μL of H2O and incubated for 20 minutes, before adding 450 μL of running buffer. The samples were analyzed using Miltenyi MACSQuant 10 (Miltenyi Biotec). The flow cytometry data were analyzed using FlowJo Software for Windows (version 10.8.1, BD Life Sciences). Leukocyte-platelet aggregates (LPAs) were identified as CD45+CD61+ events and monocyte-platelet aggregates as CD45+CD14+CD61+ events. Neutrophil-platelet aggregates (NPAs) and lymphocyte-platelet aggregates were identified as CD45+CD14−CD61+ and were distinguished by differences in size and granularity.
Statistical analysis
Data are presented as mean ± SD. The transcriptomic analysis utilized nominal P values, unless otherwise indicated. Association between continuous values was determined using Pearson correlation coefficient (r). Two-sided parametric paired Student's t-tests were used for within-group comparisons unless otherwise specified. Normality of data was assessed with the Shapiro–Wilk normality test. One-way analysis of variance was performed when 3 or more groups were compared for 1 variable, followed if significant by Dunnett post hoc multiple comparisons test. P values <0.05 were considered statistically significant. Statistical analyses were performed with GraphPad Prism (version 9.2.0 for Windows, GraphPad Software).
Results
P2Y12 inhibition, but not ASA suppresses IFN signaling pathways in MKs
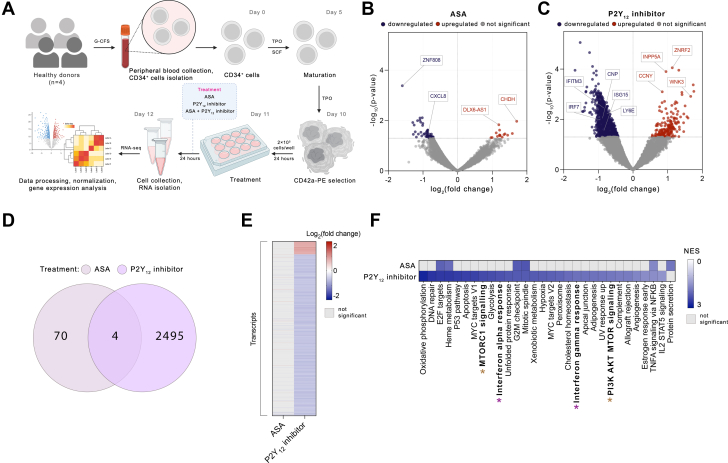
To investigate the global platelet-mediated effect of antiplatelet therapy, we used human MKs as platelet surrogates. Human CD34+-derived MKs were treated with ASA (100 μmol/L), the P2Y12 inhibitor, AZD1283 (5 μmol/L), or both drugs together for 24 hours; RNA was collected; and the transcriptome was assessed by RNA-seq (Figure 1A).
Figure 1.
P2Y12 Inhibitor, But Not ASA Suppresses IFN Signaling Pathways in MKs
(A) Experimental workflow: CD34+ cells were differentiated into megakaryocytes (MKs) for 10 days in the presence of human thrombopoietin (TPO). MKs were purified by selection of CD42a-positive cells. Purified MKs were treated with 100 μmol/L aspirin (ASA), 5 μmol/L AZD1283 (P2Y12 inhibitor), or both drugs together for 24 hours. After 24 hours, cells were collected, RNA was isolated, and RNA-sequencing (RNA-seq) performed. (B, C) Volcano plots of differentially expressed transcripts of MKs treated with ASA (B) or P2Y12 inhibitor (C). Colored dots are P < 0.05; red dots represent up-regulated and blue dots down-regulated genes. (D) Venn diagram and (E) heatmap of differentially expressed genes identified in MKs treated with ASA or P2Y12 inhibitor (P < 0.05). Red color represents up-regulated and blue down-regulated genes. (F) Gene set enrichment analysis of hallmark pathways of MKs treated with ASA or P2Y12 inhibitor (P < 0.05). The purple asterisk highlights interferon (IFN)-related pathways and the brown asterisk MTOR-related pathways. G2M = Gap 2 phase mitosis; G-CSF = granulocyte colony-stimulating factor; NES = Normalized Enrichment Score; NFKB = nuclear factor κB; SCF = stem cell factor. STAT5 = signal transducer and activator of transcription 5; TNFA = tumor necrosis factor α; UV = ultraviolet.
Following incubation of MKs with ASA, 74 transcripts were differentially expressed, with 59 transcripts down- and 15 up-regulated (P < 0.05) (Figures 1B, 1D, and 1E, Supplemental Table 4). Treatment of MKs with the P2Y12 inhibitor had a more robust effect on the MK transcriptome with 2,499 transcripts differentially expressed (2,309 down- and 190 up-regulated; P < 0.05) (Figures 1C to 1E, Supplemental Table 5). We validated the inhibitory effect of AZD1283 on the messenger RNA level using ticagrelor (Supplemental Figure 2). Because DAPT comprising both ASA and a P2Y12 inhibitor is frequently prescribed after myocardial infarction or coronary stent placement for cardiovascular event prevention,33,34 MKs were incubated with both ASA and a P2Y12 inhibitor. Treatment of MKs with DAPT led to the differential expression of 4,192 genes (2,689 down- and 1,503 up-regulated; P < 0.05) (Supplemental Figures 3A to 3C, Supplemental Table 6).
To understand the biological pathways altered by each treatment, we performed gene set enrichment analysis (Figure 1F, Supplemental Figure 3D). Treatment of MKs with ASA led to down-regulation of only 6 pathways (P < 0.05). In contrast, P2Y12 inhibition significantly affected multiple biological pathways, with 29 down-regulated pathways (P < 0.05), and no pathways significantly up-regulated. Among the most down-regulated pathways by P2Y12 inhibition were those associated with immune regulation, including IFNα and IFNγ response pathways (Normalized Enrichment Score [NES] = −2.30, P = 0.002 and NES = −1.95, P = 0.003, respectively) (Figure 1F). DAPT down-regulated 17 biological pathways (P < 0.05), including fatty acid metabolism, estrogen response, and myogenesis (Supplemental Figure 3D).
MK and platelet P2Y12 inhibitor-responsive genes
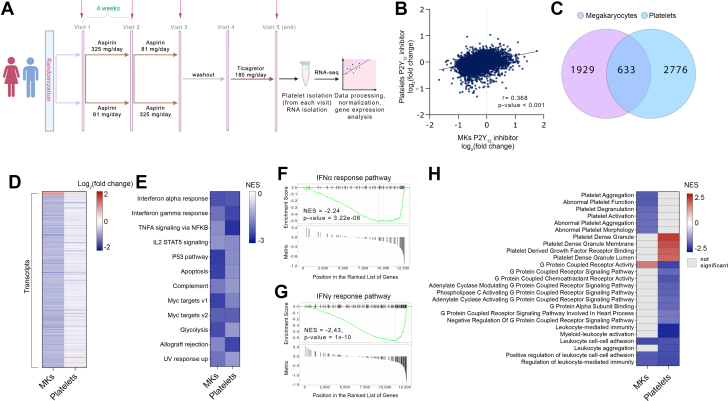
To address whether primary MKs can serve as a transcriptomic surrogate for human circulating platelets, we integrated our in vitro data with platelet sequencing data from a human cohort taking either ASA or ticagrelor (P2Y12 inhibitor) (Figure 2A, Supplemental Tables 4 and 5).30,31
Figure 2.
MK and Platelet P2Y12 Inhibition Responsive Genes
(A) The clinical cohort of healthy subjects (n = 70) began with a baseline visit (visit 1), randomized to either low- or high-dose ASA for 4 weeks each (visit 2), crossed over to the other ASA dose for 4 weeks (visit 3), ASA washout (visit 4), and 4 weeks of P2Y12 inhibitor exposure (visit 5). Red arrows indicate time points of blood collection. (B) Scatterplot of fold change differences comparing MKs treated with P2Y12 inhibitor and P2Y12-inhibited platelets from the clinical cohort. (C) Venn diagram and (D) heatmap of overlapping and differentially expressed genes observed in MKs and platelets (P < 0.05). Red color represents up-regulated and blue down-regulated genes. (E) Gene set enrichment analysis of hallmark pathways, comparison between P2Y12-inhibited pathways in MKs and platelets (P < 0.05). (F, G) Enrichment plots of (F) IFNα- and (G) IFNγ-response pathways in P2Y12-inhibited platelets. Relative gene positions in the ranked list of genes are indicated. P values were determined from the gene set enrichment analysis output. Abbreviations as in Figure 1.
Consistent with our in vitro MK data, P2Y12 inhibition induced a more robust effect than ASA with 3,409 P2Y12-responsive genes differentially regulated (1,589 down- and 1,820 up-regulated).30,31 Integration of our in vitro MK and ex vivo human platelet data yielded a significant correlation between the MK and platelet transcriptomes in response to P2Y12 inhibition (r = 0.368, P < 0.001) (Figure 2B), but no significant correlation between the in vitro MK and human platelet data in response to ASA (Supplemental Figure 4). There was a significant gene overlap of P2Y12 inhibition across MKs (in vitro) and platelets (ex vivo) with 633 overlapping genes (Figures 2C and 2D). Gene set enrichment analysis pathway analysis of overlapping genes revealed that these genes are associated with a negative enrichment in pathways associated with IFNα, IFNγ, and inflammation (Figure 2E). Consistent with data in MKs, the top pathways suppressed by P2Y12 inhibition were IFNα and IFNγ responses (P = 3.22 × 10−8 and P = 1.00× 10−10, respectively) (Figures 2F and 2G, respectively) in platelets. Gene ontology pathway analysis of the MK and platelet transcriptomes following P2Y12 inhibition identified down-regulation in leukocyte-related pathways, including leukocyte cell-cell adhesion, leukocyte aggregation, and leukocyte-mediated immunity (Supplemental Tables 7 and 8). P2Y12 inhibition in the MK also noted down-regulation of pathways associated with platelet aggregation and activation (Supplemental Table 7).
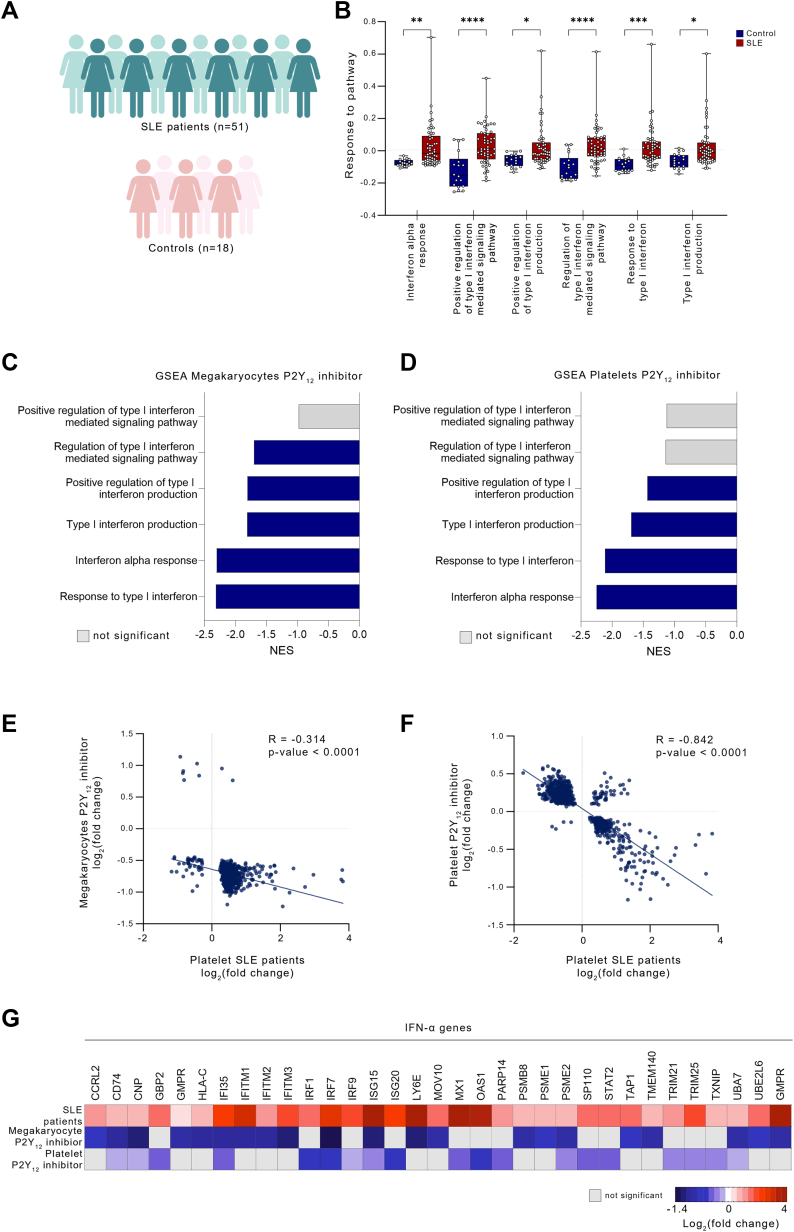
P2Y12 inhibition attenuates genes up-regulated in SLE
Previously, our group and others reported that platelet activation contributes to the pathogenesis of SLE, in part, via enrichment of platelet type I IFN signaling.13,14,35 To investigate whether previously identified IFN-associated transcripts up-regulated in platelets from patients with SLE could be suppressed by P2Y12 treatment, we integrated the platelet transcriptome of patients with SLE13 with our in vitro and ex vivo transcriptomic studies of P2Y12 inhibition. Pathway analysis of the SLE cohort (Figure 3A) confirmed that IFN-related pathways were significantly up-regulated in SLE (P < 0.05) (Figure 3B).14 Of note, P2Y12 inhibition down-regulated the same IFNα- and type I IFN-related pathways (up-regulated in SLE platelets) in both MKs and platelets (Figures 3C and 3D). Integration of the platelet transcriptome of patients with SLE vs control patients with differential expression analysis of P2Y12 inhibitors vs control solution/at baseline, in both MKs and platelets, demonstrated a significant negative correlation (r = −0.314 and r = −0.842, respectively; P < 0.001 for each comparison) (Figures 3E and 3F, Supplemental Figure 5), suggesting that IFN-associated transcripts up-regulated in SLE could be attenuated by P2Y12 inhibition. Specifically, among 72 up-regulated IFNα platelet SLE genes reported by El Bannoudi et al,13 33 (46%) were significantly down-regulated by P2Y12 inhibition, including ISG15, IRF3, IRF7, and IFITM3 (Figure 3G). Reactome analysis of the overlapping genes differentially regulated in both SLE and P2Y12 therapy found that 26.4% of genes are related to immunity and inflammation (Supplemental Table 9).
Figure 3.
P2Y12 Inhibition Attenuates IFN Genes Up-Regulated in Patients With SLE
(A) Experimental workflow: Platelets were collected from patients with systemic lupus erythematosus (SLE) and healthy control subjects, RNA was isolated, and RNA-sequencing was performed. (B) Hallmark and Gene Ontology pathway analysis, focused on IFNα and type I IFN in platelets from patients with SLE (P < 0.05). Data presented as mean ± SD; P values were determined from the gene set enrichment analysis (GSEA) output. ∗P < 0.05; ∗∗P < 0.01; ∗∗∗P < 0.001. (C, D) Overlapping pathways in P2Y12-inhibited MKs and platelets (blue = P < 0.05; gray = P = NS). (E, F). Scatterplots of fold change differences comparing platelets of patients with SLE vs control subjects and P2Y12-inhibited MKs (E) and platelets (F); P < 0.05. (G) Heatmap of differentially expressed genes in platelets from patients with SLE and P2Y12-inhibited MKs and platelets. The genes represent IFNα-related genes, up-regulated in patients with SLE, and down-regulated on P2Y12 inhibition in MK and platelet groups. Abbreviations as in Figure 1.
The clinical application of the MK and platelet transcriptomic in vitro findings were similarly evaluated in a second cohort of patients with COVID-19 with available platelet RNA-seq (Supplemental Figures 6A and 6B).6,32 Consistent with our findings in SLE, there was a significant negative correlation between the differential expression of COVID-19 vs control patients and P2Y12 inhibition of both the MK and platelet transcriptome, respectively (P < 0.001 in both) (Supplemental Figures 6C and 6D). We identified 15 up-regulated IFNα-associated transcripts in patients with COVID-19, which were down-regulated by P2Y12 inhibition in platelets and MKs (Supplemental Figure 6E).
P2Y12 inhibition suppresses IFN-induced platelet activation
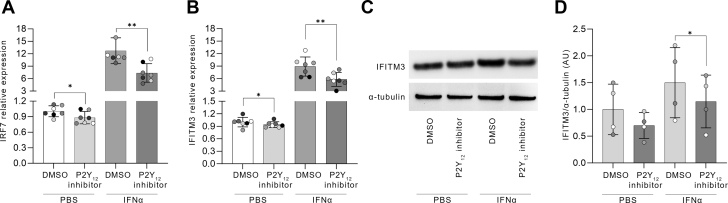
To validate our findings, we investigated whether P2Y12 inhibition could suppress IFN signaling following an inflammatory challenge. We reasoned that in addition to the potent antithrombotic effects of P2Y12 inhibition, suppression of P2Y12 receptor-mediated inflammation may be beneficial in the context of IFN-associated immunothrombosis.36 MKs were pretreated with or without P2Y12 inhibition and then stimulated with recombinant IFNα. As expected, ISGs IRF7 and IFITM3, identified in our P2Y12-inhibited MK transcripts, were increased 11- and 8.5-fold, respectively, following IFNα stimulation (Figures 4A and 4B). Pretreatment with the P2Y12 inhibitor significantly abrogated this effect, by reducing IRF7 and IFITM3 expression by 42% and 34%, respectively. These results were confirmed at the protein level, with IFITM3 expression significantly reduced following P2Y12 pretreatment and subsequent IFNα exposure (P < 0.001) (Figures 4C and 4D, Supplemental Figure 7).
Figure 4.
P2Y12 Inhibition Suppresses IFN-Associated MK Activation
MKs were pretreated with a P2Y12 inhibitor for 20 hours, stimulated with IFNα for another 4 hours, and lysed. (A-D) Levels of IFNα proteins were measured using real-time polymerase chain reaction (n = 7) (A, B) and immunoblot densitometry (n = 4) (C, D). Data presented as mean ± SD and compared using 2-sided paired Student's t-tests. ∗P < 0.05; ∗∗P < 0.01. DMSO = dimethyl sulfoxide; PBS = phosphate-buffered saline; other abbreviations as in Figure 1.
Next, we assessed the contribution of IFNα to LPA formation in whole blood. LPA is a marker of platelet and leukocyte activation and is commonly increased in thromboinflammatory IFN-associated diseases, including SLE and COVID-19.6,37 Treatment of whole blood with IFNα resulted in increased LPA formation (Supplemental Figures 8A to 8C and 8E). Consistent findings were observed for monocyte-platelet aggregates, NPAs, and lymphocyte platelet aggregates (Supplemental Figures 8D and 8F to 8H). Pretreatment of whole blood with P2Y12 inhibitor prior to stimulation with IFNα significantly suppressed LPA formation (P = 0.001) (Supplemental Figures 8C and 8E). A similar P2Y12 effect was seen with a significant reduction in monocyte-platelet aggregates, NPAs, and lymphocyte platelet aggregates (Supplemental Figures 8F to 8H).
P2Y12 inhibition suppresses MTOR signaling pathway
We next sought to evaluate mechanisms by which P2Y12 inhibition affects ISGs and protein expression in MKs and platelets. It is well known that a key pathway that regulates the translation of proteins in MKs and platelets is the MTOR signaling pathway.38,39 Activation of MTOR signaling has been reported to regulate the expression of ISGs, which are downstream effectors of IFN signaling.40,41 Deletion of MTOR in a MK model resulted in the loss of up-regulated expression of IFITM3 when cells were stimulated with IFNα, suggesting that the translation of IFITM3, one of the ISGs in MKs, is regulated by the MTOR pathway.42
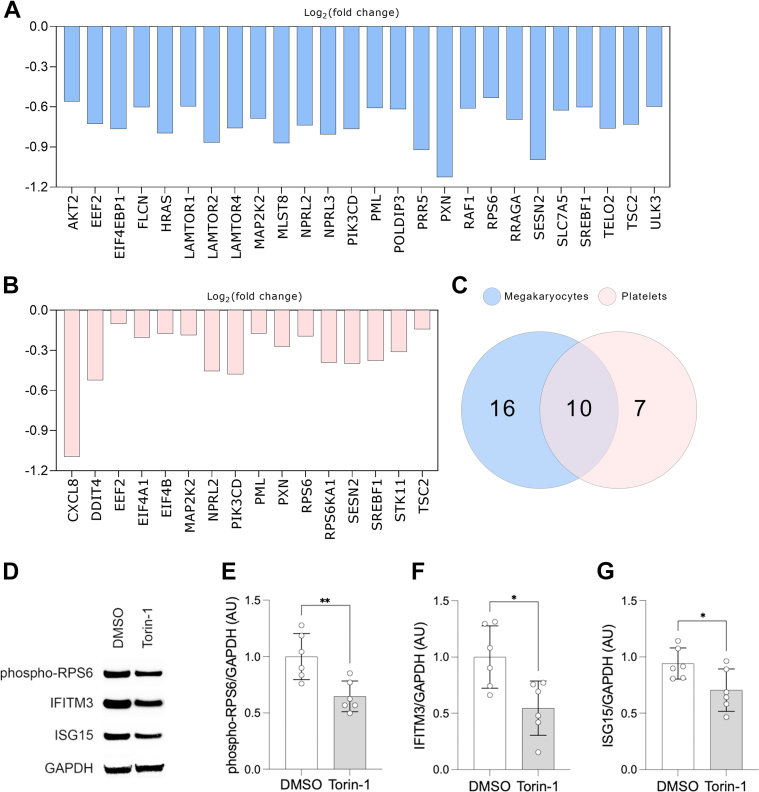
Pathway analysis demonstrated that MTOR complex 1, an MTOR protein complex, was down-regulated in MKs by P2Y12 inhibition (Figure 1F). To understand the significance of MTOR, we analyzed key MTOR signaling pathway genes. EIF4EBP1 and RPS6 are the 2 main MTOR downstream effectors of the eukaryotic translation initiation.43 Activation of MTOR signaling promotes the release of EIF4EBP1, allowing EIF4E to initiate translation, whereas phosphorylation of RPS6 enhances ribosomal function and protein synthesis.44 Among the MTOR-related genes, we identified 26 genes (Figure 5A), which were significantly down-regulated in P2Y12-inhibited MKs, including EIF4EBP1 (Log2FC = −0.765; P = 0.017) and RPS6 (Log2FC = −0.53; P = 0.046). Moreover, in the platelet transcriptome following treatment with P2Y12 inhibition, we identified 17 down-regulated MTOR-related genes (Figure 5B). Between platelets and MKs, we found 10 overlapping genes (Figure 5C).
Figure 5.
P2Y12 Inhibition Suppresses MTOR Signaling Pathway
(A, B) Bar graphs of differentially expressed MTOR-related genes identified in MKs (A) and platelets (B) treated with P2Y12 inhibitor (P < 0.05). (C) Venn diagram of overlapping MTOR-related genes between MKs and platelets. Heatmap of differentially expressed MTOR pathway genes identified in MKs and platelets treated with P2Y12 inhibitor (P < 0.05). (D to G) MEG-01 cells were treated with 2.5 μmol/L torin-1 for 24 hours and lysed. Protein levels (D) of (E) phospho-RPS6 (F) IFITM3 and (G) ISG15 were measured using immunoblot densitometry (n = 6). Data presented as mean ± SD and compared using 2-sided unpaired Student's t-tests. ∗P < 0.05; ∗∗P < 0.01. Abbreviations as in Figures 1 and 4.
To understand the role of the MTOR pathway in the translation of ISGs into proteins, we treated MKs with MTOR inhibitor, torin-1. As expected, phosphorylation of RPS6 was significantly down-regulated (Figures 5D and 5E). Of note, we observed down-regulation of IFITM3 and ISG15 protein expression (Figures 5D, 5F, and 5G). Altogether, these data suggest that P2Y12 inhibition has a significant effect on the MTOR pathway in both MKs and platelets.
Discussion
APTs, including ASA and P2Y12 inhibitors, are widely used in the treatment and prevention of cardiovascular events.2,45,46 Whereas the primary targets of these therapies have been identified, increasing evidence suggests a nonhemostatic effect of APTs.47, 48, 49 This study provides a comprehensive characterization of MK and platelet transcriptomes in response to ASA and/or P2Y12 inhibitors and investigates their potential role in inflammatory and IFN-mediated diseases.
To investigate the effects of ASA and P2Y12 inhibition on the gene expression of MKs, we employed an in vitro model of MKs differentiated from CD34+ hematopoietic progenitor cells. The use of CD34+-derived MKs as a surrogate model for studying platelet biology offers several advantages. It allows the investigation of platelet gene function and the assessment of platelet-like responses, including surface expression of P-selectin, PAC-1 (the activated form of glycoprotein IIb/IIIa), fibrinogen binding in response to agonists, adhesion, and spreading on substrates like fibrinogen or collagen, which mediate platelet behavior in hemostasis and thrombosis.50 Moreover, CD34+-derived MKs are permissive to viral infections7 and can be subjected to CRISPR/Cas9 gene editing.42,50 Even though MKs provide a useful model for studying platelet biology, limitations exist when compared to platelets. Notably, MKs cannot aggregate like platelets, and their granule packaging differs.51,52 Additionally, the translation and post-translational modifications in MKs may not accurately reflect those in platelets.53 Despite these limitations, the overlapping phenotype and significant proportion of overlapping genes expressed in platelets and MKs,54,55 support their utility as a model. Our transcriptomic analysis of MKs exposed to APT showed that ASA had a negligible impact on the MK transcriptome. A consistent small effect of ASA was found in the platelet transcriptome from a cohort of volunteers who received either low- or high-dose ASA daily, with no difference between doses.30 In contrast, P2Y12 inhibition had a more pronounced effect on both MK and platelet transcriptomes.31 The difference observed between ASA and a P2Y12 inhibitor on the transcriptome of platelets and MKs is likely due to their distinct mechanisms of action. ASA irreversibly inhibits COX-1, an enzyme that converts arachidonic acid into prostaglandins, which are precursors of thromboxane A2. Thromboxane A2 is a potent platelet agonist that stimulates platelet aggregation.56 P2Y12 inhibitors block the P2Y12 receptor, a subtype of ADP receptor that mediates platelet activation and amplification. By blocking the P2Y12 receptor, expression of the glycoprotein IIb/IIIa complex is reduced, which is the final common pathway for platelet aggregation.57
Our study used the CD34+-derived MK as a model of platelet function and transcriptomics. Both platelets and MKs were treated with APT. Our data suggest a consistent transcriptomic response for 633 genes in both in vitro MKs and ex vivo platelets from healthy subjects treated with APT. Of note, we identified 1,929 and 2,776 genes in MKs or platelets whose expression was altered solely in MKs or platelets following P2Y12 treatment, respectively. This difference in gene expression profile underscores the unique molecular pathways that are activated in MKs and platelets in response to P2Y12 inhibition. It highlights the intricate complexity of platelet signaling and function, which may not be fully replicated in an in vitro MK setting. Nevertheless, the overlap of 633 genes provides a valuable core set of genes affected by P2Y12 inhibition. Platelets and MKs mediate inflammation and immune regulation through cytokines.58, 59, 60, 61 They can sense inflammatory signals via various receptors and modulate cytokine secretion.62,63 Moreover, MKs contribute to the immune landscape by releasing IFNs, which are central players in immune activation.7 The interaction among platelets, MKs, and the immune system provides an opportunity for potential treatments. In the context of SLE, it is known that patients with SLE have higher levels of type I IFN activity.11,12 This is also seen in platelet transcriptome, where patients with SLE display increased expression of IFN-related genes and enrichment of IFN-response pathways.13,14 Additionally, dysregulation in various immune-related pathways is observed, emphasizing platelets' multifaceted involvement. Notably, patients with proteinuria, a severe SLE manifestation, exhibit a distinct platelet transcriptomic profile, where oxidative phosphorylation and platelet activity modules were decreased, suggesting shifts in pathways related to coagulation and immune response.14,64,65 Then, targeting the platelet IFN pathway could be beneficial in SLE severity. Several therapeutics have been developed to target type I IFN in the treatment of SLE. Unfortunately, many of these interventions failed to meet their primary endpoint.66 Development of novel therapies or different strategies targeting IFN in patients with SLE is needed. Whereas inhibiting IFN signaling broadly may affect both pathogenic and protective responses, targeting the platelet IFN pathway may allow for a more nuanced approach. Data from our group and others demonstrate a hyper-reactive platelet phenotype in SLE and a differentially expressed platelet transcriptome enriched in IFN-mediated pathways.13,14 Selectively targeting the platelet IFN pathway may minimize off-target effects and reduce the risk of systemic side effects. Moreover, combining therapies that target the platelet IFN pathway with existing or emerging treatments for SLE may provide a synergistic approach. Our study found that P2Y12 inhibition suppresses IFNα and IFNγ pathways in both platelets and MKs. In fact, we observed a strong negative correlation between the gene expression of patients with SLE (vs control subjects) and that of platelets and MKs treated with a P2Y12 inhibitor. In support of our findings, P2Y12 inhibition reduces lupus-like disease severity in a lupus mouse model15 and platelet activation markers in patients with SLE.67 Altogether, our data suggest that P2Y12 inhibitor may have therapeutic potential in patients with SLE by attenuating platelet-induced IFNα responses.
In the MK population, type I IFN increases the expression of ISGs, such as IRF7 and IFITM3.7,13,68 IRF7 is a key transcription factor that enhances the synthesis of type I IFN through TLR signaling.69 Production of type I IFN requires phosphorylation and translocation of IRF7 into the nucleus through interaction with MYD88.70 Our transcriptomic data demonstrated that MK P2Y12 receptor inhibition down-regulates expression of IRF7, which is elevated in SLE and COVID-19.6,13 In addition, MKs stimulated with IFNα showed up-regulation of IRF7, which was attenuated due to P2Y12 inhibition. IFITM3 is an antiviral protein that limits virus entry and replication in cells and its expression is increased in platelets and MKs during viral and nonviral infections.7,42 P2Y12 inhibition reduces IFITM3 expression in MKs at baseline and IFNα-stimulated conditions. Our results suggest that P2Y12 inhibition may attenuate IFN responses during inflammatory conditions.
In addition to the effect of drugs on gene expression and protein translation after 24 hours, we investigated the short-term response of P2Y12 inhibition. Platelets can interact with leukocytes (monocytes and neutrophils) through the P-selectin–P-selectin glycoprotein ligand 1 axis, leading to the maturation of monocytes to antigen-presenting cells and the priming of neutrophils. An increase in circulating LPA has been linked to proinflammatory conditions.71,72 Additionally, LPA is increased in SLE37 and COVID-19.6,73 We showed that IFNα increased LPA formation in the whole blood. Moreover, when whole blood was pretreated with a P2Y12 inhibitor and stimulated with IFNα, we noted the reduction of the formed LPA. Our results highlight the complexity of platelet responses to APT, where immediate pharmacodynamic effects are observed alongside transcriptional and translational changes. It suggests that whereas P2Y12 inhibition quickly attenuates platelet activity, it may also provoke long-term alterations in the transcriptome and proteome, suggesting different mechanisms of action. Previously, it has been reported that P2Y12 inhibition reduces the formation of LPA induced by ADP74 and U-46619.17 Moreover, patients with sickle cell disease, which are characterized by increased levels of NPA, had decreased NPA number on treatment with P2Y12 inhibitor.75 Thus, there is a compelling rationale supporting that P2Y12 inhibition may reduce IFN-mediated LPA formation.
MTOR is a serine/threonine protein kinase known to regulate cell growth and differentiation at the level of protein translation. MTOR signaling can regulate the translation of proteins in cells by controlling the activity of 2 major downstream effectors, EIF4EBP1 and RPS6 kinase.43 Phosphorylation of 4EBP1 and RPS6 by MTOR initiates translation. It has been reported that MTOR regulates the translation of proteins in MKs and platelets.38,39 Prior data also suggest that P2Y12 inhibition can affect the MTOR pathway.76 Our pathway analysis of MKs suppressed by P2Y12 inhibitor demonstrated the down-regulation of MTORC1 and PI3K/AKT/MTOR pathways. Moreover, both, EIF4EBP1 and RPS6 were significantly down-regulated at the messenger RNA level. Campbell et al42 showed in the CD34+-derived MK model that deletion of MTOR abolished the up-regulation of IFITM3 expression in the cells stimulated with IFNα, suggesting that IFITM3 translation is controlled by the MTOR pathway. Additionally, treatment of MKs with torin-1, an inhibitor of the MTOR pathway, demonstrated down-regulation in the expression of IFN genes, IFITM3 and ISG15. Moreover, we demonstrated that CD34+-derived MKs pretreated with P2Y12 inhibition (at baseline) and additionally stimulated with IFNα, reduced IFITM3 expression on transcriptomic and proteomic levels. As P2Y12 inhibition has pleiotropic effects beyond thrombosis and hemostasis, our data suggest that it may inhibit additionally the translation of the proteins, including IFN-related proteins, via the MTOR signaling pathway. This can have therapeutic implications for preventing thrombosis and inflammation in various diseases.
Study limitations
A major goal of this study was to investigate the impact of APT on the nonhemostatic effector properties of MKs and platelets. Whereas our study integrates the effects of both ASA and P2Y12 inhibitors on MKs to platelet responses, we did not study the effect of less commonly used APTs (eg, PAR-1 antagonist). Although we observed a robust and consistent anti-inflammatory effect of P2Y12 inhibition on both MKs and platelets, the translation of these findings to clinical outcomes requires further investigation. Moreover, due to the small sample size in our MK groups (n = 4), we used nominal P values for the analysis of differentially expressed genes and this approach could increase the false discovery rate. Whereas our findings may not be unique to the SLE population, because there is no direct evaluation of P2Y12 inhibitors use in SLE, our study provides exciting preliminary data on the potential therapeutic implications of P2Y12 inhibition in an understudied patient population at high cardiovascular risk. Finally, a model of CD34+-derived MKs exhibits limitations in aggregation, granule packaging, and environmental interactions, which may not fully replicate platelet processes in the body.
Conclusions
Targeting the P2Y12 receptor modulates MK and platelet inflammatory signaling pathways. P2Y12-mediated suppression of platelet IFNα signaling may benefit proinflammatory and autoimmune diseases such as SLE.
Funding Support and Author Disclosures
This work was supported by National Institutes of Health grants R01HL139909 (to Drs Berger and Buyon), R35HL144993 (to Dr Berger), 1OT2HL156812-01 (to Dr Berger), R01HL167917 (to Dr Barrett), and R01HL118049 (to Dr Voora). The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Perspectives.
COMPETENCY IN MEDICAL KNOWLEDGE: The comprehensive analysis of MK and platelet transcriptomes suppressed by P2Y12 inhibitors and their role in modulating immune reactions adds insights into understanding immune-mediated disorders and their therapeutic potential. P2Y12 inhibitors might synergize with existing treatments to provide more comprehensive control over immune dysregulation and thrombotic tendencies.
TRANSLATIONAL OUTLOOK: Further investigation into the precise molecular mechanisms linking P2Y12 inhibition, platelet function, and immune modulation would provide a deeper understanding of the nonhemostatic effects of platelet directed therapies. This knowledge could uncover novel drug targets within platelet-mediated pathways. In addition to SLE, the role of platelet activation and its modulation by P2Y12 inhibitors could be investigated in other inflammatory conditions, expanding the potential applications of these agents.
Acknowledgment
Figures were created with BioRender and Adobe Photoshop (version 23.0.2).
Footnotes
The authors attest they are in compliance with human studies committees and animal welfare regulations of the authors’ institutions and Food and Drug Administration guidelines, including patient consent where appropriate. For more information, visit the Author Center.
Appendix
For supplemental figures and tables, please see the online version of this paper.
Appendix
References
- 1.Golebiewska E.M., Poole A.W. Platelet secretion: from haemostasis to wound healing and beyond. Blood Rev. 2015;29(3):153–162. doi: 10.1016/j.blre.2014.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Majithia A., Bhatt D.L. Novel antiplatelet therapies for atherothrombotic diseases. Arterioscler Thromb Vasc Biol. 2019;39(4):546–557. doi: 10.1161/ATVBAHA.118.310955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pariser D.N., Hilt Z.T., Ture S.K., et al. Lung megakaryocytes are immune modulatory cells. J Clin Invest. 2021;131(1) doi: 10.1172/JCI137377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.De Wispelaere K., Freson K. The analysis of the human megakaryocyte and platelet coding transcriptome in healthy and diseased subjects. Int J Mol Sci. 2022;23(14):7647. doi: 10.3390/ijms23147647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Machlus K.R., Italiano J.E., Jr. The incredible journey: from megakaryocyte development to platelet formation. J Cell Biol. 2013;201(6):785–796. doi: 10.1083/jcb.201304054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Barrett T.J., Cornwell M., Myndzar K., et al. Platelets amplify endotheliopathy in COVID-19. Sci Adv. 2021;7(37) doi: 10.1126/sciadv.abh2434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Campbell R.A., Schwertz H., Hottz E.D., et al. Human megakaryocytes possess intrinsic antiviral immunity through regulated induction of IFITM3. Blood. 2019;133(19):2013–2026. doi: 10.1182/blood-2018-09-873984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.De Kock L., Ver Donck F., Thys C., et al. Combined transcriptome and proteome profiling of SRC kinase activity in healthy and E527K defective megakaryocytes. Haematologica. 2021;106(12):3206–3210. doi: 10.3324/haematol.2021.279248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assi M., Thon-Hon V.G., Jaffar-Bandjee M.C., Martinez A., Gasque P. Regulation of type I-interferon responses in the human epidermal melanocyte cell line SKMEL infected by the Ross River alphavirus. Cytokine. 2015;76(2):572–576. doi: 10.1016/j.cyto.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Kopitar-Jerala N. The role of interferons in inflammation and inflammasome activation. Front Immunol. 2017;8:873. doi: 10.3389/fimmu.2017.00873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bengtsson A.A., Ronnblom L. Role of interferons in SLE. Best Pract Res Clin Rheumatol. 2017;31(3):415–428. doi: 10.1016/j.berh.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Ronnblom L., Leonard D. Interferon pathway in SLE: one key to unlocking the mystery of the disease. Lupus Sci Med. 2019;6(1) doi: 10.1136/lupus-2018-000270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.El Bannoudi H., Cornwell M., Luttrell-Williams E., et al. Platelet LGALS3BP induces myeloid inflammation in systemic lupus erythematosus. Arthritis Rheumatol. 2022;75(5):711–722. doi: 10.1002/art.42382. [DOI] [PubMed] [Google Scholar]
- 14.Cornwell M.G., Bannoudi H.E., Luttrell-Williams E., et al. Modeling of clinical phenotypes in systemic lupus erythematosus based on the platelet transcriptome and FCGR2a genotype. J Transl Med. 2023;21(1):247. doi: 10.1186/s12967-023-04059-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duffau P., Seneschal J., Nicco C., et al. Platelet CD154 potentiates interferon-alpha secretion by plasmacytoid dendritic cells in systemic lupus erythematosus. Sci Transl Med. 2010;2(47) doi: 10.1126/scitranslmed.3001001. [DOI] [PubMed] [Google Scholar]
- 16.Yokoyama H., Ito N., Soeda S., et al. Prediction of antiplatelet effects of aspirin in vivo based on in vitro results. Clin Appl Thromb Hemost. 2013;19(6):600–607. doi: 10.1177/1076029613484084. [DOI] [PubMed] [Google Scholar]
- 17.Rolling C.C., Sowa M.A., Wang T.T., et al. P2Y12 inhibition suppresses proinflammatory platelet-monocyte interactions. Thromb Haemost. 2023;123(2):231–244. doi: 10.1055/s-0042-1758655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.RNAseq_PE: Paired-end RNAseq data processing workflow designed for execution on the BigPurple HPC https://github.com/mgildea87/RNAseq_PE
- 19.Andrews S. 2010. FastQC: A Quality Control Tool for High Throughput Sequence Data.http://www.bioinformatics.babraham.ac.uk/projects/fastqc/ [Google Scholar]
- 20.Ewels P., Magnusson M., Lundin S., Kaller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32(19):3047–3048. doi: 10.1093/bioinformatics/btw354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen S., Zhou Y., Chen Y., Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–i890. doi: 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dobin A., Davis C.A., Schlesinger F., et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29(1):15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liao Y., Smyth G.K., Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics. 2014;30(7):923–930. doi: 10.1093/bioinformatics/btt656. [DOI] [PubMed] [Google Scholar]
- 24.Love M.I., Huber W., Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mootha V.K., Lindgren C.M., Eriksson K.F., et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34(3):267–273. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- 26.Subramanian A., Tamayo P., Mootha V.K., et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liberzon A., Birger C., Thorvaldsdottir H., Ghandi M., Mesirov J.P., Tamayo P. The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wu T., Hu E., Xu S., et al. clusterProfiler 4.0: A universal enrichment tool for interpreting omics data. Innovation (Camb) 2021;2(3) doi: 10.1016/j.xinn.2021.100141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Yu G., Wang L.G., Han Y., He Q.Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. doi: 10.1089/omi.2011.0118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Myers R.A., Ortel T.L., Waldrop A., Dave S., Ginsburg G.S., Voora D. Aspirin effects on platelet gene expression are associated with a paradoxical, increase in platelet function. Br J Clin Pharmacol. 2022;88(5):2074–2083. doi: 10.1111/bcp.15127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Myers R.A., Ortel T.L., Waldrop A., et al. Platelet RNA biomarker of ticagrelor-responsive genes is associated with platelet function and cardiovascular events. Arterioscler Thromb Vasc Biol. 2024;44(2):423–434. doi: 10.1161/ATVBAHA.123.319759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barrett T.J., Bilaloglu S., Cornwell M., et al. Platelets contribute to disease severity in COVID-19. J Thromb Haemost. 2021;19(12):3139–3153. doi: 10.1111/jth.15534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bouget J., Balusson F., Maignan M., et al. Major bleeding risk associated with oral anticoagulant in real clinical practice. A multicentre 3-year period population-based prospective cohort study. Br J Clin Pharmacol. 2020;86(12):2519–2529. doi: 10.1111/bcp.14362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kamran H., Jneid H., Kayani W.T., et al. Oral antiplatelet therapy after acute coronary syndrome: a review. JAMA. 2021;325(15):1545–1555. doi: 10.1001/jama.2021.0716. [DOI] [PubMed] [Google Scholar]
- 35.Lood C., Amisten S., Gullstrand B., et al. Platelet transcriptional profile and protein expression in patients with systemic lupus erythematosus: up-regulation of the type I interferon system is strongly associated with vascular disease. Blood. 2010;116(11):1951–1957. doi: 10.1182/blood-2010-03-274605. [DOI] [PubMed] [Google Scholar]
- 36.Mansour A., Bachelot-Loza C., Nesseler N., Gaussem P., Gouin-Thibault I. P2Y(12) inhibition beyond thrombosis: effects on inflammation. Int J Mol Sci. 2020;21(4):1391. doi: 10.3390/ijms21041391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nhek S., Clancy R., Lee K.A., et al. Activated platelets induce endothelial cell activation via an interleukin-1beta pathway in systemic lupus erythematosus. Arterioscler Thromb Vasc Biol. 2017;37(4):707–716. doi: 10.1161/ATVBAHA.116.308126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lindemann S., Tolley N.D., Eyre J.R., Kraiss L.W., Mahoney T.M., Weyrich A.S. Integrins regulate the intracellular distribution of eukaryotic initiation factor 4E in platelets. A checkpoint for translational control. J Biol Chem. 2001;276(36):33947–33951. doi: 10.1074/jbc.M104281200. [DOI] [PubMed] [Google Scholar]
- 39.Weyrich A.S., Denis M.M., Schwertz H., et al. mTOR-dependent synthesis of Bcl-3 controls the retraction of fibrin clots by activated human platelets. Blood. 2007;109(5):1975–1983. doi: 10.1182/blood-2006-08-042192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Platanias L.C. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5(5):375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- 41.Livingstone M., Sikstrom K., Robert P.A., Uze G., Larsson O., Pellegrini S. Assessment of mTOR-dependent translational regulation of interferon stimulated genes. PLoS One. 2015;10(7) doi: 10.1371/journal.pone.0133482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Campbell R.A., Manne B.K., Banerjee M., et al. IFITM3 regulates fibrinogen endocytosis and platelet reactivity in nonviral sepsis. J Clin Invest. 2022;132(23) doi: 10.1172/JCI153014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Laplante M., Sabatini D.M. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Showkat M., Beigh M.A., Andrabi K.I. mTOR signaling in protein translation regulation: implications in cancer genesis and therapeutic interventions. Mol Biol Int. 2014;2014 doi: 10.1155/2014/686984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aggarwal D., Bhatia K., Chunawala Z.S., et al. P2Y(12) inhibitor versus aspirin monotherapy for secondary prevention of cardiovascular events: meta-analysis of randomized trials. Eur Heart J Open. 2022;2(2) doi: 10.1093/ehjopen/oeac019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Passacquale G., Sharma P., Perera D., Ferro A. Antiplatelet therapy in cardiovascular disease: current status and future directions. Br J Clin Pharmacol. 2022;88(6):2686–2699. doi: 10.1111/bcp.15221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Siasos G., Mourouzis K., Tousoulis D. Pleiotropic effects of antiplatelet treatment in patients with coronary artery disease. Hellenic J Cardiol. 2018;59(6):344–346. doi: 10.1016/j.hjc.2018.06.016. [DOI] [PubMed] [Google Scholar]
- 48.Schnorbus B., Jurk K., Lackner K.J., Welk C., Munzel T., Gori T. Effects of clopidogrel, prasugrel and ticagrelor on microvascular function and platelet reactivity in patients with acute coronary syndrome undergoing coronary artery stenting. A randomized, blinded, parallel group trial. Front Cardiovasc Med. 2021;8 doi: 10.3389/fcvm.2021.780605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alaaeddine R.A., AlZaim I., Hammoud S.H., et al. The pleiotropic effects of antithrombotic drugs in the metabolic-cardiovascular-neurodegenerative disease continuum: impact beyond reduced clotting. Clin Sci (Lond) 2021;135(8):1015–1051. doi: 10.1042/CS20201445. [DOI] [PubMed] [Google Scholar]
- 50.Montenont E., Bhatlekar S., Jacob S., et al. CRISPR-edited megakaryocytes for rapid screening of platelet gene functions. Blood Adv. 2021;5(9):2362–2374. doi: 10.1182/bloodadvances.2020004112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Battinelli E.M., Thon J.N., Okazaki R., et al. Megakaryocytes package contents into separate α-granules that are differentially distributed in platelets. Blood Adv. 2019;3(20):3092–3098. doi: 10.1182/bloodadvances.2018020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen Y., Yuan Y., Li W. Sorting machineries: how platelet-dense granules differ from α-granules. Biosci Rep. 2018;38(5) doi: 10.1042/BSR20180458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang B., Zheng J. Platelet generation in vivo and in vitro. Springerplus. 2016;5(1):787. doi: 10.1186/s40064-016-2384-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kammers K., Taub M.A., Rodriguez B., et al. Transcriptional profile of platelets and iPSC-derived megakaryocytes from whole-genome and RNA sequencing. Blood. 2021;137(7):959–968. doi: 10.1182/blood.2020006115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nersisyan S., Montenont E., Loher P., et al. Characterization of all small RNAs in and comparisons across cultured megakaryocytes and platelets of healthy individuals and COVID-19 patients. J Thromb Haemost. 2023;21(11):3252–3267. doi: 10.1016/j.jtha.2023.07.028. [DOI] [PubMed] [Google Scholar]
- 56.Warner T.D., Nylander S., Whatling C. Anti-platelet therapy: cyclo-oxygenase inhibition and the use of aspirin with particular regard to dual anti-platelet therapy. Br J Clin Pharmacol. 2011;72(4):619–633. doi: 10.1111/j.1365-2125.2011.03943.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Dorsam R.T., Kunapuli S.P. Central role of the P2Y12 receptor in platelet activation. J Clin Invest. 2004;113(3):340–345. doi: 10.1172/JCI20986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Bakogiannis C., Sachse M., Stamatelopoulos K., Stellos K. Platelet-derived chemokines in inflammation and atherosclerosis. Cytokine. 2019;122 doi: 10.1016/j.cyto.2017.09.013. [DOI] [PubMed] [Google Scholar]
- 59.von Hundelshausen P., Koenen R.R., Sack M., et al. Heterophilic interactions of platelet factor 4 and RANTES promote monocyte arrest on endothelium. Blood. 2005;105(3):924–930. doi: 10.1182/blood-2004-06-2475. [DOI] [PubMed] [Google Scholar]
- 60.Cunin P., Penke L.R., Thon J.N., et al. Megakaryocytes compensate for Kit insufficiency in murine arthritis. J Clin Invest. 2017;127(5):1714–1724. doi: 10.1172/JCI84598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kohler A., De Filippo K., Hasenberg M., et al. G-CSF-mediated thrombopoietin release triggers neutrophil motility and mobilization from bone marrow via induction of Cxcr2 ligands. Blood. 2011;117(16):4349–4357. doi: 10.1182/blood-2010-09-308387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Du Y., Ah Kioon M.D., Laurent P., et al. Chemokines form nanoparticles with DNA and can superinduce TLR-driven immune inflammation. J Exp Med. 2022;219(7) doi: 10.1084/jem.20212142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Cunin P., Nigrovic P.A. Megakaryocytes as immune cells. J Leukoc Biol. 2019;105(6):1111–1121. doi: 10.1002/JLB.MR0718-261RR. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Katz G., Smilowitz N.R., Blazer A., Clancy R., Buyon J.P., Berger J.S. Systemic lupus erythematosus and increased prevalence of atherosclerotic cardiovascular disease in hospitalized patients. Mayo Clin Proc. 2019;94(8):1436–1443. doi: 10.1016/j.mayocp.2019.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Moulton V.R., Suarez-Fueyo A., Meidan E., Li H., Mizui M., Tsokos G.C. Pathogenesis of human systemic lupus erythematosus: a cellular perspective. Trends Mol Med. 2017;23(7):615–635. doi: 10.1016/j.molmed.2017.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Paredes J.L., Niewold T.B. Type I interferon antagonists in clinical development for lupus. Expert Opin Investig Drugs. 2020;29(9):1025–1041. doi: 10.1080/13543784.2020.1797677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Vial G., Gensous N., Savel H., et al. The impact of clopidogrel on plasma-soluble CD40 ligand levels in systemic lupus erythematosus patients: the CLOPUS phase I/II pilot study. Joint Bone Spine. 2021;88(2) doi: 10.1016/j.jbspin.2020.105097. [DOI] [PubMed] [Google Scholar]
- 68.Negrotto S., De Giusti C.J., Lapponi M.J., et al. Expression and functionality of type I interferon receptor in the megakaryocytic lineage. J Thromb Haemost. 2011;9(12):2477–2485. doi: 10.1111/j.1538-7836.2011.04530.x. [DOI] [PubMed] [Google Scholar]
- 69.Hong Y., Bai M., Qi X., et al. Suppression of the IFN-alpha and -beta induction through sequestering IRF7 into viral inclusion bodies by nonstructural protein NSs in severe fever with thrombocytopenia syndrome bunyavirus infection. J Immunol. 2019;202(3):841–856. doi: 10.4049/jimmunol.1800576. [DOI] [PubMed] [Google Scholar]
- 70.Ning S., Pagano J.S., Barber G.N. IRF7: activation, regulation, modification and function. Genes Immun. 2011;12(6):399–414. doi: 10.1038/gene.2011.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Neumann F.J., Marx N., Gawaz M., et al. Induction of cytokine expression in leukocytes by binding of thrombin-stimulated platelets. Circulation. 1997;95(10):2387–2394. doi: 10.1161/01.cir.95.10.2387. [DOI] [PubMed] [Google Scholar]
- 72.Celi A., Pellegrini G., Lorenzet R., et al. P-selectin induces the expression of tissue factor on monocytes. Proc Natl Acad Sci U S A. 1994;91(19):8767–8771. doi: 10.1073/pnas.91.19.8767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Manne B.K., Denorme F., Middleton E.A., et al. Platelet gene expression and function in patients with COVID-19. Blood. 2020;136(11):1317–1329. doi: 10.1182/blood.2020007214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Storey R.F., Judge H.M., Wilcox R.G., Heptinstall S. Inhibition of ADP-induced P-selectin expression and platelet-leukocyte conjugate formation by clopidogrel and the P2Y12 receptor antagonist AR-C69931MX but not aspirin. Thromb Haemost. 2002;88(3):488–494. [PubMed] [Google Scholar]
- 75.Polanowska-Grabowska R., Wallace K., Field J.J., et al. P-selectin-mediated platelet-neutrophil aggregate formation activates neutrophils in mouse and human sickle cell disease. Arterioscler Thromb Vasc Biol. 2010;30(12):2392–2399. doi: 10.1161/ATVBAHA.110.211615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Moore S.F., Hunter R.W., Hers I. Protein kinase C and P2Y12 take center stage in thrombin-mediated activation of mammalian target of rapamycin complex 1 in human platelets. J Thromb Haemost. 2014;12(5):748–760. doi: 10.1111/jth.12552. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.