Abstract
The first differentiation event of the mammalian embryo is thought to occur during blastulation and results in two populations of cells, the inner cell mass (ICM) and the trophectoderm. Most embryonic stem (ES) cell lines have been derived from the ICM or a further subset of ICM cells known as the epiblast. There appears to be a limited period of embryonic development during which pluripotent ES cells can be adapted from the cells of the blastocyst to culture. A method is presented here that allows ES cell lines to be isolated from preblastocyst mouse embryos. These lines were derived from 129S2/SvHsd mouse morulae and earlier cleavage stages with high efficiency. The lines expressed genes and antigens characteristic of pluripotent ES cells. XY cell lines remained karyotypically stable through extensive passaging and produced germ-line-competent chimeras upon blastocyst injection. These results suggest that true ES cells can be derived from embryos explanted at any stage of preimplantation development in the mouse. This finding raises the interesting question of whether ES cell lines derived from embryos at different stages of preimplantation development possess the same potential.
Keywords: morula
Whether from man, mouse, mink, or monkey, putative embryonic stem (ES) cell lines have shown great diversity both in their differentiation potential and in their ability to participate in development to form chimeras. Even in the mouse, for which there are the most data, ES cells with germ-line potential have only been derived from a limited number of strains. It is far from clear as to why certain strains are more amenable to ES cell derivation than others. Previous work established that ES cells derived from the late inner cell mass (ICM) originate exclusively from the epiblast rather than the primitive endoderm or trophectoderm (1). Although starting with epiblast isolated from advanced blastocysts greatly increased the ease of obtaining ES cells, certain strains such as ICR and the NOD strain derived therefrom still proved to be largely refractory with this method (2). The possibility exists that mouse strains may differ in the period of development that readily yields ES cell lines. The stage of developmental from which ES cells are derived might therefore prove crucial to their use as models of differentiation and in regenerative medicine.
Studies in the mouse have shown that it is possible to isolate ES cells from a limited window of early development. No validated ES cell lines have yet been isolated from either preblastocyst or postimplantation stage embryos. Putative ES cell lines have been obtained from eight-cell through compacted morula stage mouse embryos (3, 4). Only limited analysis was carried out on these cell lines, and in no instance was germ-line transmission demonstrated. Postimplantation embryos have not, so far, yielded ES cell lines. The loss of potential to form ES cells therefore seems to coincide with the transition from implanting blastocyst to early egg cylinder. Interestingly, and possibly significantly, this corresponds with when epiblast cells cease to yield chimerism after blastocyst injection (5). The lack of validated ES cell lines from either preblastocyst or postimplantation stage embryos has led to the view that ES cells are cells of the ICM/epiblast that have been adapted to culture.
The origin of ES cells has received added interest because human ES (hES) cells, unlike their murine counterparts, are reported to differentiate into the extraembryonic trophectodermal lineage in vitro. Trophoblast cells seem to form spontaneously at low levels in hES cell cultures, and the growth factor BMP4 is thought to enhance this differentiation (6). Mouse ES cells do not generate trophectoderm without genetic manipulation (7) and show distinct features from mouse cell lines that are capable of generating trophectodermal tissue (8). Because early mouse ICM cells still retain the ability to form trophectoderm if externalized secondarily (9), it is possible that hES cells originate at this early blastocyst stage. However, production of trophoblasts by clonal hES cell lines does not seem to have been reported, leaving open the possibility that hES cell-derived trophoblasts stem from a low level of contaminating trophoblast precursors. Regardless, differences between human and mouse embryos and ES cell lines prompt the question of whether mouse and human embryos yield ES cells at different stages of embryonic development and, more generally, whether ES cells derived from different stages show distinct differences.
The preblastocyst period in mouse development constitutes a time when blastomeres are not yet committed to either the ICM or trophectodermal lineage (10). It is therefore conceivable that ES cell lines derived from morulae or earlier cleavage stage embryos might originate before cells were committed to one or other of these two lineages and thus exhibit a wider developmental potential than those of ICM or epiblast origin. Alternatively, however, permissiveness for ES cell derivation could be restricted to a specific stage of early development to which embryos must progress irrespective of when they are explanted. In either case, the properties of ES cells derived from preblastocyst embryos would provide new insight into the ES cell state. Described here is a method for deriving ES cell lines from preblastocyst mouse embryos. Expression of pluripotent markers, extensive contribution to chimeras, and germ-line competence validates these preblastocyst-derived lines as true ES cells. A more detailed comparison of preblastocyst- and blastocyst-derived ES cell lines may now determine whether different stages of preimplantation development yield distinct ES cell lines and whether this contributes to the dramatic differences in developmental potential between seemingly similar ES cell lines (11).
Materials and Methods
Mice and Embryos. Mice were kept on a 14 h light/10 h dark regime with the dark period either from 7:00 p.m. to 5:00 a.m. or from 11:00 a.m. to 9:00 p.m. Natural matings were used for all experiments. Twelve hours from the middle of the dark period was termed embryonic day (E) 0.5. Zygotes, two-cell, four-cell, eight-cell, and morula stage embryos were obtained from the PO closed-bred strain, the 129S2/SvHsd inbred strain, or a cross between 129S2/SvHsd females and males bred on a 129 background that express eGFP ubiquitously (12). E13.5 fetuses from the PO strain were used to obtain primary embryonic fibroblasts (PEFs). Blastocysts for ES cell injections were obtained from the MF1 strain on E3.5. PO females made pseudopregnant by mating with vasectomized males were used as recipients for embryo transfer on E2.5. All embryos were flushed from the oviducts or uterine horns into Hepes-buffered potassium simplex optimized medium (mKSOM) (13).
Derivation and Culture of ES Cell Lines. Zygote, two-cell, four-cell, eight-cell, morula, and blastocyst stage embryos were recovered on E0.5, E1.5, E2.0, E2.5, E3.0, and E3.5, respectively. Zonae pellucidae were removed with brief exposure to Tyrode's saline acidified to pH 2.5 (14). These denuded embryos were either explanted individually into separate 1.9-cm2 wells that had been seeded 24–72 h earlier with PEFs irradiated with 30 gray (feeders) or into a microdrop of Ca2+, Mg2+-free PBS in 1.9-cm2 wells. Feeders were plated at a density of 7.5 × 104 cells per cm2. Embryos that were deliberately “stuck” to the dish in the PBS microdrops were carefully flooded with a feeder cell suspension after an interval of 5 min. PEFs and feeders were maintained with DMEM (Sigma) supplemented with 2.2 g/liter NaHCO3, 10% heat-inactivated FCS (HyClone), 2 mM l-glutamine (Sigma), and 0.1 mM 2-mercaptoethanol (Sigma). ES cell medium, which consisted of Knockout DMEM (Invitrogen) supplemented with 15% FCS (92K3384, Sigma), 103 units/ml recombinant murine leukemia inhibitory factor (LIF) (ESGRO, Chemicon), 2 mM l-glutamine, 0.1 mM 2-mercaptoethanol, and 1× nonessential amino acids (Sigma), was used for culture of the embryos and any ES-like cell colonies or lines derived therefrom. Antibiotics at concentrations of 50 units/ml and 50 μg/ml for penicillin and streptomycin, respectively, were used only for primary explants of PEFs and embryos.
Entire colonies formed by the attached embryos were dissected away from the feeder layer with glass needles and dissociated into a single-cell suspension by a 10-min incubation in 0.5% pronase (Calbiochem) in PBS at room temperature, followed by 30 min at 37°C in Ca2+-free OC medium with 0.8 mg/ml EGTA. The resulting suspension was distributed onto a fresh feeder layer. After 3–4 days, wells were passaged with 0.25% trypsin/EDTA onto a fresh feeder layer. Resulting ES cell colonies were propagated for two to four passages before being cryopreserved in N2 (l).
Nomenclature of ES Cell Lines. Mouse ES cells were coded to indicate the developmental stage of the embryo from which they were derived. For example, ES cells derived from a mouse embryo explanted at the morula stage were termed mm (mouse morula) followed by a number. Sublines were denoted by a letter (i.e., mm4a), and passages were denoted with a “P” followed by the passage number. The culture was denoted as P0 upon the first appearance of ES cell colonies.
Immunostaining. Cells and embryos were fixed for 30 min in 4% paraformaldehyde that had been prepared from 16% stock (Electron Microscopy Sciences) and refrigerated until immediately before use. They were then permeabilized with 0.2% Triton-X in PBS (PBT). Blocking solution consisted of 10% normal goat or donkey serum (Abcam) in PBT and was used for either 2 h at room temperature or overnight at 4°C. Primary antibodies were diluted in blocking solution and incubated with the cells overnight at 4°C. Fluorescent secondary antibodies (Molecular Probes; 1:500) were incubated with the cells for 1 h at room temperature. Primary antibodies were omitted from control samples. Antibodies used were as follows: mouse monoclonal Oct4 (Santa Cruz Biotechnology; 1:400), rabbit polyclonal Oct4 (Santa Cruz Biotechnology; 1:400), mouse monoclonal Cdx2 (BioGenex; 1:50), rabbit polyclonal Nanog (kindly provided by Ian Chambers, Institute for Stem Cell Research, Edinburgh, U.K.; 0.625 μg/ml), rabbit polyclonal Gata4 (Santa Cruz Biotechnology; 1:100), mouse monoclonal SSEA1 (Developmental Studies Hybridoma Bank, Iowa City, IA; 1:20), mouse monoclonal SSEA3 (Developmental Studies Hybridoma Bank; 1:20), and mouse monoclonal SSEA4 (Developmental Studies Hybridoma Bank; 1:20). Nuclei were visualized with DAPI (Sigma; 1 μg/ml).
Oct4 is a POU domain transcription factor expressed in ES cells and all cells of the morula, and is predominantly restricted to the ICM at the late blastocyst stage (15). Nanog is a homeodomain protein implicated in maintaining pluripotency with expression beginning in the compacted morula and maintained in the ICM and epiblast, and in ES cells (16–18). Cdx2 is a transcription factor initially expressed at the morula stage that becomes restricted to the trophectoderm of the blastocyst (19). Gata4 is one of the transcription factors initially expressed in the primitive endoderm (20). Blastocyst-derived murine ES cells express SSEA1 and lack expression of SSEA3 and SSEA4, whereas hES cells exhibit a reciprocal expression pattern (21).
Karyotyping. Chromosome preparation and G-banding was carried out as described in ref. 22. At least 4 × 106 ES cells were plated into a 25-cm2 feeder flask the night before analysis. The next morning, cells were mitotically arrested with colcemid, scraped from the flask, suspended in 0.56% KCl, fixed with methanol:glacial acetic acid (3:1), and spread onto slides. The spreads were G-banded 1 week later with Giemsa stain (Sigma).
Chimera Analysis. Blastocyst injection was performed as described in ref. 23. In brief, 10–15 ES cells were injected with a beveled pipette into MF1 host blastocysts in hanging drops of mKSOM-Hepes on a cooled stage. Embryos were allowed to recover for 45–90 min in mKSOM at 37°C and then transferred to E2.5 pseudopregnant PO females.
For making eight-cell/ES aggregation chimeras, depression wells were created in the floor of 50-mm Petri dishes with a darning needle, and drops of mKSOM were then placed over them (24). A small clump of 5–15 ES cells was placed with each denuded eight-cell PO embryo in separate depressions and cultured for 24 h under mineral oil (Sigma). Resulting aggregates were transferred to E2.5 pseudopregnant PO females. All male offspring showing chimerism in coat color were mated to strain MF1 females to test for germ-line transmission.
The developmental potential of morula-derived cells was also assessed in vitro and in chimeras at E6.5. MF1 host blastocysts were injected with eGFP+ morula-derived cells and either cultured in DMEM supplemented with 15% FCS or transferred to pseudopregnant recipients. In vitro-cultured outgrowths were analyzed for eGFP expression after 5 days. Transferred embryos were removed from their deciduae, and Reichert's membrane was reflected on E6.5. Embryos were transferred to 18-well μ-Slides (ibidi), and the contribution of eGFP+ donor cells was assessed by using fluorescent microscopy.
Results
Embryo Attachment and Outgrowth. Denuded morulae were carefully placed in direct contact with the feeder layer (Fig. 1I). One hundred forty-three of 283 (50.5%) such morulae attached to the feeders within 15 h, and each formed a single colony with no evidence of blastulation (Fig. 1J). The nearly 50% ratio of attaching to nonattaching morulae was consistent from experiment to experiment with a standard deviation of 18%. Within 2 days, attached morula colonies were obviously bipartite with morphologically distinct inner core and outer ring cells (Fig. 1K), and most notably, at no stage was cavitation evident between the core and ring. Rather, it appeared that the outer cells of the compacted morulae outgrew into the ring structure and that the inner cells stayed tightly attached to each other, forming the core. This core varied in size, possibly reflecting the number of inside cells in the morula at the time of its attachment. Some colonies were observed through 5 days of culture during which time no trophoblast giant cells or extraembryonic endoderm-like cells could be seen growing out of them (data not shown). Attached morulae colonies on 0.1% gelatin or tissue culture plastic without feeders failed to form this core/ring structure, flattened out, and differentiated with trophoblast giant cells evident by day 3 (Fig. 1 M–P). This finding shows that the feeder layer is restricting differentiation of the attached morulae outgrowths.
Fig. 1.
Comparison of embryo outgrowths from different preimplantation stages. Zygote (A–D), eight-cell (E–H), and morula (I–L) stage embryos that initially attached to a feeder layer outgrew into a characteristic core/ring structure with no evidence of blastulation. (M–P) In the absence of feeders, attached morulae flattened out and differentiated. (Q–T) Morulae that failed to attach cavitated to form a blastocyst. All images are to scale. (Scale bar, 100 μm.)
The morphogenesis of nonattaching morulae was drastically different from the ≈50% of denuded morulae in each experiment that attached to the feeders (Fig. 1 Q–T and Fig. 2). Nonattaching morulae invariably cavitated to form blastocyst-like structures (Fig. 1S and Fig. 2B) and only subsequently attached to the feeder layer, after which they formed an ICM-like outgrowth around which trophoblast giant cells were evident by day 4–5. Phytohemagglutinin treatment (25) was used in further trials in an attempt to increase the percentage of morulae that attached to the feeder layer but failed to significantly increase the rate of attachment.
Fig. 2.
Effect of initial attachment to a feeder layer on the morphogenesis of morula outgrowths. Stereomicroscopic view on day 3 of culture of morulae that initially attached (A) and a morula that failed to initially attach and subsequently cavitated to form a blastocyst (B). Note the larger core in the attached morulae compared with the ICM of the blastocyst. (Scale bars, 100 μm.)
It remains unclear why only about half of the morulae consistently attach before the stage of blastulation, and why this should invariably result in suppression of blastocoel formation. Attachment to the feeder layer was never observed in premorula stage embryos (1–8 cell) without modifying the method. Premorula embryos could be “stuck” to the dish by placing them in a small drop of protein-free medium immediately after acidic Tyrode's treatment to remove the zona pellucida. After 5 min, a feeder cell suspension was dispensed over the stuck embryos and most remained loosely stuck until the morula stage, at which time they became firmly attached to the feeder layer and subsequently spread out to form a core/ring structure similar to attached morulae. The timing of the formation of the characteristic core/ring colony morphology differed depending on the developmental stage of the explanted embryos. Three days of culture was required for attached morulae (Fig. 1 I–L), but the core/ring structure was only apparent after 6–7 days of culture from stuck zygotes (Fig. 1 A–D), 4–5 days from stuck four-cell embryos (data not shown), and 3–4 days from stuck eight-cell embryos (Fig. 1 E–H).
The identity of the attached embryo colonies was explored by immunohistochemical analysis of transcription factors expressed in the early embryo. Oct4 and Nanog were found in cells of the core, Gata4 in a shell immediately adjacent to the core, and Cdx2 in the entire outer ring structure (Fig. 3; see Materials and Methods for a description of the proteins analyzed). Cdx2, Oct4, and Nanog expression also was examined in morulae and blastocysts. All three proteins were expressed in all blastomeres of the 16-cell morula (Fig. 4D and data not shown). Oct4 was found primarily in the ICM of the E3.5 blastocyst, but expression was also observed in the polar trophectoderm (Fig. 4 A and E). Cdx2 was expressed exclusively in the trophectoderm (Fig. 4 B, C, and E), and Nanog was expressed in most, but not all, ICM cells (Fig. 4C) of the E3.5 blastocyst. Gata4 was not detected in any of the morulae or E3.5 blastocysts examined. From this analysis it appears that the attached morula colonies continue a pseudonormal developmental progression to a tri-layered (Cdx2-positive trophectoderm, Oct4- and Nanog-positive ICM/epiblast, and Gata4-positive primitive endoderm) structure even in the absence of cavitation.
Fig. 3.
Expression of early lineage markers in attached morulae. Attached morulae outgrowths on day 3 of culture express Oct4 (A and D) and Nanog (G) in the core cells, Cdx2 in the cells of the outer ring (B), and Gata4 in the cells surrounding the core (E). Nuclei were visualized with DAPI (C, F, and I; blue). The Nanog-positive cells were sitting just above the outer ring's focal plane in G–I, causing the Cdx2-positive outer ring to be out of focus in this specimen. All images are to scale. (Scale bar, 100 μm.)
Fig. 4.
Expression of early lineage markers in morulae and blastocysts. Laser scanning confocal (A–C) and fluorescent microscopic (D–E) analysis revealed expression of Oct4 in all cells of the morula and primarily in the ICM of the blastocyst (A, D, and E; red), Cdx2 in all cells of the morula and exclusively in the trophectoderm of the blastocyst (B–E; green), and Nanog in most but not all cells of the ICM at the blastocyst stage (C; red). Nuclei were stained with DAPI (A and B; blue).
Derivation of Cell Lines from Embryo Outgrowths. Entire colonies obtained from attached morulae on day 3 of culture were peeled off the feeder layer with a pair of fine needles and dissociated into a single-cell suspension (see Materials and Methods). The resulting suspension was distributed onto a fresh feeder layer. After 3–4 days, wells with ES-like cell colonies were passaged with trypsin onto a fresh feeder layer. ES cell lines were derived from 48% of strain 129 attached morulae that were treated with these methods (n = 23). The techniques have yet to be applied to morulae of other strains. All morula-derived cell lines showed varying degrees of differentiation into extraembryonic endoderm-like cells at early passage number. Because whole-well passaging was used to propagate the cells for 1 or 2 passages initially, sublines were derived by dissociation of single ES cell colonies with 0.125% trypsin and dispersal onto a fresh feeder layer.
Putative ES cell lines were also derived from 5 of 6 day-4 colonies from stuck four-cell embryos and 8 of 19 day-3.5 colonies from stuck eight-cell embryos. These lines were classified by morphology and growth characteristics only, and they have yet to be analyzed further.
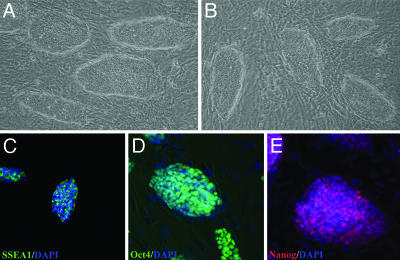
Characterization of Cell Lines. Morula-derived lines grow with morphologies typical of ES cells (Fig. 5 A and B). Immunohistochemistry showed that the cells expressed SSEA1 (Fig. 5C), Oct4 (Fig. 5D), and Nanog (Fig. 5E). Lines were negative for SSEA3 and SSEA4. In early passages, Cdx2 was coexpressed with Oct4, but expression was highly variable and never observed after continued passage, subcloning, or cryopreservation. Although all morula-derived lines were Nanog-positive, expression was not uniform within individual colonies (Fig. 5E). The same pattern of expression was seen in germ-line-competent, epiblast-derived ES cell lines (data not shown), and the basis for this mosaic pattern is currently unclear.
Fig. 5.
Expression of molecular markers characteristic of ES cells in morula-derived cell lines. Phase contrast photomicrographs of morula-derived ES cell lines mm1c (A) and mm10 (B) as well as representative expression of SSEA1 (C), Oct4 (D), and Nanog (E) in morula-derived ES cell lines. Nuclei were stained with DAPI (C–E; blue).
G-banded metaphase spreads were analyzed for 11 morula-derived cell lines to determine their genetic stability. Nine lines were found to be XX and only two were found to be XY. Typically, the bias is toward XY ES cell lines, but during the period when these experiments were undertaken, XX lines predominated even with conventional methods of derivation from ICM/epiblast (P.J.T. and F. Brook unpublished observations). XX lines tended to lose one X between passages 6 and 14 (mm1, mm2, and mm3). Line mm9 had the only autosomal aneuploidy as it was 41, XX trisomy 8 at passage 6. The two XY lines, mm10 and mm11, were both euploid at all passages tested (P5–P21), and this shows that morula-derived cell lines can maintain genomic stability in culture.
The developmental potential of the two XY lines, mm10 P12 and mm11e P10, was tested by their contribution to chimeras. Ten to 15 cells were injected into MF1 strain host blastocysts, and the resulting embryos were transferred to E2.5 pseudopregnant PO strain females and analyzed at E6.5 or carried to term. Chimerism was judged by coat color, as morula-derived lines were agouti and MF1 or PO host embryos were albino, or by eGFP expression in the case of line mm11e. Of 25 embryos analyzed at E6.5, 21 showed chimerism in the epiblast. In 17 of these chimeras, the morula-derived cells (mm11e) contributed in high proportions to the entire epiblast, whereas the remaining 4 chimeras had weak donor contribution or contribution to only part of the epiblast. Most of the embryos analyzed had not commenced gastrulation, but the two early primitive streak embryos showed chimerism in the nascent mesoderm. The morula-derived cells did not contribute significantly to the extraembryonic ectoderm, visceral endoderm, or to cells of Reichert's membrane. One embryo had a small patch of two to three donor cells in the extraembryonic ectoderm, whereas another had a few cells with weak eGFP expression in Reichert's membrane. Additional analysis of the extraembryonic potential of morula-derived cells was carried out in vitro. Blastocysts and morulae were injected with 10 mm11e cells and were cultured for 5 days in DMEM with 15% FCS. Donor contribution to the outgrowths was analyzed by eGFP expression. Unmanipulated eGFP positive blastocysts were used as a control to ensure that the eGFP transgene expression was maintained in all cell types of the outgrowths. All injected morulae and blastocyst outgrowths had eGFP-positive mm11e cells in the ICM-derived portion, but none of the easily distinguishable trohectoderm giant cells were of mm11e donor origin.
Of the injected blastocysts that were carried to term, 6 of 8 pups born from line mm10 were chimeras and 3 of the chimeras were male, whereas 12 of 17 pups born from line mm11e were chimeras and 9 of the chimeras were male. Aggregation chimeras were generated from line mm11e P14. Eight-cell stage PO strain embryos were aggregated with 5–15 mm11e cells in depression wells covered with mKSOM. The following day, mm11e cells were mainly located in the center of the resulting compacted morulae, which were transferred to E2.5 pseudopregnant PO females. Of 11 embryos transferred, only 3 produced live pups, and all were chimeras (2 males and 1 female). All chimeras produced from both blastocyst injections and eight-cell/mm aggregations showed, on average, >70% donor contribution to coat color. Interestingly, 1 male chimera from eight-cell/mm11e aggregations was entirely agouti (Fig. 6B). This finding was confirmed by eGFP expression and supports the robust developmental capacity of morula-derived cell lines. Line mm11e P10 has transmitted through the germ line (Fig. 6A).
Fig. 6.
Contribution of morula-derived ES cells to chimeras. (A) Germ-line-competent male chimera (right) from morula-derived ES cell line mm11e P10 with strain MF1 female mate (left) and pups. (B) One entirely agouti chimera was derived from eight-cell/mm11e aggregations. The host eight-cell embryo was from the albino PO strain.
Discussion
Until now, derivation of germ-line-competent ES cells in the mouse has only been demonstrated by using blastocysts or their tissue derivatives as the starting material. Although cells resembling ES cells in both morphology and growth characteristics have been obtained previously from preblastocyst stages (3, 4), their pluripotency was never thoroughly demonstrated, and germ-line-competent chimeras were never obtained. The results presented here show that preblastocyst stage embryos can be used to derive germ-line-competent mouse ES cells with high efficiency.
ES cell lines were readily obtained from denuded embryos explanted before blastulation. Consistently, ≈50% of denuded morulae attached within hours to the feeder cell layer. Attached morulae exhibited a strikingly different pattern of morphogenesis from those that did not attach. Most obviously, formation of a blastocoel was completely suppressed in attached morulae. In the early embryo, the blastocoel forms when the initial trophectoderm cells differentiate. Molecular markers characteristic of cell populations of the early embryo indicated that differentiation into trophectoderm and ICM tissue had not been suppressed. Lack of trophoblast giant cells in attached morulae outgrowths, however, suggests that the feeder layer imposed some restraints on further differentiation. Attached morula outgrowths that were cultured for 3 days yielded ES cell lines in 48% of cases. This is remarkably greater than the 25% success rate that is typically considered high when using conventional methods of ES cell derivation from blastocysts. Blastocyst injection experiments demonstrated that morula-derived lines contributed in high proportions to resulting chimeras and were, most notably, also able to colonize the germ line of those mice. These findings show that genuine pluripotent cell lines can de derived from preblastocyst stage mouse embryos, and they provide a simple and efficient alternative to the blastocyst as sources of such cells. Putative ES cells have also been obtained recently from human morulae by initially culturing the morulae underneath a feeder layer (26).
Although denuded premorula stages did not spontaneously attach to the feeders, they could be made to exhibit the same core/ring outgrowths as attached morulae by “sticking” them to the culture dish and then adding feeder cells. With this method, ≈78% of premorula embryos remained attached to the feeders and outgrew into the core/ring colony morphology characteristic of attached morulae. Putative ES cell lines were obtained from >50% of stuck four-cell and eight-cell outgrowths after 4 and 3.5 days of culture, respectively.
The possibility exists that ES cells derived during the preblastocyst period may have a wider differentiation potential than those derived from blastocysts. Blastocyst-derived ES cells rarely, if ever, contribute to trophectoderm or extraembryonic endoderm derivatives in chimeras (27). Investigations into the extraembryonic potential of morula-derived cells provided no evidence that they are able to colonize the trophectodermal lineage. This finding suggests that regardless of the stage of explantation, embryos must progress to an equivalent period before ES cells can be obtained. Attached embryos from progressively earlier stages required longer periods in culture before they reached an equivalent state that yielded ES cells. It remains unclear whether this state represents the sole time point from which ES cells can be obtained or whether it demarcates the beginning of a window of time during which the derivation of ES cells is possible. Notably, preblastocyst embryos divide through cleavage divisions that increase the number of cells without growth. It may not be until cleavage divisions cease that cells can be adapted from the embryo (or embryo outgrowth) to propagate in culture. Recent evidence suggests that cleavage divisions are maintained throughout preimplantation development (28), and further studies may define distinct changes in the control of cell division that occur between cells dividing by cleavage and growth-based mechanisms that are required for the extraordinary ability of ES cells to proliferate in vitro. The data presented here suggest that this transition routinely occurs in culture and may define the stage at which ES cells can be derived.
While given substantial attention for their potential in regenerative medicine, ES cells also provide a powerful tool to aid researchers in understanding the mechanisms that control early developmental processes. The stage of embryonic development at which ES cells are derived may be crucial to their subsequent function in experimental models and cell therapy. The findings presented here afford the possibility to compare the developmental potential of ES cell lines derived from embryos at any stage of preimplantation development. Even in the mouse, ES cell lines can vary considerably in their developmental potential. This may be due, at least in part, to epigenetic modifications that are known to occur throughout preimplantation development (29). Epigenetic abnormalities of ES cells are not corrected in differentiated tissue derivatives (30), and the epigenetic status of the embryo used to derive ES cells may influence the developmental potential of the ES cells obtained. Additionally, in vitro fertilization, embryo culture, nuclear transfer, and superovulation are known to cause methylation defects that can result in abnormal development (31, 32), and further consideration must be afforded to how these manipulations are affecting the normalcy of resulting ES cell lines, especially in the human. The paucity of human embryos available for research and the lack of an analogous chimera assay impede rigorous characterization of the normal developmental potential of hES cells. A significant amount of further investigation must be undertaken in the mouse and non-human primate to potentiate the use of hES cells in the laboratory and the clinic.
Acknowledgments
I thank Prof. Richard Gardner and Dr. Ron McKay for advice, guidance, and review of the manuscript; Dr. Frances Brook, Tim Davies, and Dr. Ted Evans for assistance with blastocyst injection and karyotyping; Prof. Chris Graham, Dr. Paul Fairchild, and members of the McKay and Gardner laboratories for technical assistance and discussion; and Dr. Karen Downs for instruction on postimplantation embryo dissections. This work was supported by the National Institutes of Health. P.J.T. is a National Institutes of Health–University of Oxford Biomedical Research Scholar.
Abbreviations: En, embryonic day n; ES, embryonic stem; hES, human ES; ICM, inner cell mass; mKSOM, potassium simplex optimized medium; PEF, primary embryonic fibroblast.
References
- 1.Brook, F. A. & Gardner, R. L. (1997) Proc. Natl. Acad. Sci USA 94, 5709-5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brook, F. A., Evans, E. P., Lord, C. J., Lyons, P. A., Rainbow, D. B., Howlett, S. K., Wicker, L. S., Todd, J. A. & Gardner, R. L. (2003) Diabetes 52, 205-208. [DOI] [PubMed] [Google Scholar]
- 3.Eistetter, H. R. (1989) Dev. Growth Differ. 31, 275-282. [DOI] [PubMed] [Google Scholar]
- 4.Delhaise, F., Bralion, V., Schuurbiers, N. & Dessy, F. (1996) Eur. J. Morphol. 34, 237-243. [DOI] [PubMed] [Google Scholar]
- 5.Gardner, R. L., Lyon, M. F., Evans, E. P. & Burtenshaw, M. D. (1985) J. Embryol. Exp. Morphol. 88, 349-363. [PubMed] [Google Scholar]
- 6.Xu, R. H., Chen, X., Li, D. S., Li, R., Addicks, G. C., Glennon, C., Zwaka, T. P. & Thomson, J. A. (2002) Nat. Biotechnol. 20, 1261-1264. [DOI] [PubMed] [Google Scholar]
- 7.Niwa, H., Miyazaki, J. & Smith, A. G. (2000) Nat. Genet. 24, 372-376. [DOI] [PubMed] [Google Scholar]
- 8.Tanaka, S., Kunath, T., Hadjantonakis, A. K., Nagy, A. & Rossant, J. (1998) Science 282, 2072-2075. [DOI] [PubMed] [Google Scholar]
- 9.Rossant, J. & Lis, W. T. (1979) Dev. Biol. 70, 255-261. [DOI] [PubMed] [Google Scholar]
- 10.Ziomek, C. A., Johnson, M. H. & Handyside, A. H. (1982) J. Exp. Zool. 221, 345-355. [DOI] [PubMed] [Google Scholar]
- 11.Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. & Roder, J. (1993) Proc. Natl. Acad. Sci. USA 90, 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hadjantonakis, A. K., Gertsenstein, M., Ikawa, M., Okabe, M. & Nagy, A. (1998) Mech. Dev. 76, 79-90. [DOI] [PubMed] [Google Scholar]
- 13.Summers, M. C., Bhatnagar, P. R., Lawitts, J. A. & Biggers, J. D. (1995) Biol. Reprod. 53, 431-437. [DOI] [PubMed] [Google Scholar]
- 14.Nicolson, G. L., Yanagimachi, R. & Yanagimachi, H. (1975) J. Cell Biol. 66, 263-274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scholer, H. R., Ruppert, S., Suzuki, N., Chowdhury, K. & Gruss, P. (1990) Nature 344, 435-439. [DOI] [PubMed] [Google Scholar]
- 16.Hatano, S. Y., Tada, M., Kimura, H., Yamaguchi, S., Kono, T., Nakano, T., Suemori, H., Nakatsuji, N. & Tada, T. (2005) Mech. Dev. 122, 67-79. [DOI] [PubMed] [Google Scholar]
- 17.Mitsui, K., Tokuzawa, Y., Itoh, H., Segawa, K., Murakami, M., Takahashi, K., Maruyama, M., Maeda, M. & Yamanaka, S. (2003) Cell 113, 631-642. [DOI] [PubMed] [Google Scholar]
- 18.Chambers, I., Colby, D., Robertson, M., Nichols, J., Lee, S., Tweedie, S. & Smith, A. (2003) Cell 113, 643-655. [DOI] [PubMed] [Google Scholar]
- 19.Beck, F., Erler, T., Russell, A. & James, R. (1995) Dev. Dyn. 204, 219-227. [DOI] [PubMed] [Google Scholar]
- 20.Arceci, R. J., King, A. A., Simon, M. C., Orkin, S. H. & Wilson, D. B. (1993) Mol. Cell. Biol. 13, 2235-2246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Henderson, J. K., Draper, J. S., Baillie, H. S., Fishel, S., Thomson, J. A., Moore, H. & Andrews, P. W. (2002) Stem Cells 20, 329-337. [DOI] [PubMed] [Google Scholar]
- 22.Nagy, A. (2003) Manipulating the Mouse Embryo: A Laboratory Manual (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY).
- 23.Robertson, E. J. (1987) Teratocarcinomas and Embryonic Stem Cells: A Practical Approach (IRL, Oxford/Washington, DC).
- 24.Nagy, A., Gocza, E., Diaz, E. M., Prideaux, V. R., Ivanyi, E., Markkula, M. & Rossant, J. (1990) Development (Cambridge, U.K.) 110, 815-821. [DOI] [PubMed] [Google Scholar]
- 25.Mintz, B., Gearhart, J. D. & Guymont, A. O. (1973) Dev. Biol. 31, 195-199. [DOI] [PubMed] [Google Scholar]
- 26.Strelchenko, N., Verlinsky, O., Kukharenko, V. & Verlinsky, Y. (2004) Reprod. Biomed. Online 9, 623-629. [DOI] [PubMed] [Google Scholar]
- 27.Beddington, R. S. & Robertson, E. J. (1989) Development (Cambridge, U.K.) 105, 733-737. [DOI] [PubMed] [Google Scholar]
- 28.Aiken, C. E., Swoboda, P. P., Skepper, J. N. & Johnson, M. H. (2004) Reproduction 128, 527-535. [DOI] [PubMed] [Google Scholar]
- 29.Santos, F., Hendrich, B., Reik, W. & Dean, W. (2002) Dev. Biol. 241, 172-182. [DOI] [PubMed] [Google Scholar]
- 30.Dean, W., Bowden, L., Aitchison, A., Klose, J., Moore, T., Meneses, J. J., Reik, W. & Feil, R. (1998) Development (Cambridge, U.K.) 125, 2273-2282. [DOI] [PubMed] [Google Scholar]
- 31.Shi, W. & Haaf, T. (2002) Mol. Reprod. Dev. 63, 329-334. [DOI] [PubMed] [Google Scholar]
- 32.Young, L. E., Fernandes, K., McEvoy, T. G., Butterwith, S. C., Gutierrez, C. G., Carolan, C., Broadbent, P. J., Robinson, J. J., Wilmut, I. & Sinclair, K. D. (2001) Nat. Genet. 27, 153-154. [DOI] [PubMed] [Google Scholar]