Abstract
Cell therapy is investigated as a powerful intervention to ameliorate the consequences of coronary artery disease. Among the different stem cell options, mesenchymal stem cells (MSCs) are particularly attractive due to their high availability, as well as immune privileged status. However, it is still unclear whether mesenchymal stem cells can acquire cardiomyogenic characteristics after they are transplanted to the myocardium. In this manuscript, we outlined a protocol that illustrates the plasticity of MSCs and their ability to acquire cardiogenic characteristics when they are in an ischemic-like environment, as typically encountered after transplantation into the ischemic heart.
Basic protocol 1: Isolation of mesenchymal stem cells
Support basic protocol 1: Characterization of MSCs
Basic protocol 2: Isolation of neonatal cardiomyoctes (NCMs)
Support basic protocol 1: Characterization of NCMs
Basic protocol 3: Cardiogenic plasticity MSCs when under ischemic like conditions
Support basic protocol 3: Characterization of the cardiogenic potential of MSCs
Keywords: cardiogenic, mesenchymal stem cells, neonatal cardiomyocytes, luciferase
Introduction
There is significant interest in the use of stem cells (SCs) as an alternative therapy for myocardial repair after myocardial infarction (MI) (Leeper, Hunter, & Cooke, 2010; Segers & Lee, 2008). Mesenchymal stem cells (MSCs) are amongst the most common adult stem cells used for this purpose (Le Blanc & Pittenger, 2005; Pittenger, 2008; Pittenger et al., 2019; Pittenger et al., 1999; Pittenger & Martin, 2004; Shake et al., 2002). MSCs are adult stem cells that originate from the bone marrow or adipose tissue and can differentiate into three main lineages: chondrogenic, osteogenic or adipogenic (Pittenger et al., 1999; Pittenger, Mosca, & McIntosh, 2000). Furthermore, there has been significant effort to elucidate whether MSCs can acquire cardiomyogenic characteristics. Specifically, different research groups have shown the potential of MSCs to acquire cardiomyogenic characteristics, mostly using ex-vivo approaches (Behfar & Terzic, 2006, 2007; Shi et al., 2016). However, the plasticity of MSCs after transplantation has been a topic of debate. Elucidating whether MSCs can acquire cardiogenic characteristics could provide a better understanding of the phenotypic changes that MSCs may undergo after transplantation to the myocardium, and their potential therapeutic efficacy.
In this manuscript we demonstrate how MSCs can express cardiomyogenic characteristics when they are under conditions resembling those that are encountered in the ischemic myocardium. Furthermore, we used state-of-the-art reporter gene molecular imaging to monitor the expression of these myogenic characteristics using conditional transgenes whose expression is regulated by pathway-specific promoters (Troponin T in this study).
Basic Protocol 1:
Isolation of Mesenchymal Stem Cells (MSCs)
Figure 1 depicts the main steps in sequence for the isolation of MSCs from the bone marrow of a rat femur in this protocol. Femurs are excised from young rats and the bone marrow is flushed out, filtered, centrifuged, re-suspended and plated in plastic tissue culture plates. MSCs adhere to the plastic surface and grow rapidly in the culture medium, while off-target cells are outcompeted within 2–3 passages.
Figure 1.
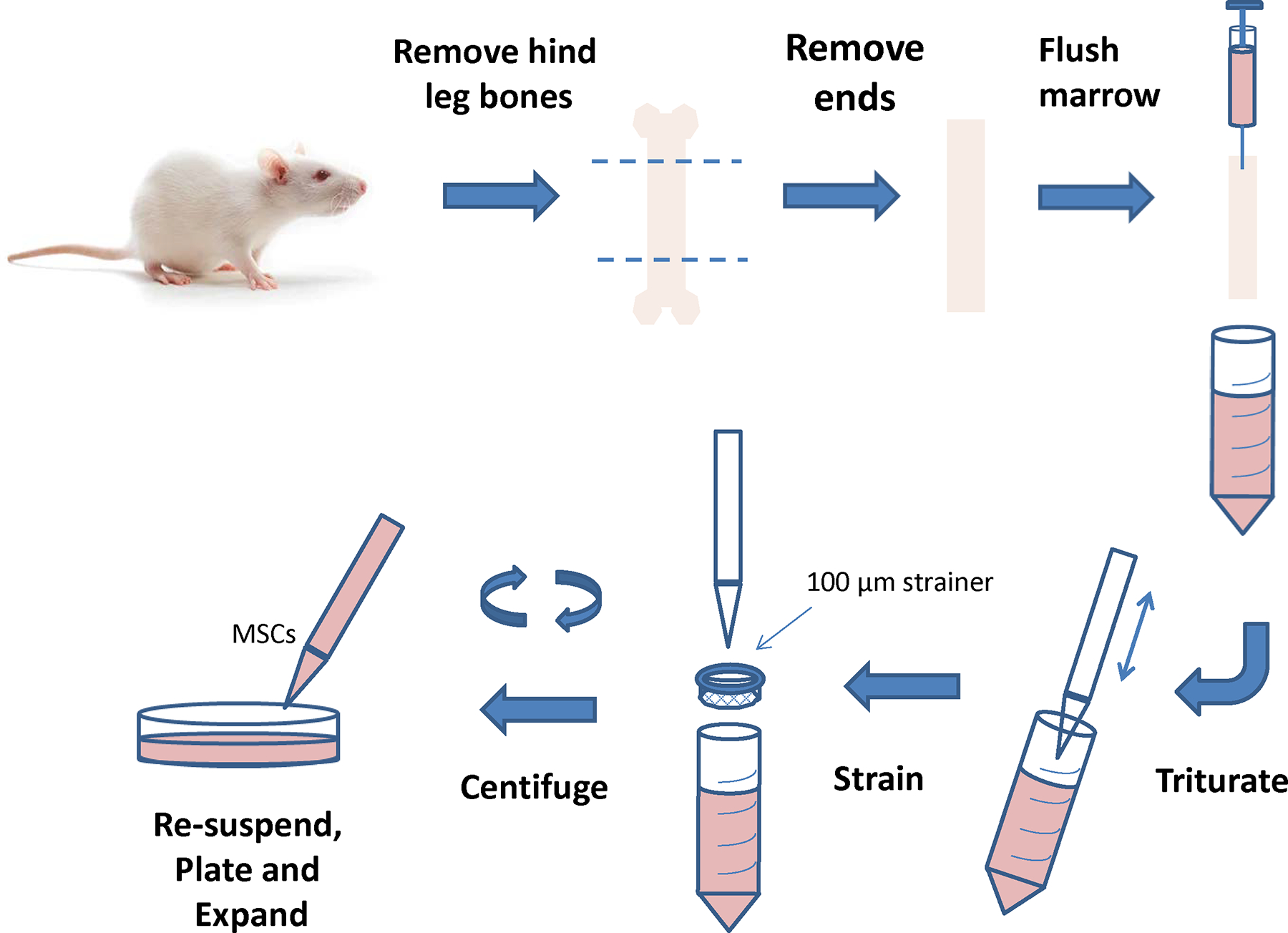
Cartoon depicting the process for isolation of mesenchymal stem cells (MSCs) from bone marrow.
Reagents
Ca2+-Mg2+ free Hank’s Balanced Salt Solution (CMF-HBSS) (Mediatech, #21–022-CV)
Fetal bovine serum (FBS) (Mediatech, #35–010-CV)
70% Alcohol
Phosphate buffered saline (PBS) (Invitrogen, #10010023)
Dimethyl sulfoxide (DMSO, Sigma Aldrich, #D5879)
Materials/Equipment
Tissue culture vessels: T75 flasks (Corning, #353136), 10 cm dishes (Fisher Scientific, NC9385690), 6-well plates (Sigma Aldrich, #CLS3516)
Dissection tools and reagent: forceps, scissors, 70% ethanol, CO2 euthanasia chamber, sterile gauze, 6 ml syringe with 21G needle, sterile razor blades.
General: 50 ml Falcon tubes (Corning, #352098), 100 μm cell strainers (Fisher Scientific, #08–771-19), serological pipettes, micropipettes and tips, tissue culture shaker/rotator, petri dish, refrigerated centrifuge
Steps and annotations
In Surgical Procedure Room:
Euthanize one 4–6 weeks old rat in CO2 euthanasia chamber.
Spray 70% alcohol on the fur around hind legs and abdomen to sterilize the area and reduce contamination risk.
Place rat in the supine position.
Spray gloves with 70% alcohol.
Dislocate and detach long bones (femur, tibia/fibula) by pulling from extremities.
Remove any muscle that remains on lower leg bones with gauze dampened with alcohol.
Put the bones into a well of a 6-well plate filled with PBS or CMF-HBSS and place on ice.
In Tissue Culture Room (under sterile conditions, in hood):
Using large scissors or garden shears remove both ends of one bone to expose marrow.
Set scissors or shear aside on sterile paper towel or gauze. Fill the syringe with isolation medium.
Flush bone marrow cavity with the syringe, collecting the medium in the tube. Repeat as needed until marrow is removed and bone appears white.
Pipet the medium containing the bone marrow into 100μm strainer until all has been strained. The goal is to remove remaining clumps.
Centrifuge the strained medium containing bone marrow at 1,250 rpm in 4°C for 5–10 minutes.
Re-suspend pellet in 10 ml fresh medium and plate in 10 cm tissue culture plate or T75 flask.
Add extra medium up to 10 ml in 10 cm plate or 20 ml in T75 flask.
Do not disturb for 3 days.
Change medium every 2–3 days until tissue culture plate/flask is 90% confluent.
Cells should be ready to freeze or Flow Cytometry by passage 3.
Freeze cells in 90% FBS and 10% DMSO.
Support Basic Protocol 1: Characterization of MSCs by Flow Cytometry
Reagents
Phosphate buffered saline (PBS, Invitrogen, #10010023)
0.25% Trypsin-EDTA (Invitrogen, #25200056)
Fetal bovine serum (FBS, Mediatech, #35–010-CV)
Normal donkey serum (Abcam, #ab7475)
Antibodies as listed in Table 1
4% paraformaldehyde (PFA, Biotium, #22023)
Table 1.
FACS antibodies used for characterization of MSCs
| Antibody | Company | Volume per million cells |
|---|---|---|
| Mouse anti-rat/mouse CD90.1-APC | Biolegend | 5 μl |
| Hamster anti-mouse/rat CD29-PEcy7 | Biolegend | 5 μl |
| Mouse anti-rat CD11b/c-APC | Biolegend | 5 μl |
| Mouse anti-rat CD45-FITC | ABD Serotec (BioRad) | 10 μl |
Materials/Equipment
Tissue culture vessels: T75 flasks (Corning, #353136), 10 cm dishes (Fisher Scientific, #NC9385690)
5 ml polystyrene round-bottom flow cytometry tubes (Falcon, #352058)
General: 15 ml Falcon tubes (Corning, #352097), serological pipettes, micropipettes and tips, tissue culture shaker/rotator, refrigerated centrifuge, tissue culture incubator
Analytical (non-sorting) digital flow cytometer running FACSDiva v6.1.3 software (FACSCanto, BD Biosciences)
BD CellQuest™ Pro analysis software (BD Biosciences)
Steps and annotations
Remove media and wash cells with 5 ml PBS.
Add 2 ml 0.25% Trypsin-EDTA and incubate at 37 °C for 3 minutes.
Gently tap the flask to allow detachment of cells from the plastic surface.
Add 200 μl of FBS to inactivate the Trypsin-EDTA.
Add 3 ml PBS and transfer the solution containing the cells in a 15 ml Falcon tube.
Centrifuge at 1,250 rpm for 5 min.
Remove the supernatant, carefully resuspend the pellet in PBS and aliquot the cells in flow cytometry tubes: 5×105 cells per tube.
Centrifuge at 1,250 rpm for 5 min.
Remove the supernatant and resuspend the pellet in 90μl PBS + 10 μl normal donkey serum (blocking solution). Incubate at room temperature for 15 min.
Add the antibodies at the desired concentration (see Table 1). Incubate at room temperature for 30 min in the dark.
Add 3 ml PBS and centrifuge at 1,250 rpm for 5 min.
Remove the supernatant and resuspend the pellet in 200μl 0.4% PFA.
Run the samples in the flow cytometer and analyze the data.
After isolation, MSCs isolated from bone marrow were characterized by Flow Cytometry as detailed in Figure 2. Specifically, isolated MSCs were negative for CD45 (hematopoietic marker) and CD11b (leukocyte marker) and positive for CD90 and CD29, confirming that they are MSCs (Dominici et al., 2006).
Figure 2.
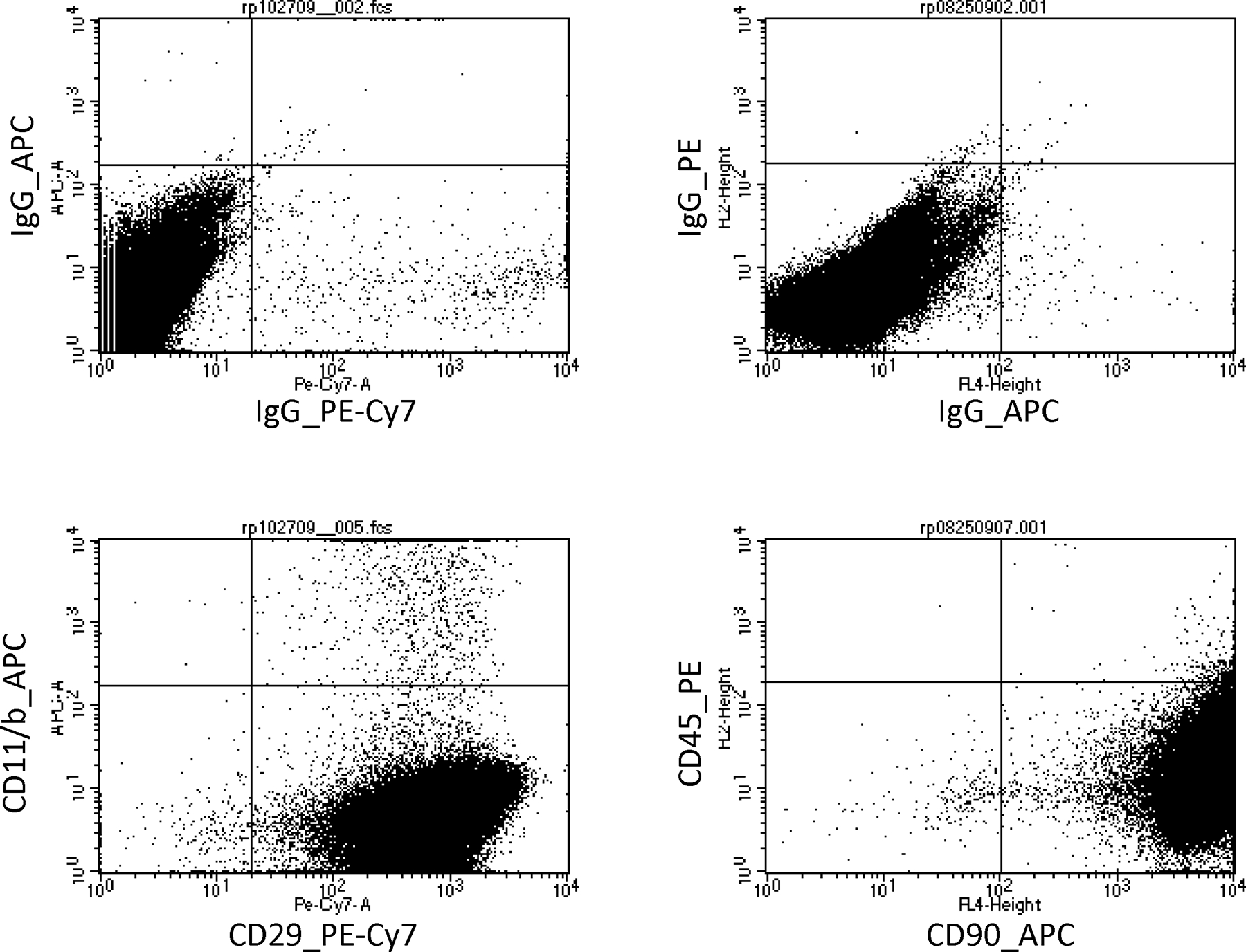
Characterization of isolated MSCs. Top panels, show the negative controls (IgG) of the CD29, CD45, CD11b/c and CD90. Bottom panels show that isolated MSCs were positive for CD29 (99.4%) and CD90 (99.98%) and negative for CD45 (0.32%) and CD11b/c (0.93%).
Basic protocol 2: Isolation of Neonatal Cardiomyocytes (NCM)
There is significant interest in the phenotype that MSCs may acquire after transplantation to the injured myocardium. To simulate some of the ischemic conditions that MSCs may encounter in that setting, cardiomyocytes that have been serum-deprived can be used. The 1st step is to isolate cardiomyocytes from rat. Steps are described below and in Figure 3.
Figure 3.
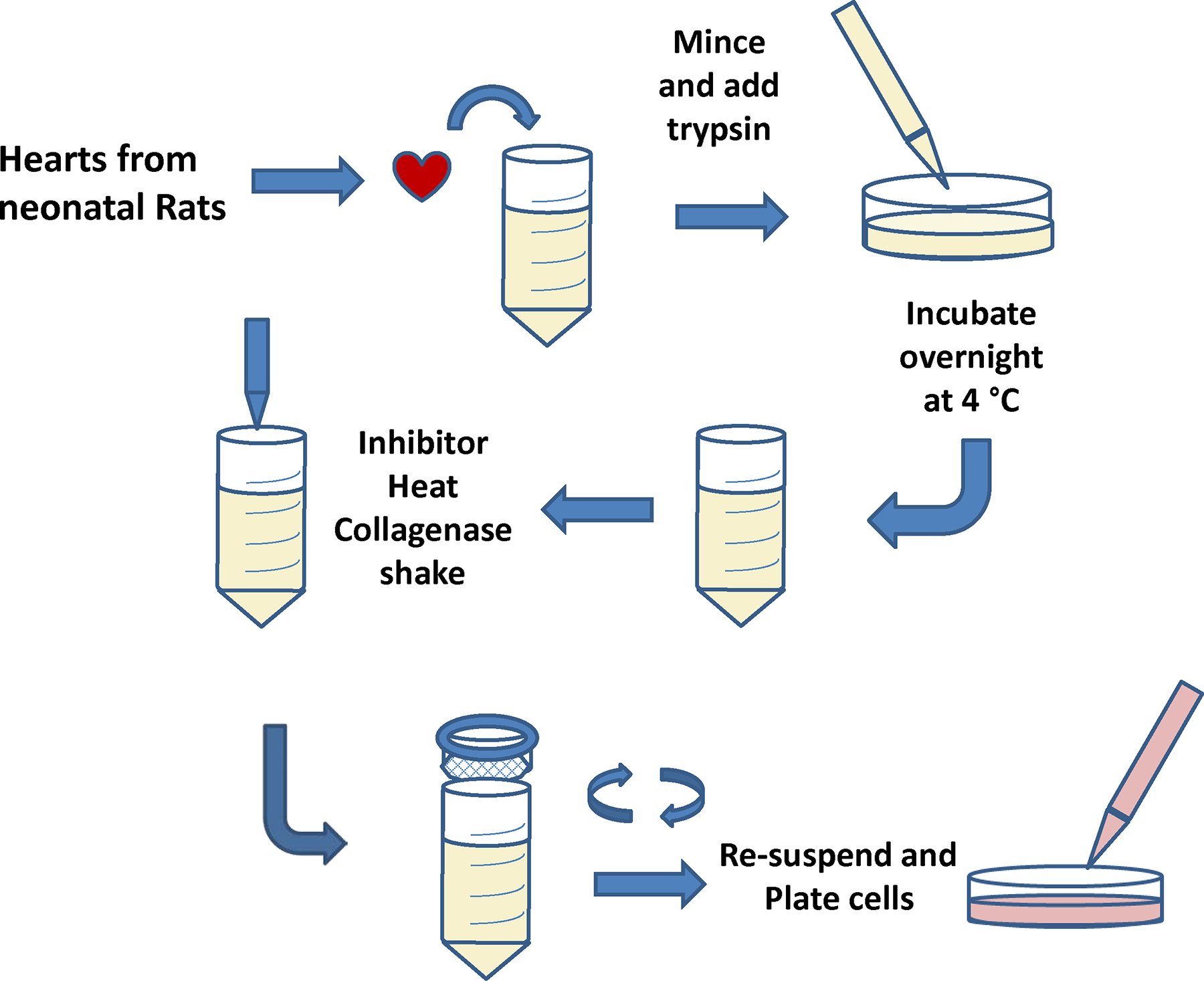
Cartoon depicting the process for isolation of neonatal cardiomyocytes (NCM).
Reagents
High glucose Dulbecco’s modified Eagle’s medium (DMEM, ThermoFisher Scientific, #11965092) supplemented with 10% fetal bovine serum (FBS, Mediatech, #35–010-CV) and 1% penicillin/streptomycin (ThermoFisher Scientific, #15140122)
Liebovitz-L-15 serum free medium (ThermoFisher Scientific, 11415064)
Ca2+ Mg2+ free Hank’s Balanced Salt Solution (CMF-HBSS, Mediatech, #21–022-CV)
Phosphate Buffered saline (PBS, Invitrogen, #10010023)
0.25% Trypsin-Ethylenediaminetetraacetic acid (EDTA) (ThermoFisher Scientific, #25200056)
100 mM 5-Bromo-2’-deoxyuridine (5-BrdU, Sigma Aldrich, #B5002) dissolved in PBS and filtered through 0.2μm syringe filter (Corning, #431229)
Tissue digestion enzymes (Worthington Biochemical Corp.): Trypsin (1mg), Trypsin (2mg), Trypsin soybean inhibitor, Collagenase (1.5mg)
Materials/Equipment
Tissue culture vessels: T75 flasks (Corning, #353136), 10 cm dishes (Fisher Scientific, NC9385690), 6-well plates (Sigma Aldrich, #CLS3516)
Dissection tools and reagent: forceps, scissors, 70% ethanol, CO2 euthanasia chamber, sterile gauze, 6 ml syringe with 21G needle, razor blades
General: 50 ml Falcon tubes (Corning, #352098), 100 μm cell strainers (Fisher Scientific, #08–771-19), serological pipets, micropipettes and tips, tissue culture shaker/rotator, petri dish, refrigerator, cell culture incubator, water bath, refrigerated centrifuge
Steps and annotations
Euthanize one rat pup by beheading.
Cut open chest and rib cage, to expose the beating heart, and excise.
Place heart in falcon tube of cold CMF-HBSS (35 mL) and keep on ice.
Mince hearts well with sterile razor blades.
Add 9 ml of fresh CMF-HBSS to petri dish containing minced hearts.
Add 2 ml CMF-HBSS to trypsin vial.
Add 1 ml trypsin solution to minced hearts and swirl the dish gently to mix.
Transfer dish to refrigerator at 4 °C and digest overnight (16–20 hours).
Transfer digested tissue and trypsin solution to a 50 ml falcon tube.
Add 1 ml of room temperature CMF-HBSS to vial of soybean trypsin inhibitor.
Transfer contents of vial to falcon tube containing digested hearts and place in 37 °C water bath.
Add 5 ml of pre-warmed L-15 medium to collagenase vial, cap and invert several times to dissolve enzymes.
Transfer contents of collagenase vial to pre-warmed falcon tube containing digested hearts and place in shaker at 37 °C set at 40–50 rpm (if using a rotator set to 4–5 rpm) for 40 minutes.
Using 10 ml pipet, triturate digested tissue suspension at least 10 times to disrupt clumps.
Allow undigested tissue to settle for 2–3 minutes.
Place the cell strainer on a 50 ml falcon tube, rinse with 2–3 ml of L-15 medium, and put the suspension through the strainer retaining settled material. Add 5 ml of L-15 medium and repeat trituration. Filter remaining suspension through strainer. Allow strained material to digest for another 50 minutes undisturbed.
Centrifuge at 50–100g for 5 minutes and re-suspend in 20ml of L-15 medium.
Repeat centrifugation and re-suspend in complete DMEM.
Plate cells in 10 cm tissue culture plates at a density of approximately 1.5–2 hearts per plate. Note: NCM cannot be trypsinized and re-plated after attachment.
Add 100 μl of 10mM 5-BrdU per ml of complete DMEM to hinder growth of any off-target cells that may be present.
Place plates in a tissue culture incubator (37°C, 5% CO2) and leave undisturbed for 2–3 days.
Change medium every 2–3 days thereafter. Make sure that NCM are beating.
Note: volumes given are appropriate for digestion of 10–12 hearts from pups less than 5 days old.
Note: NCMs do not proliferate in culture. Furthermore, they cannot be detached and re-plated in a fresh culture vessel post isolation. However, with proper care NCMs can be maintained for several weeks in the same culture dish without loss of function.
Note: NCMs may stop beating temporarily after media changes (or when calcium in medium is exhausted), thus their condition and function should be evaluated before, or a few hours after, changing culture medium. Only NCM that are actively beating should be used to produce conditioned medium. The same plate of NCM may be treated with low serum medium more than once, so long as they are still beating.
Note: Using a minimal volume of low serum medium when treating NCMs will yield higher concentration of cytokines and will therefore be more effective in the induction of the cardiogenic potential of MSCs.
Support basic protocol 2: Characterization of NCMs
Immunofluorescence of cardiac proteins in NCMs.
Reagents
4% paraformaldehyde (Biotium, 22023)
Phosphate Buffered Saline (PBS, Invitrogen, 10010023)
Goat serum (Sigma Aldrich, #G9023)
4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific, #D3571)
Materials/Equipment
-
1.
4-well chamber slides (Fisher Scientific, #12–565-2)
-
2.
Inverted microscope
-
5.
Pipettes
-
6.
Cover slides (Fisher Scientific, #12–545-FP) and ProLong Gold antifade mountant (Thermofisher Scientific, #P36930)
Steps and annotations
Plate cells in 4-well chamber slides, 10,000 cells/well.
Treat twice with conditioned medium, 2.5 ml/well for 24 hours.
Repeat procedure twice.
Fix cells with 4% paraformaldehyde at room temperature for 30 minutes.
Rinse with PBS three times.
Permeabilize as indicated in chart, then rinse with PBS three times.
Block for one hour with PBS containing 10% serum from the host in which the secondary antibody was produced.
Dilute antibody in blocking buffer as indicated in chart and incubate for one hour at room temperature or overnight at 4 °C.
Rinse with PBS three times.
Dilute desired secondary antibody (Table 1) 1:200 in blocking buffer and incubate for 45 minutes at room temperature.
Rinse with PBS three times.
Counterstain with 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes.
Coverslip with ProLong Gold mountant and photograph on confocal microscope at 20x magnification.
Table 1.
IHC Antibodies used for cardiogenic characterization
| Primary Antibody | Dilution | Permeabilization |
|---|---|---|
| Anti-MYH6 (MHC, Abcam, #ab207926) | 1:100 or 10 ug/ml | 0.1% Triton X-100 in PBS |
| Anti-Cardiac Troponin T (Abcam, #ab8295) | 1:400 or 4 ug/ml | 0.1% Triton X-100 in PBS |
| Anti-Connexin 43 / GJA1 (Abcam, #ab11370) | 1:100 or 6 ug/ml | 0.1% Triton X-100 in PBS (optional) |
| Secondary Antibody | Dilution | - |
| Goat anti-mouse IgG-FITC (Santa Cruz Biotech, #sc-2010) | 1:200 | - |
| Goat anti-mouse IgG-TR (Santa Cruz Biotech, #sc-3916) | 1:200 | - |
| Goat anti-rabbit IgG-TR (Santa Cruz Biotech, #sc-2780) | 1:200 | - |
Real Time Quantitative Polymerase Chain Reaction
Reagents
RNEasy Plus Mini kit (Qiagen, #74134), 2-mercaptoethanol (BioRad, #161–0710)
Moloney Murine Leukemia Virus Reverse Transcriptase kit (M-MLV, Life Technologies, #28025–013), 100 mM dNTP Set (Invitrogen, #10297–018), ribonuclease inhibitor (RNaseOut, Life Technologies, #10777–019), Oligo dT (Life Technologies, #18418012), nuclease free water
Light cycler probes master mix 2x (Life Technologies, #4304437), pre-designed PrimeTime® qPCR assays (Integrated DNA Technologies—see Table 2 below).
Table 2.
Cardiogenic primers for PCR
| Gene | Forward sequence | Reverse sequence | Probe sequence |
|---|---|---|---|
| Actb | GGATGTCAACGTCACACTTCA | CAGGTCATCACTATCGGCAA | CCACAGGATTCCATACCCAGGAAGG |
| Mef2c | CCTCCCATTCCTTGTCCTG | CAAGAACACAATGCCATCAGTG | AATAACTCCCAGTCGGCCCAGTC |
| Myl2 | TGTCCCTTAGGTCATTCTTGTC | CTCCAACGTGTTCTCCATGT | AAGCCGTCTCTGTTCTGGTCCATG |
| Gja1 | GGTGGAGTAGGCTTGGAC | CCTTTGACTTCAGCCTCCAA | AGTGAAAGAGAGGTGCCCAGACATG |
| TropT | CTGCCTTTCCTTCTCTCGTTC | CTCTGATCGAGGCTCACTTC | AGCTCCTCTTCCTCCTTCTTCCTGT |
| Nkx 2.5 | CTGTCTCGGCTTTGTCCAG | CTACATTTTATCCGCGAGCCTA | CGCACAGCTCTTTCTTATCCGCC |
Materials/Equipment
2720 Thermal Cycler (Applied Biosystems) for retrotranscription
Thermal Cycler, AB #7900HT Fast Real-Time PCR System for gene expression analysis
Nanodrop (NanoDrop One, ThermoFisher Scientific) for RNA quantification
General: microcentrifuge, micropipettes, tips, microcentrifuge tubes, MicroAmp Optical 96/384-Well Reaction Plate (Applied Biosystems, #N8010560/4309849), MicroAmp Optical Adhesive Film (Applied Biosystems, #4311971)
Steps and annotations
Extract the RNA using the RNeasy Mini plus extraction kit, according to the manufacturer’s instructions.
-
Perform cDNA synthesis using M-MLV Reverse Transcriptase kit and random primers (Oligo d(T)):
Reaction mix (total volume 20 μl): 10 μl of RNA (100 ng/μl from RNA extraction) + 1 μl Oligo d(T) + 1 μl dNTP (stock: 10 mM) + 4 μl RT buffer + 2 μl DTT (0.1 M) + 1 μl RNase out + 1 μl M-MLV RT.
Reaction conditions in 2720 Thermal Cycler: 23 °C for 10 min, 37 °C for 60 min, 95 °C for 5 min and 4 °C for 10 min.
-
Real Time RT-PCR in AB 7900HT Fast Real-Time PCR System:
Reaction mix (total volume 12 μl/well): 6 μl LightCycler 480 Probes Master mix + 1.2 μl of 500 nM of each primer and 250 nM of probe + 1 μl cDNA + 3.8 μl H2O.
Reaction conditions in Real-Time PCR System: initial denaturation at 95 °C for 10 min, followed by 40 cycles of these 2-steps: 95 °C for 15 sec and 60 °C for 1 min.
Normalize gene expression levels using Actb as housekeeping gene (see Table 2).
Analyze data and present results in terms of relative expression using the comparative cycle threshold (Ct) method (2−ΔΔCt).
Table 2.
Antibodies used for cardiogenic characterization
| Primary Antibody | Dilution | Permeabilization |
|---|---|---|
| Anti-MYH6 (MHC, Abcam, #ab207926) | 1:100 or 10 ug/ml | 0.1% Triton X-100 in PBS |
| Anti-Connexin 43 / GJA1 (Abcam, #ab11370) | 1:100 or 6 ug/ml | 0.1% Triton X-100 in PBS (optional) |
| Anti-Cardiac Troponin T (Abcam, #ab8295) | 1:400 or 4 ug/ml | 0.1% Triton X-100 in PBS |
| Secondary Antibody | Dilution | - |
| Goat anti-mouse IgG-FITC (Santa Cruz Biotech, #sc-2010) | 1:200 | - |
| Goat anti-mouse IgG-TR (Santa Cruz Biotech, #sc-3916) | 1:200 | - |
| Goat anti-rabbit IgG-TR (Santa Cruz Biotech, #sc-2780) | 1:200 | - |
After isolation, NCMs were characterized using polymerase chain reaction (PCR) and histology. Isolated NCMs showed significant expression of cardiomyogenic markers Myosin and Troponin (Figure 4).
Figure 4.
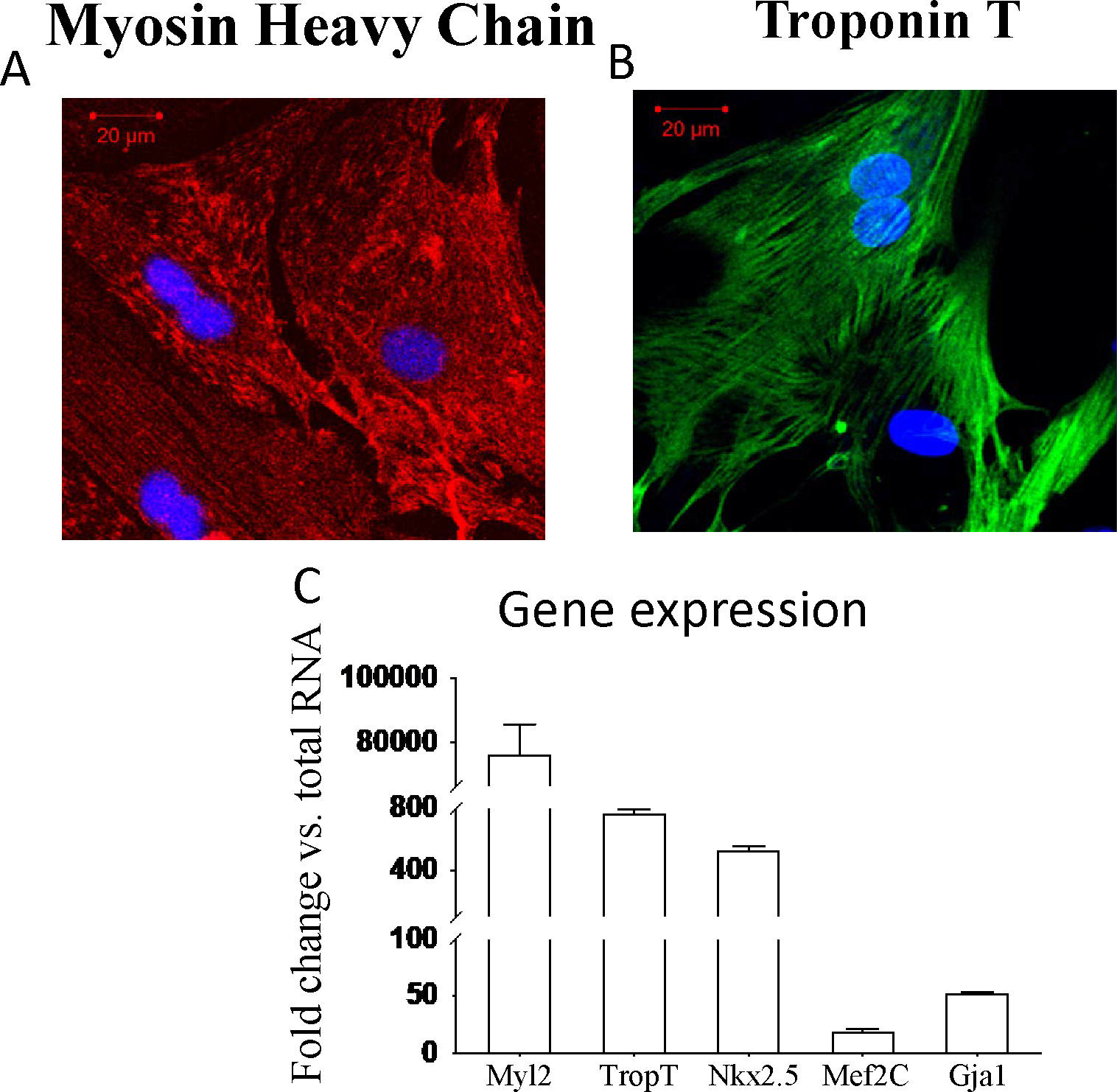
Characterization of NCM. A and B show that isolated NCM are strongly positive for MHC (Texas Red) and Trop2 (FITC). Panel C depicts the RT-qPCR with increased expression of cardiac proteins of Myl2, TropT, Nkx2.5, Mef2C, and Gja-1, compared to total RNA.
Basic protocol 3: Cardiogenic plasticity of MSCs when under ischemic-like conditions
The goal of this protocol (Figure 5) is to establish the cardiogenic plasticity of MSCs under conditions similar to the ones they will encounter under ischemia. For that, MSCs were exposed to the supernatant obtained from low-serum neonatal cardiomyocytes. Subsequently, conditioned MSCs were studied for cardiomyogenic characteristics.
Figure 5.
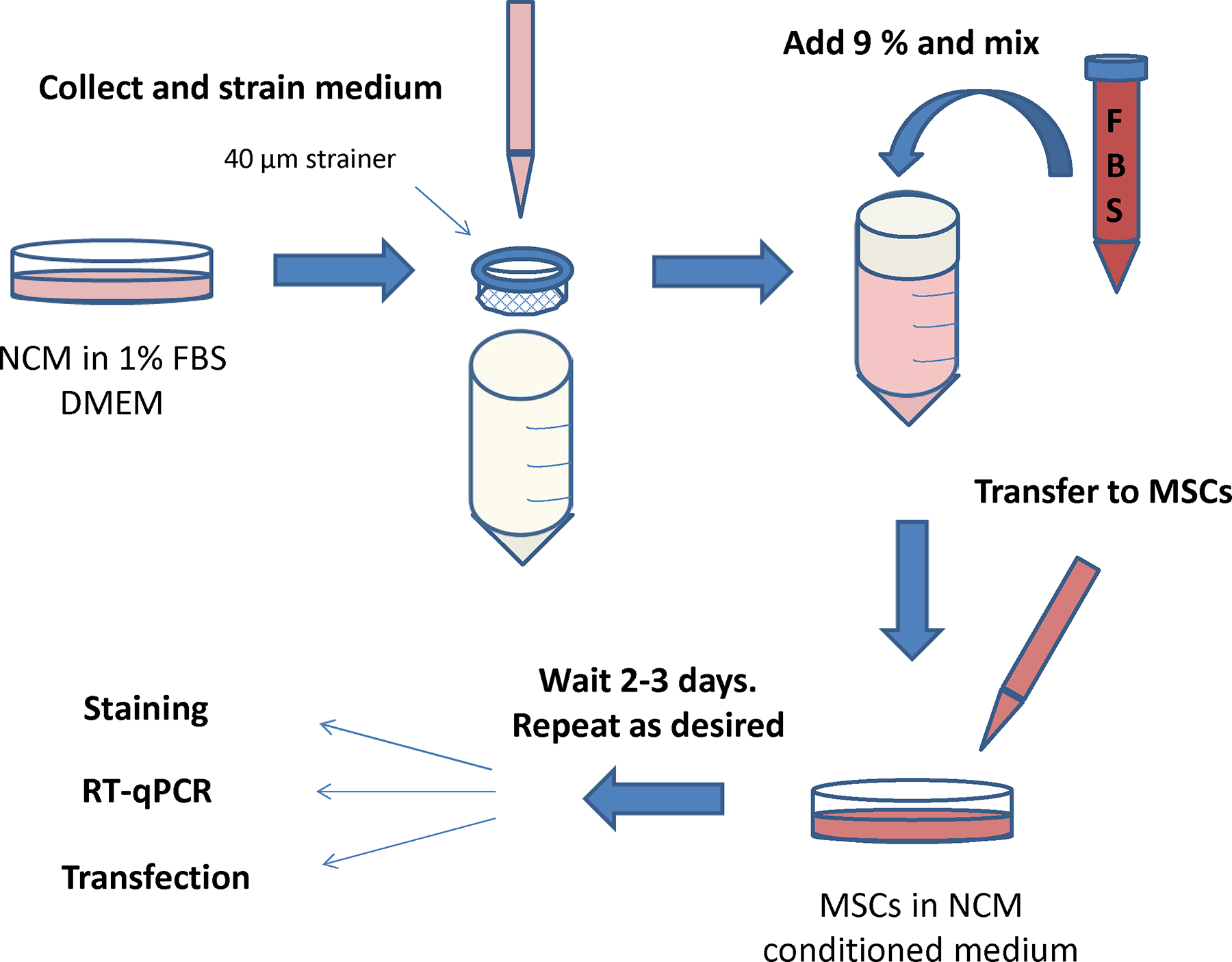
Step-by-step process for the induction of MSCs towards a cardio phenotype.
Cell Culture Reagents
High glucose Dulbecco’s modified Eagle’s medium (DMEM, ThermoFisher Scientific, #11965092)
10% fetal bovine serum (FBS, Mediatech, #35–010-CV)
1% penicillin/streptomycin (ThermoFisher Scientific, #15140122)
Cell culture materials
10 cm dishes (Fisher Scientific, #NC9385690)
6-well plates (Sigma Aldrich, #CLS3516)
40 μm Falcon strainer (Fisher Scientific, #08–771-1)
Hypoxic chamber
Steps and annotations
Plate 400,000 rat MSCs (per flask) into T75 flasks.
Prepare low serum DMEM: DMEM + 1% FBS and 1% antibiotics
Use two 10 cm plates of NCM for each flask of MSCs to be tested.
Aspirate medium from NCM, replace with 7–8 ml low serum DMEM, and return to incubator for 24 hours.
Collect and pool conditioned medium from NCM plates. Rinse plates with PBS and replace with normal culture medium. Return plates to incubator.
If desired, centrifuge pooled medium at high speed to pellet any remaining cell debris.
Filter through 40 μm cell strainer, then adjust the serum back up to 10% by adding fresh FBS.
Aspirate medium from flasks of MSCs to be treated with conditioned medium and rinse with PBS. Add 15 ml conditioned medium per flask. Return flask to incubator for 2–3 days.
Repeat procedure with conditioned medium every 2–3 days as desired. MSCs should begin expressing cardiomyocyte proteins after the second or third treatment.
Support basic protocol 3: Characterization of the cardiomyogenic potential of MSCs
Immunofluorescence of cardiac proteins in NCMs.
Reagents:
4% paraformaldehyde (Biotium, 22023)
Phosphate Buffered Saline (PBS, Invitrogen, 10010023)
Goat serum (Sigma Aldrich, #G9023)
4′,6-diamidino-2-phenylindole (DAPI, ThermoFisher Scientific, #D3571)
ProLong Gold antifade mountant (Thermofisher Scientific, #P36930)
Materials:
4-well chamber slides (Fisher Scientific, #12–565-2)
Microscope
Pipettes
Steps and annotations:
Plate cells in 4-well chamber slides, 10,000 cells/well.
Treat twice with conditioned medium, 2.5 ml/well for 24 hours.
Repeat procedure twice.
Fix cells with 4% paraformaldehyde at room temperature for 30 minutes.
Rinse with PBS three times.
Permeabilize as indicated in chart, then rinse with PBS three times.
Block for one hour with PBS containing 10% serum from the host in which the secondary antibody was produced.
Dilute antibody in blocking buffer as indicated in chart and incubate for one hour at room temperature or overnight at 4 °C.
Rinse with PBS three times.
Dilute desired secondary antibody (Table 2) 1:200 in blocking buffer and incubate for 45 minutes at room temperature.
Rinse with PBS three times.
Counterstain with 4′,6-diamidino-2-phenylindole (DAPI) for 5 minutes.
Coverslip with Prolong Gold and photograph on confocal microscope at 20x magnification.
Real Time Quantitative Polymerase Chain Reaction
Reagents
RNEasy Plus Mini kit (Qiagen, #74134), 2-mercaptoethanol (BioRad, #161–0710)
Moloney Murine Leukemia Virus Reverse Transcriptase kit (M-MLV, Life Technologies, #28025–013), 100 mM dNTP Set (Invitrogen, #10297–018), ribonuclease inhibitor (RNaseOut, Life Technologies, #10777–019), Oligo dT (Life Technologies, #18418012), nuclease free water
Light cycler probes master mix 2x (Life Technologies, #4304437), pre-designed PrimeTime® qPCR assays (Integrated DNA Technologies—see Table 2 below).
Table 3.
Cardiogenic probe sequences for PCR
| Gene | Forward sequence | Reverse sequence | Probe sequence |
|---|---|---|---|
| Actb | GGATGTCAACGTCACACTTCA | CAGGTCATCACTATCGGCAA | CCACAGGATTCCATACCCAGGAAGG |
| Mef2c | CCTCCCATTCCTTGTCCTG | CAAGAACACAATGCCATCAGTG | AATAACTCCCAGTCGGCCCAGTC |
| Myl2 | TGTCCCTTAGGTCATTCTTGTC | CTCCAACGTGTTCTCCATGT | AAGCCGTTCTGTTCTGGTCCATG |
| Gja1 | GGTGGAGTAGGCTTGGAC | CCTTTGACTTCAGCCTCCAA | AGTGAAAGAGAGGTGCCCAGACATG |
| TropT | CTGCCTTTCCTTCTCTCGTTC | CTCTGATCGAGGCTCACTTC | AGCTCCTCTTCCTCCTTCTTCCTGT |
Materials/Equipment
2720 Thermal Cycler (Applied Biosystems) for retrotranscription
Thermal Cycler, AB 7900HT Fast Real-Time PCR System for gene expression analysis
Nanodrop (NanoDrop One, ThermoFisher Scientific) for RNA quantification
General: microcentrifuge, micropipettes, tips, microcentrifuge tubes, MicroAmp Optical 96/384-Well Reaction Plate (Applied Biosystems, #N8010560/4309849), MicroAmp Optical Adhesive Film (Applied Biosystems, #4311971)
Steps and annotations
Extract the RNA using the RNeasy Mini plus extraction kit, according to the manufacturer’s instructions.
-
Perform cDNA synthesis using M-MLV Reverse Transcriptase kit and random primers (Oligo d(T)):
Reaction mix (total volume 20 μl): 10 μl of RNA (100 ng/μl from RNA extraction) + 1 μl Oligo d(T) + 1 μl dNTP (stock: 10 mM) + 4 μl RT buffer + 2 μl DTT (0.1 M) + 1 μl RNase out + 1 μl M-MLV RT.
Reaction conditions in 2720 Thermal Cycler: 23 °C for 10 min, 37 °C for 60 min, 95 °C for 5 min and 4 °C for 10 min.
-
Real Time RT-PCR in AB 7900HT Fast Real-Time PCR System:
Reaction mix (total volume 12 μl/well): 6 μl LightCycler 480 Probes Master mix + 1.2 μl of 500 nM of each primer and 250 nM of probe + 1 μl cDNA + 3.8 μl H2O.
Reaction conditions in Real-Time PCR System: initial denaturation at 95 °C for 10 min, followed by 40 cycles of these 2-steps: 95 °C for 15 sec and 60 °C for 1 min.
Normalize gene expression levels using Actb as housekeeping gene (see Table 2).
Analyze data and present results in terms of relative expression using the comparative cycle threshold (Ct) method (2−ΔΔCt).
Reporter Gene Activity
Reagents:
Plasmid firefly luciferase gene (TropT-fluc, made in-house)
Effectene transfection reagent (Qiagen, #301425)
Luciferase Assay System (Promega, #E1501)
Phosphate Buffered saline (PBS, Invitrogen, #10010023)
Passive Lysis buffer (Promega, #E1941)
Equipment:
12-well plates (Fisher Scientific, #0720082), pipettes, tilt rocker, refrigerator, ice, refrigerated centrifuge, microcentrifuge tubes, luminometer 20/20n (Turner Biosystems, Sunnyvale, CA)
Steps and annotations:
MSCs stably express a reporter gene firefly luciferase driven by the TropT promoter
Plate rat MSCs (treated and untreated) at a density of 50,000 cells per well in 12-well plates.
After 24 hours, transfect with plasmid with Effectene transfection reagent, using 800 ng/well of reporter gene plasmid, 75 μl/well buffer EC, 6.4 μl/well enhancer and 8 μl/well hyperfect reagent. Change medium after 6 hours.
After 24 hours, thaw Luciferase Assay Reagent on ice. Meanwhile, aspirate medium from transfected cells, rinse with 500 μl PBS per well, and add 200 μl Passive Lysis buffer per well to lyse cells. Place plate on rocker in refrigerator at 4 °C for 30 minutes to allow complete lysis.
Collect lysate in microcentrifuge tubes, centrifuge at 4 °C in microcentrifuge at 13,000 rpm for 15 minutes, and place on ice.
Combine 100 μl Luciferase Assay Reagent and 20 μl undiluted cell lysate, vortex for 1s and measure on luminometer under steady state conditions, using Luciferase Assay Reagent alone as a blank.
Note: Correct all readings for blank. If desired, measure protein concentration in all samples and normalize luminometer readings to protein concentrations.
Characterization of MSCs exposed to NCM conditioned media: results
When MSCs were exposed to conditioned medium from serum deprived NCM they showed significant upregulation of myocyte enhancer factor 2C (Mef2c, early), Myosin Light Chain 2 (Myl2, early), Gap junction alpha-1 (Gja1, early) and TropT (late) markers of cardiogenic plasticity (Figure 6). Immunofluorescence staining verified increased expression of the cardiac proteins MHC, TropT and connexin 1 (CXA1) compared to control (non-induced) MSCs (Figure 7) with 85.1±3.5% of cells having some degree of TropT and 80.7±2.5% of cells having some degree of MHC expression. Lastly, MSCs transfected with the TropT-two step amplification strategy (TSTA)-Fluc reporter gene plasmid demonstrated a time-related increased in TropT-TSTA-Fluc expression compared to control (non-treated) MSCs (Figure 8).
Figure 6.
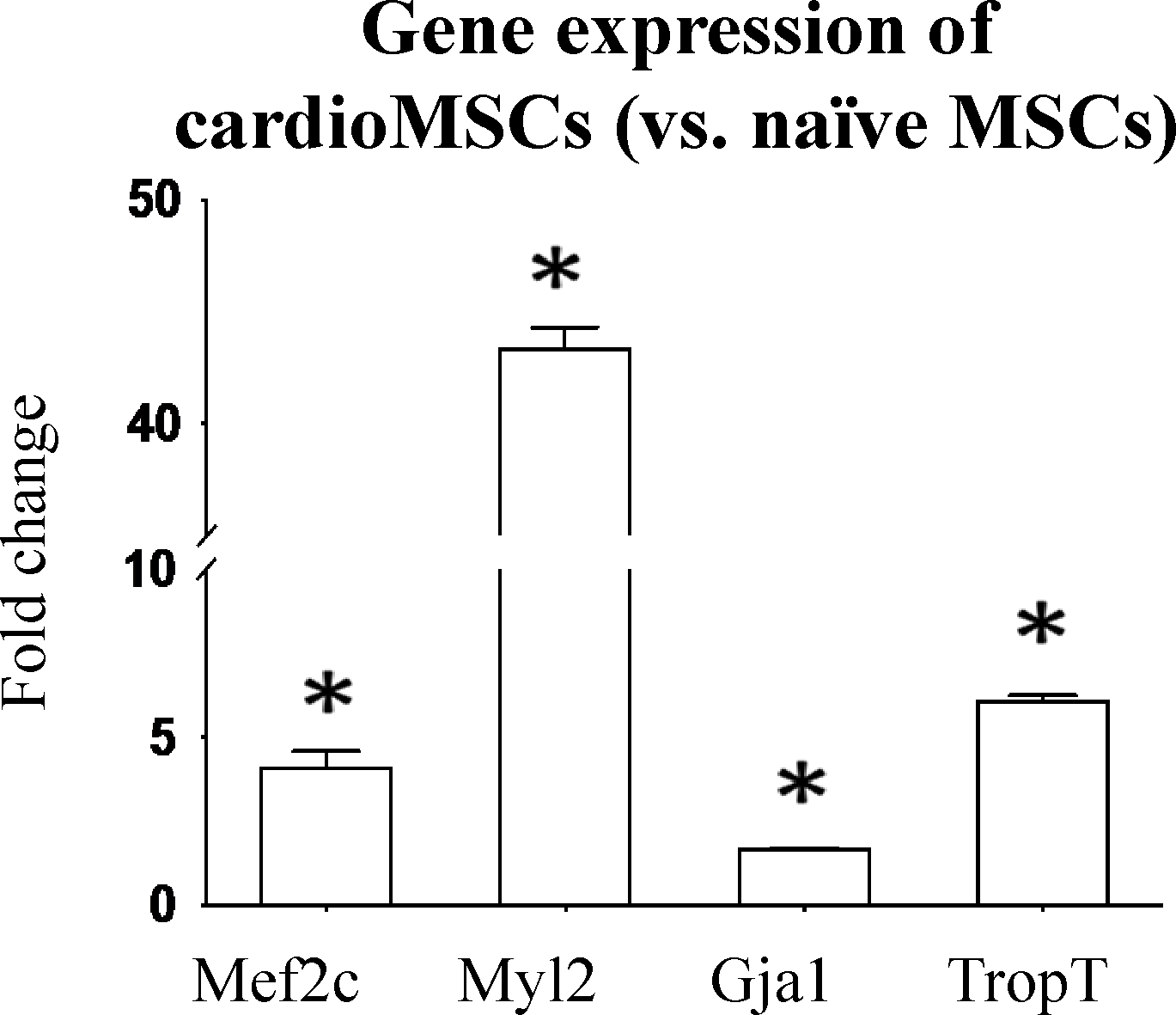
Real Time Quantitative PCR showing the increased mRNA expression of Mef2c, Myl2, Gja1 (early) and TropT (late) markers in cardio MSCs compared to naïve MSCs. Data is expressed as fold change compared to naïve MSCs. P<0.05 vs. control for all genes.
Figure 7.
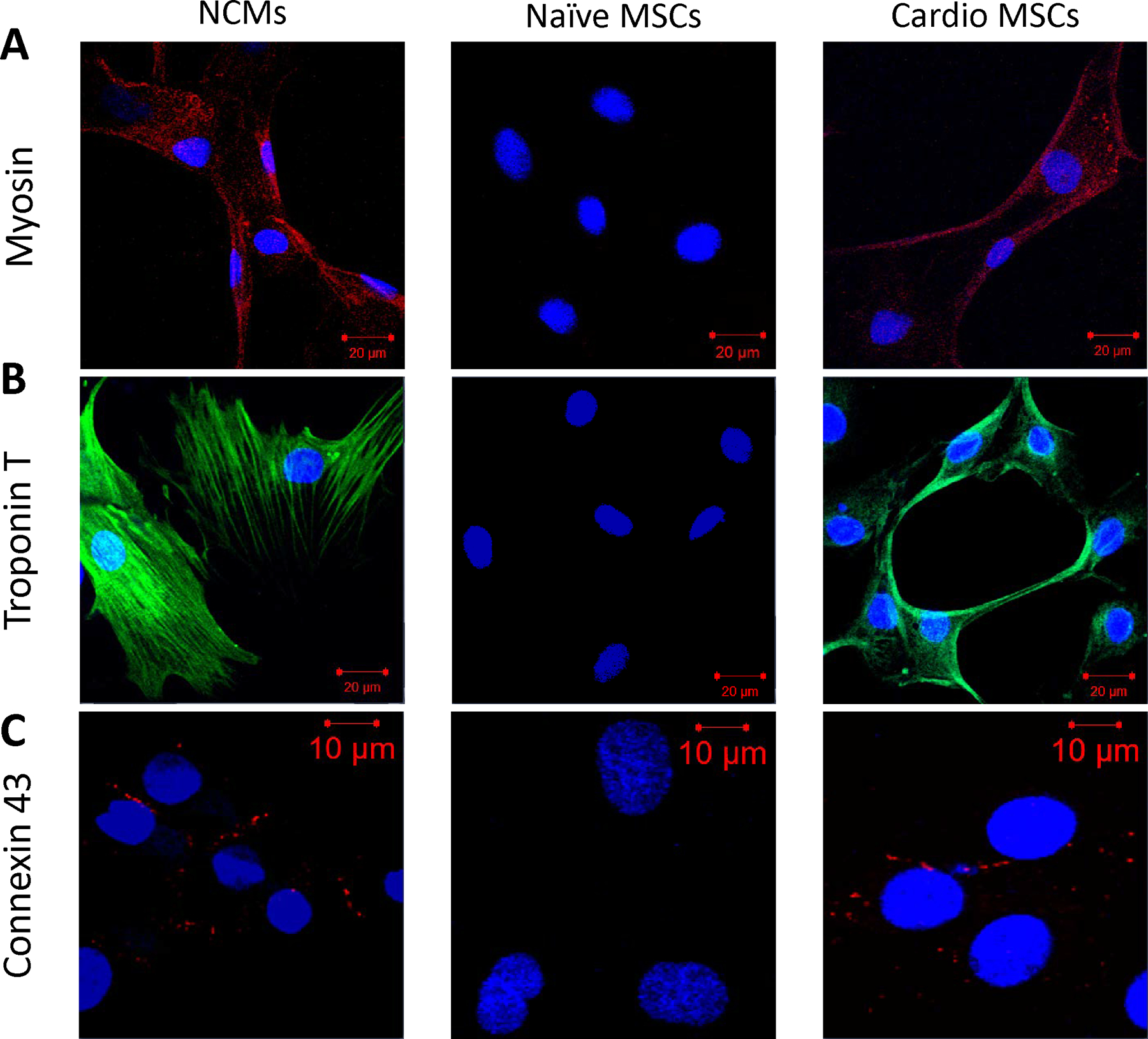
Immunofluorescence staining for cardiac proteins. NCM (left), naïve MSCs (middle) and cardio MSCs (right column) were stained for (A) MHC (Texas Red), (B) TropT (FITC) and (C) Cx43 (Texas Red) and counterstained with DAPI. Cardio MSCs showed increased expression of cardiac proteins, indicating a shift to a cell with cardiomyogenic characteristics. Slides were photographed on a laser scanning confocal microscope at 20x magnification.
Figure 8.
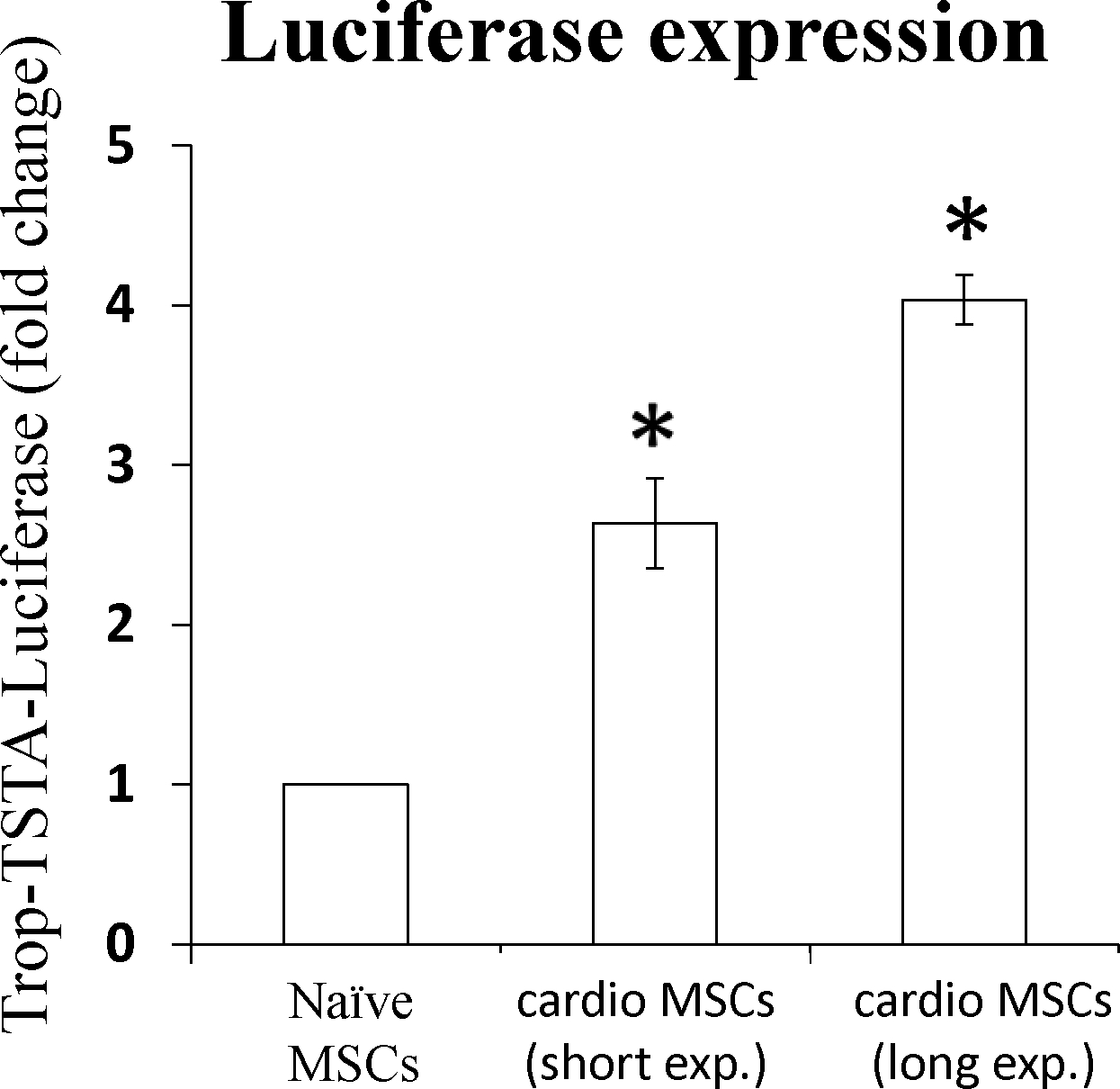
MSCs, carrying the cTnT-TSTA-Fluc reporter gene, were exposed to NCM-conditioned medium in a time dependent fashion, and their phenotype assessed using bioluminescence. Data is expressed as fold change compared to (non-treated) control MSCs. Both groups of cardio MSCs (increasing exposure to ischemic-like conditioned media) were significantly different (p<0.05) compared to naïve MSCs.
Commentary and Troubleshooting
In this study, we described a process to modulate the phenotype of MSCs, showing that they can acquire cardiomyogenic characteristics, when under conditions similar to what MSCs may encounter when transplanted to the ischemic myocardium.
The approach of this study, using the supernatant of hypoxic neonatal cardiomyocytes, is an attempt to partially reproduce the hostile environment that MSCs encounter after transplantation. We have previously shown that the post-ischemic myocardium is a noxious environment characterized by hypoxia and ischemia in the border zone, leading to nutrient deprivation (Psaltis et al., 2013). Due to the complexity of the stimuli involved, and the dynamic nature of the changes, we believe that the post-ischemic microenvironment cannot be accurately reproduced in-vitro. Hence the critical importance of our in vivo studies in providing evidence of the feasibility of monitoring stem cell biology directly in the living subject. Importantly, the degree of the changes in the gene expression of cardiac proteins that illustrated the plasticity of MCS in our study was similar compared to other approaches previously validated in the literature (Behfar et al., 2010; Sun, Sun, & Liu, 2018).
To monitor the change in phenotype, we elected to use the Fluc reporter gene system, which has been a cornerstone of BLI, to evaluate cell fate in small animal studies. We recently published a detailed process on how to use a reporter gene approach to monitor cell biology (Peterson et al., 2020). The identified endogenous promoter, derived from Troponin T gene, was efficient in driving the expression of the reporter gene Fluc to monitor changes to a specific cell phenotype.
In conclusion, this study demonstrates that MSCs can acquire cardiomyogenic characteristics. Furthermore, this phenotypic change happened after they were exposed to conditioned media from cardiomyocytes in an ischemic environment, a condition likely similar to what MSCs would encounter in vivo. In addition, we provide feasibility of using a conditional reporter gene approach to monitor the phenotype of transplanted progenitor cells. When combined, the protocols described here provide a platform to study the phenotypic changes of stem cells ex-vivo, under conditions similar to what they may encounter in the in vivo setting.
Time considerations
Isolation of MSCs: takes approx. ½ day, depending on the number of animals used. It takes about 14–21 days until cells can be frozen at passage 3. As a general rule, MSCs should be passaged 1:4 up to 1:6 for expansion after the 1st passage following isolation. Because they grow very quickly, by passage 2 (which is labeled passage 3 on freezing) there should be few off-target cells.
Isolation of NCM: is a two day process-about 2–3 hrs. in the afternoon on the first day, then about half a day the next day (usually in the early afternoon because of the incubation time for the trypsin). The cells are usable within 1 week, as they can’t be amplified or frozen. Generally the isolation was done on Thurs-Friday, change the medium on Monday and can use them within a day or two after that. In general, we give them a couple days after the 1st medium change because they can double after isolation. They are usable for several weeks, if they are cultured carefully so as not to get them overgrown by off-target cells or to have them begin to lift off form the plate.
To induce MSC plasticity using NCMs (Protocol 3), there are a number of steps to be considered. It takes approximately 2 days for the NCMs in low serum. Subsequently, the conditioned media of NMCs should stay at least 2–3 days on MSCs, and that timing should be multiplied by the number of challenges desired based on study design. In our study we performed either 2–3 challenges, resulting in 4–6 days of MSCs being exposed to NCM-conditioned media.
Acknowledgements
This work was supported in part by R56 HL113371 (MR-P), R01 HL119795 (MR-P) and RO1CA209888 (PR). The luminometer used in this study was obtained through a grant from Turner Biosystems, Sunnyvale, CA.
Bibliography
- Behfar A, & Terzic A (2006). Derivation of a cardiopoietic population from human mesenchymal stem cells yields cardiac progeny. Nat Clin Pract Cardiovasc Med, 3 Suppl 1, S78–82. doi: 10.1038/ncpcardio0429 [DOI] [PubMed] [Google Scholar]
- Behfar A, & Terzic A (2007). Optimizing adult mesenchymal stem cells for heart repair. J Mol Cell Cardiol, 42(2), 283–284. doi: 10.1016/j.yjmcc.2006.11.003 [DOI] [PubMed] [Google Scholar]
- Behfar A, Yamada S, Crespo-Diaz R, Nesbitt JJ, Rowe LA, Perez-Terzic C, . . . Terzic A (2010). Guided cardiopoiesis enhances therapeutic benefit of bone marrow human mesenchymal stem cells in chronic myocardial infarction. J Am Coll Cardiol, 56(9), 721–734. doi: 10.1016/j.jacc.2010.03.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominici M, Le Blanc K, Mueller I, Slaper-Cortenbach I, Marini F, Krause D, . . . Horwitz E (2006). Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy, 8(4), 315–317. doi: 10.1080/14653240600855905 [DOI] [PubMed] [Google Scholar]
- Le Blanc K, & Pittenger M (2005). Mesenchymal stem cells: progress toward promise. Cytotherapy, 7(1), 36–45. doi: 10.1080/14653240510018118 [DOI] [PubMed] [Google Scholar]
- Leeper NJ, Hunter AL, & Cooke JP (2010). Stem cell therapy for vascular regeneration: adult, embryonic, and induced pluripotent stem cells. Circulation, 122(5), 517–526. doi: 10.1161/CIRCULATIONAHA.109.881441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peterson KM, Franchi F, Olthoff M, Chen IY, Paulmurugan R, & Rodriguez-Porcel M (2020). Pathway-specific reporter genes to study stem cell biology. Stem cells. doi: 10.1002/stem.3167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF (2008). Mesenchymal stem cells from adult bone marrow. Methods Mol Biol, 449, 27–44. doi: 10.1007/978-1-60327-169-1_2 [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Discher DE, Peault BM, Phinney DG, Hare JM, & Caplan AI (2019). Mesenchymal stem cell perspective: cell biology to clinical progress. NPJ Regen Med, 4, 22. doi: 10.1038/s41536-019-0083-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittenger MF, Mackay AM, Beck SC, Jaiswal RK, Douglas R, Mosca JD, . . . Marshak DR (1999). Multilineage potential of adult human mesenchymal stem cells. Science, 284(5411), 143–147. doi: 10.1126/science.284.5411.143 [DOI] [PubMed] [Google Scholar]
- Pittenger MF, & Martin BJ (2004). Mesenchymal stem cells and their potential as cardiac therapeutics. Circ Res, 95(1), 9–20. doi: 10.1161/01.RES.0000135902.99383.6f [DOI] [PubMed] [Google Scholar]
- Pittenger MF, Mosca JD, & McIntosh KR (2000). Human mesenchymal stem cells: progenitor cells for cartilage, bone, fat and stroma. Curr Top Microbiol Immunol, 251, 3–11. doi: 10.1007/978-3-642-57276-0_1 [DOI] [PubMed] [Google Scholar]
- Psaltis PJ, Peterson KM, Xu R, Franchi F, Witt T, Chen IY, . . . Rodriguez-Porcel M (2013). Noninvasive monitoring of oxidative stress in transplanted mesenchymal stromal cells. JACC. Cardiovascular imaging, 6(7), 795–802. doi: 10.1016/j.jcmg.2012.11.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segers VF, & Lee RT (2008). Stem-cell therapy for cardiac disease. Nature, 451(7181), 937–942. doi: 10.1038/nature06800 [DOI] [PubMed] [Google Scholar]
- Shake JG, Gruber PJ, Baumgartner WA, Senechal G, Meyers J, Redmond JM, . . . Martin BJ (2002). Mesenchymal stem cell implantation in a swine myocardial infarct model: engraftment and functional effects. Ann Thorac Surg, 73(6), 1919–1925; discussion 1926. doi: 10.1016/s0003-4975(02)03517-8 [DOI] [PubMed] [Google Scholar]
- Shi S, Wu X, Wang X, Hao W, Miao H, Zhen L, & Nie S (2016). Differentiation of Bone Marrow Mesenchymal Stem Cells to Cardiomyocyte-Like Cells Is Regulated by the Combined Low Dose Treatment of Transforming Growth Factor-beta1 and 5-Azacytidine. Stem Cells Int, 2016, 3816256. doi: 10.1155/2016/3816256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HH, Sun PF, & Liu WY (2018). MiR-98–5p regulates myocardial differentiation of mesenchymal stem cells by targeting TBX5. Eur Rev Med Pharmacol Sci, 22(22), 7841–7848. doi: 10.26355/eurrev_201811_16409 [DOI] [PubMed] [Google Scholar]


