Abstract
To investigate the functional domains of the coat protein (CP; 189 amino acids) of Brome mosaic virus, a plant RNA virus, 19 alanine-scanning mutants were constructed and tested for their infectivity in barley and Nicotiana benthamiana. Despite its apparent normal replicative competence and CP production, the C-terminal mutant F184A produced no virions. Furthermore, virion-forming C-terminal mutants P178A and D182A failed to move from cell to cell in both plant species, and mutants D181A and V187A showed host-specific movement. These results indicate that the C-terminal region of CP plays some important roles in virus movement and encapsidation. The specificity of certain mutations for viral movement in two different plant species is evidence for the involvement of host-specific factors.
One viral component, coat protein (CP), encoded by several positive-strand RNA plant viruses, is multifunctional. As well as protecting viral RNAs from degradation, the CP plays a major role in symptom modulation (12, 36, 43), replication (6, 20, 21), long-distance movement, and cell-to-cell movement (7, 29) in some viruses. To function in these roles, the CP is thought to interact physically with putative host-derived factors, as well as with viral components such as viral RNAs and CP itself (7, 29). However, not all RNA viruses share the requirement for CP in order to spread systemically, because deletion of the CP gene from either Tomato bushy stunt virus (44) or Barley stripe mosaic virus (35) has no significant effect on viral spread in plants. Interestingly, the CP is one of the most abundant proteins in virus-infected plants. The ability of CP to accumulate to high levels in host cells suggests that the CP may suppress or evade some plant defense responses, perhaps through interaction with host factors.
Brome mosaic virus (BMV) is a well-studied, tripartite, single-stranded, positive-sense RNA plant virus (3). RNA1 and RNA2 encode viral replicase proteins 1a and 2a, respectively, whereas RNA3 codes for the 3a movement protein (MP) and CP. The CP gene is expressed through a subgenomic mRNA, RNA4 (3). BMV replicase proteins have been characterized extensively by genetic and biochemical approaches (2, 46), and the 3a MP has also been well examined by several mutation analyses linked to phenotypic investigations (17–19, 31, 32, 36, 41) and by biochemical and cytological techniques (16, 23). On the other hand, the study of CP has progressed mostly in terms of capsid architecture (28), although information about its roles in viral infection has recently been accumulating. Flasinski et al. (15) reported that mutations, mainly introduced into the N-terminal and hydrophobic domains of BMV CP (BCP), affect multiplication as well as movement of the virus in barley and in a variety of Chenopodium hybridum. More detailed examinations of the N-terminal region were performed (8, 36, 37, 39) and indicate that the N-terminal region, especially the arginine-rich domain, is important for virus infection in barley and C. quinoa. The seven N-terminal residues of BCP have significant effects on lesion formation in Chenopodium species. Whereas these studies have predominantly revealed some roles of the N-terminal and hydrophobic regions of BCP, the C-terminal and internal hydrophilic regions have been less well studied. These regions may be particularly important because they are probably displayed on the surface of the BCP molecule (45) and are therefore likely to interact with putative host plant factors and/or viral components. A deletion mutant study of BCP suggested that the loss of 12 C-terminal residues affected encapsidation, as well as virus infectivity, in barley and Chenopodium species (36). Moreover, all of the BCP-interacting barley proteins that we have recently identified (Y. Okinaka, K. Mise, and I. Furusawa, Abstr. 9th Internatl. Cong. Mol. Plant-Microbe Interact., abstr. 136, 1999) require at least the C-terminal portion of BCP for binding. Therefore, in this study, we investigated the roles of putative C-terminal and internal surface regions of BCP in virus infectivity by using BCP mutants, each of which bears a single or double consecutive amino acid substitution with alanine (9).
The plasmids used in this study are summarized in Table 1. The cDNA clones of wild-type BMV strain M1 (pB1TP3, pB2TP5, and pB3TP8) (22) were kindly provided by P. Ahlquist (University of Wisconsin—Madison). All BMV RNA3 mutants were made by site-directed mutagenesis (5) of pB3TP8 with sets of mutagenized forward primers (Table 1) and reverse primers that completely matched the corresponding BMV RNA3 sequences. The PCR products amplified with these primer sets were digested with restriction enzymes. The resulting DNA fragments contained the following mutagenized sequences: 251-bp StyI-StyI fragments in pB3SK052053AA, pB3EQ110112AA, and pB3SS128129AA; 244-bp SacI-AvaIII fragments in pB3SS078079AA, pB3NK082083AA, and pB3YL155156AA; a 59-bp AvaIII-StuI fragment in pB3HV-175176AA; and 138-bp StuI-HindIII fragments in pB3P178A, pB3T179A, pB3F180A, pB3D181A, pB3D182A, pB3F183A, pB3F184A, pB3T185A, pB3P186A, pB3V187A, pB3Y188A, and pB3R189A. These DNA fragments were recovered after agarose gel electrophoresis. The corresponding restriction fragments in pB3TP8 were replaced with these fragments to produce the pB3TP8 derivatives, and the introduced mutations were then verified by automated DNA sequencing. Enzymatic DNA digestions, ligations, and transformations were performed by standard methods (40).
TABLE 1.
Summary of plasmids used in this study
| Plasmida | Encoded polypeptide | Amino acid substitutedb/relevant RNA sequencec |
|---|---|---|
| pB1TP3 | BMV wild-type 1a | |
| pB2TP5 | BMV wild-type 2a | |
| pB3TP8 | BMV wild-type 3a and wild type CP | |
| pB3TP8-derived mutants | BMV wild-type 3a and CP variant | |
| pBSK052053AA | SK052053AA | 52(S), 53 (K)/UCA AAG→GCA GCG |
| pB3SS078079AA | SS078079AA | 78(S), 79 (S)/UCU UCU→GCU GCU |
| pB3NK082083AA | NK082083AA | 82(N), 83 (K)/AAU AAG→GCU GCG |
| pB3EQ110112AA | EQ110112AA | 110(E), 112 (Q)/GAG AAA CAG→GCG AAA GCG |
| pB3SS128129AA | SS128129AA | 128(S), 129 (S)/UCC UCG→GCC GCG |
| pB3YL155156AA | YL155156AA | 155(Y), 156 (L)/UAU CUG→GCU GCG |
| pB3HV175176AA | HV175176AA | 175(H), 176 (V)/CAC GUA→GCC GCA |
| pB3P178A | P178A | 178 (P)/CCU→GCU |
| pB3T179A | T179A | 179 (T)/ACG→GCG |
| pB3F180A | F180A | 180 (F)/UUC→GCC |
| pB3D181A | D181A | 181 (D)/GAU→GCU |
| pB3D182A | D182A | 182 (D)/GAC→GCC |
| pB3F183A | F183A | 183 (F)/UUC→GCC |
| pB3F184A | F184A | 184 (F)/UUC→GCC |
| pB3T185A | T185A | 185 (T)/ACC→GCC |
| pB3P186A | P186A | 186 (P)/CCG→GCG |
| pB3V187A | V187A | 187 (V)/GUU→GCU |
| pB3Y188A | Y188A | 188 (Y)/UAU→GCU |
| pB3R189A | R189A | 189 (R)/AGG→GCG |
pB1TP3, pB2TP5, and pB3TP8 were described in detail previously (22).
The amino acid residues substituted for with alanine are shown by the standard one-letter codes in parentheses accompanied by the numbers that indicate their position in BCP.
The short RNA sequences represent codon changes that induce amino acid substitutions. As an exception, the mutant EQ110112AA accompanies the unchanged AAA codon.
Barley [Hordeum vulgare L. cv. Hinodehadaka] and Nicotiana benthamiana plants were planted in a growth room at 25°C with 16 h of illumination per day and daily watering with half-strength Hoagland's solution (13). Synthesis of capped transcripts from EcoRI-linearized full-length cDNA plasmids pB1TP3 and pB2TP5, as well as pB3TP8 and its derivatives (26); inoculation of whole plants and protoplasts with transcripts (26, 31); extraction of total nucleic acids from plant leaves (4) and protoplasts (26); and preparation of samples for tissue printing analysis (31) were performed as described previously. Each experiment was repeated at least two or three times with independently synthesized in vitro transcripts.
For the identification of virion RNAs, viral RNAs were extracted from virion fractions isolated from infected barley protoplasts by polyethylene glycol precipitation (26) 24 h after inoculation. Northern blot analysis to detect positive-sense BMV RNAs was performed as described previously (24), except that the DIG (digoxigenin) Labeling and Detection kit (Roche, Indianapolis, Ind.) was used. Northern blotting patterns were densitometrically quantified with the NIH Image program v. 1.61. Western blot analysis of BCP was performed as described previously (10) after suspension of infected protoplasts directly in Laemmli's sample buffer (27). Enzyme-linked immunosorbent assays (ELISAs) were carried out as described previously (34), and virus yields were estimated by using a serial dilution of purified BMV virion as the standard. In both assays, BCP was detected with a rabbit anti-BMV antiserum (ATCC PVAS-178; American Type Culture Collection).
Design of alanine-scanning mutagenesis for the putative external BCP regions.
Virus infection of plants requires the association of host and viral factors, which is likely to occur between the external structures of the molecules. To verify whether the external regions of BCP are required for the viral infection of plants, we searched predicted surface sites on the BCP molecule by the method of Emini et al. (14) (Fig. 1) and performed their alanine-scanning mutagenesis (9), excluding the putative N-terminal surface regions previously investigated in detail (8, 15, 30, 36, 37, 39). In this way, 19 site-directed mutations were successfully introduced into the internal and C-terminal regions of BCP (Table 1). The intensive mutagenesis at the C terminus was performed because this region was expected to be important for BMV infection (36) and to be displayed outside of the BCP β-barrel conformation (45).
FIG. 1.
Surface probability plot of BCP. The surface probability of BCP (189 amino acids) was analyzed by the method of Emini et al. (4). Arrows accompanied by numbers indicate the positions at which amino acid substitutions were introduced.
Effects of internal mutations of BCP on virus infectivity in barley.
RNA3 derivatives with mutations causing double amino acid substitutions in the internal sites of BCP were tested for their infectivity in barley. RNA transcripts of the mutants were coinoculated with wild-type BMV RNA1 and RNA2 into both barley plants and protoplasts. Transfection of barley protoplasts with all of these mutants resulted in efficient accumulation of progeny viral RNAs. In addition, both the full-length CP (CP1) and the truncated CP (CP2), which is translated from the second AUG codon of RNA4 (39), were accumulated, although the electrophoretic mobilities of the two CPs differed slightly (Fig. 2). However, encapsidation competence was reduced by 70% in B3EQ110112AA and abolished in B3SK052053AA, B3SS128129AA, B3YL155156AA, and B3HV175176AA. Northern blot analysis of the barley plants inoculated with these mutant transcripts revealed that B3SS078079AA, B3NK082083AA, and B3EQ110112AA produced progeny viral RNAs in both inoculated and systemic leaves, although no viral RNAs were observed with B3SK052053AA, B3SS128129AA, B3YL155156AA, or B3HV175176AA (Table 2). Virus accumulation was also estimated by using ELISA to measure the CP content in the systemically infected leaves of inoculated plants. In plants inoculated with B3SS078079AA, virus accumulation was found to be two-thirds that of plants inoculated with wild-type RNA3, and B3NK082083AA and B3EQ110112AA accumulated progeny viruses to a level one-third that of the wild type (Table 2). No CP accumulation was detected with B3SK052053AA, B3SS128129AA, B3YL155156AA, or B3HV175176AA. One interesting observation was that the infectivity of mutants B3SS078079AA and B3NK082083AA was significantly reduced in barley plants, although these mutants showed good encapsidation competence in protoplasts, indicating their defects in cell-to-cell and/or long-distance movement. No virus infectivity in plants inoculated with B3SK052053AA, B3SS128129AA, B3YL155156AA, or B3HV175176AA was observed, which is probably attributable to their lack of encapsidation competence. Sequence analysis of the progeny viral RNAs purified from the systemically infected leaves of barley was performed, as previously described (33), and demonstrated that all of the mutagenized positions shown in Table 1 were conserved after virus multiplication (data not shown).
FIG. 2.
(A) Replicative competence and encapsidation assays of BCP internal mutants. Barley protoplasts were transfected with the indicated wild-type (Wt) or variant RNA3 transcripts, together with wild-type RNA1 and RNA2. Progeny RNAs were extracted from the infected protoplasts (P) and purified virions (V) and subjected to Northern blot analysis. The positions of the four BMV RNAs are indicated on the left. (B) Western blot analysis of CP accumulation and integrity in the internal mutants. The positions of CP1 and CP2 (39) are indicated on the left.
TABLE 2.
Analyses of BCP mutants bearing amino aid substitution in the putative internal surface regions
| BCP mutanta | Encapsidation (arbitrary unit)b | Movementc
|
Virus yield (mg/g FW)d | |
|---|---|---|---|---|
| I | S | |||
| Mock | 0.0 | − | − | 0.0 ± 0.0 |
| B3SK052053AA | 0.0 | − | − | 0.0 ± 0.0 |
| B3SS078079AA | 1.1 | + | + | 2.5 ± 0.1 |
| B3NK082083AA | 0.8 | + | + | 1.3 ± 0.2 |
| B3EQ110112AA | 0.3 | + | + | 1.4 ± 0.0 |
| B3SS128129AA | 0.0 | − | − | 0.0 ± 0.0 |
| B3YL155156AA | 0.0 | − | − | 0.0 ± 0.0 |
| B3HV175176AA | 0.0 | − | − | 0.0 ± 0.0 |
| Wild-type B3 | 1.0 | + | + | 4.0 ± 0.3 |
Wild-type BMV RNA3 or its derivatives were used as the inoculum together with wild-type BMV RNAs 1 and 2.
The Northern blotting data in Fig. 2 and from two other experiments were densitometrically analyzed, averaged, and indicated as an aribtrary unit.
The transcripts were inoculated on 6-day-old barley seedlings, and virus infectivity was measured by the existence of viral RNAs 2 weeks after inoculation. BMV RNAs in inoculated (I) and systemic (S) leaves were detected by tissue printing assay (31).
CP accumulation in secondary leaves (systemic leaves) was measured by ELISA and expressed as virion concentration. Data are means ± standard deviation for three replicates. FW, fresh weight.
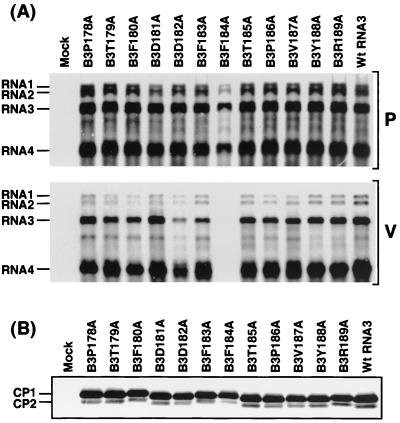
Encapsidation competence of BCP C-terminal mutants.
It was previously demonstrated that the deletion mutant of BCP that lacks the C-terminal residues 178 to 189 is deleteriously affected in its encapsidation competence and infectivity in plants of Chenopodium species (36). To further these investigations, BCP mutants were constructed by introducing single amino acid substitutions with alanine at the C terminus (Table 1). Inoculation of barley protoplasts with these mutant transcripts, together with wild-type RNA1 and RNA2, revealed that progeny RNAs and CP accumulated to levels similar to those in protoplasts inoculated with wild-type RNA3 for all of the mutants tested (Fig. 3), although the electrophoretic mobilities of the mutated CPs differed slightly from one another. Interestingly, in the encapsidation assay, progeny RNAs were not detected in the virion fraction when protoplasts were inoculated with B3F184A, whereas they were detected with all of the other mutants tested (Fig. 3A and Table 3). These results indicate that Phe184 and/or the corresponding RNA sequence is important for the encapsidation process or the stability of the virus particles. A fine X-ray crystallographic study of the CP from a closely related bromovirus, Cowpea chlorotic mottle virus (CCMV) (45), indicates that Phe184 (Phe186 in CCMV CP) plays an essential role in dimer formation, an initial step in virion assembly. In dimer formation, Phe184 may interact hydrophobically with many different amino acid residues in the parallel β structures of BCP. Furthermore, the conservation of this phenylalanine residue among four bromoviruses, BMV (1), CCMV (11), Broad bean mottle virus (38), and Spring beauty latent virus (K. Fujisaki, K. Mise, and I. Furusawa, unpublished data), suggests that this phenylalanine residue may be important in their encapsidation processes. The X-ray crystallographic data also suggest that Asp182 in BCP may contribute to hydrophilic CP dimer contacts. However, unexpectedly, the B3D182A mutant still displayed good encapsidation competence (Fig. 3, and Table 3).
FIG. 3.
(A) Replicative competence and encapsidation assays of BCP C-terminal mutants. (B) CP accumulation and integrity of BCP C-terminal mutants. All procedures were performed as described in the legend to Fig. 2.
TABLE 3.
Analyses of BCP C-terminal mutanta
| BCP mutant | Result for:
|
|||||||
|---|---|---|---|---|---|---|---|---|
| Barley
|
N. benthamiana
|
|||||||
| Encapsidation (arbitrary unit)b | Movement
|
Virus yield | Movement
|
Virus yield
|
||||
| I | S | I | S | I | S | |||
| Mock | 0.0 | − | − | 0.0 ± 0.0 | − | − | 0.0 | 0.0 ± 0.0 |
| B3P178A | 1.1 | − | − | 0.0 ± 0.0 | − | − | 0.0 | 0.0 ± 0.0 |
| B3T179A | 1.1 | + | + | 4.1 ± 1.8 | + | + | 7.6 | 2.9 ± 0.5 |
| B3F180A | 0.8 | + | + | 3.6 ± 0.5 | + | + | 4.4 | 2.6 ± 0.5 |
| B3D181A | 1.3 | + | + | 5.4 ± 2.2 | + | − | 0.4 | 0.0 ± 0.1 |
| B3D182A | 0.9 | − | − | 0.0 ± 0.0 | − | − | 0.0 | 0.0 ± 0.0 |
| B3F183A | 1.0 | + | + | 2.5 ± 0.8 | + | + | 1.5 | 2.6 ± 0.7 |
| B3F184A | 0.0 | − | − | 0.0 ± 0.0 | − | − | 0.0 | 0.0 ± 0.0 |
| B3T185A | 1.0 | + | + | 5.2 ± 1.6 | + | + | 4.7 | 4.0 ± 1.0 |
| B3P186A | 1.0 | + | + | 2.7 ± 0.1 | + | + | 2.6 | 2.9 ± 1.4 |
| B3V187A | 1.0 | + | + | 2.4 ± 0.5 | + | + | 0.3 | 0.6 ± 0.3 |
| B3Y188A | 0.9 | + | + | 3.7 ± 0.5 | + | + | 1.3 | 2.8 ± 0.3 |
| B3R189R | 0.8 | + | + | 3.4 ± 0.7 | + | + | 3.1 | 3.4 ± 1.1 |
| Wild-type B3 | 1.0 | + | + | 5.1 ± 1.4 | + | + | 6.2 | 4.5 ± 1.0 |
All of the procedures and directions follow those described in the footnotes to Table 2 with the following exceptions: encapsidation data were obtained from Fig. 3 and two other experiments, and virus yield data obtained with the inoculated leaves of N. benthamiana are means of two independent experiments.
Effects of C-terminal mutations of BCP on virus infectivity in different hosts.
The infectivity of BCP mutants, each bearing a single amino acid substitution at the C terminus, was examined in both barley and N. benthamiana plants. Data are summarized in Table 3. Among the 12 C-terminal mutants, B3T179A, B3F180A, B3F183A, B3T185A, B3P186A, B3Y188A, and B3R189A showed vigorous systemic infections in both plant species and gave 49 to 102% virus yields relative to wild-type RNA3 in barley and 57 to 89% of wild-type yields in N. benthamiana. The effects of these seven mutations on virus accumulation were relatively minor compared with the effects of the other mutations described below. Interestingly, the phenotypic symptoms observed with B3T185A infection were mild (data not shown), even though the level of accumulated virus was similar to that with wild-type RNA3 infection. In contrast, no infectivity was detected with mutant B3P178A, B3D182A, or B3F184A in either plant species, even in the inoculated leaves (data not shown in part). However, the accumulation and encapsidation of viral RNAs occurred normally in barley protoplasts after inoculation with B3P178A and B3D182A, as mentioned earlier. For further confirmation of this causation, the virion fractions prepared from protoplasts infected with either of three movement-incompetent mutants, B3P178A, B3D182A, or B3F184A, were observed under the electron microscope as previously described (36). Icosahedral virions, apparently identical to those of wild-type BMV, were observed with inoculations of B3P178A and B3D182A, whereas no virions were detected with B3F184A (data not shown). A plausible explanation for the loss of infectivity in B3F184A is that the lack of encapsidation competence mentioned above interferes with virus multiplication in the host plants because BMV has been reported to move from cell to cell in virion form (25, 39, 42). The mutants B3P178A and B3D182A perhaps lack the ability of cell-to-cell movement, due to the defective interactions of the virions with BMV proteins or with host factors that are functionally similar in the two hosts. The former could include putative virion-MP interactions during transport through tubular structures (25). Our data are also consistent with the previous observation that cell-to-cell movement of BMV requires the CP, together with the 3a protein (42). By comparison with the X-ray crystallographic data of CCMV CP (45), the C-terminal residues of BCP (positions 178 to 189) would not be displayed either on the surface of the virion particle or on the dimer molecule. Therefore, residues Pro178 and Asp182 may not mediate direct interaction of the virions with viral or plant factors, but may affect such interactions indirectly through a change in virion shape. However, our electron microscopy observations of B3P178A and B3D182A virions suggest that the change must be slight. Alternatively, nonassembled BCP monomer itself may play some role in virus movement, in which Pro178 and Asp182 interact directly with BMV RNA, BMV proteins, and/or host factors. Of these interactions, the former two may involve the formation of a putative CP-MP-BMV RNA complex during the intra- and/or intercellular movement of BMV (7, 29). Mechanisms for these interactions are suggested by the fact that BMV 3a MP binds to BMV RNAs (16, 23). Finally, it is also possible that the changes at Pro178 and Asp182 may elicit defense responses in the initially infected host cells (47).
Other noteworthy observations are that, with respect to the mutants B3D181A and B3V187A, systemic infections occurred in barley with virus yields of 106 and 47%, respectively, relative to that with wild-type RNA3, but were abolished and significantly reduced (13% wild-type virus yield), respectively, in N. benthamiana. With the mutants B3D181A and B3V187A, the virus yields were approximately 6 and 5% of that with wild-type RNA3, respectively, in the inoculated leaves of N. benthamiana. These results indicate that the B3D181A mutation does not affect virus infectivity in barley, but may inhibit both cell-to-cell and long-distance movement in N. benthamiana. Similarly, the infectivity of the B3V187A mutant was still high in barley, but its cell-to-cell movement ability in N. benthamiana may be reduced. Sequence analysis of the progeny viral RNAs purified from the systemically infected leaves (except in the case of N. benthamiana infected with B3D181, in which the inoculated leaves were analyzed) demonstrated that all of the mutagenized nucleotides listed in Table 1 were conserved after virus multiplication (data not shown). A threshold of virus concentration required for successful long-distance movement might explain this phenomenon in part, but this possibility can be eliminated here, because B3V187A, which gave virus yields in inoculated leaves similar to or lower than that of B3D181A, still exhibited systemic infection. A possible explanation for these host-specific infections with B3D181A and B3V187A is that viral infection in N. benthamiana is supported by some host-specific factors that interact with wild-type BCP, but not with the mutant CP of either B3D181A or B3V187A. Alternatively, as discussed above, some defense response may be induced by these two mutants only in N. benthamiana.
Acknowledgments
We thank Paul Ahlquist for the cDNA clones of the BMV M1 strain, Jennifer Becker for critical review of the manuscript, and Mariko Takada for technical support.
This work was supported in part by Grant-in-Aid 09NP1501 for Creative Basic Research from the Ministry of Education, Science, Sports, and Culture, Japan, and Grant-in-Aid JSPS-RFTF96L00603 from the “Research for the Future” program of the Japan Society for the Promotion of Science.
REFERENCES
- 1.Ahlquist P, Luckow V, Kaesberg P. Complete nucleotide sequence of brome mosaic virus RNA3. J Mol Biol. 1981;153:23–38. doi: 10.1016/0022-2836(81)90524-6. [DOI] [PubMed] [Google Scholar]
- 2.Ahlquist P. Bromovirus RNA replication and transcription. Curr Opin Genet Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 3.Ahlquist P. Bromoviruses. In: Granoff A, Webster R G, editors. Encyclopedia of virology. Vol. 1. San Diego, Calif: Academic Press; 1994. pp. 198–204. [Google Scholar]
- 4.Allison R, Thompson C, Ahlquist P. Regeneration of a functional RNA virus genome by recombination between deletion mutants and requirement for cowpea chlorotic mottle virus 3a and coat genes for systemic infection. Proc Natl Acad Sci USA. 1990;87:1820–1824. doi: 10.1073/pnas.87.5.1820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Barik S. Site-directed mutagenesis by double polymerase chain reaction. In: White B A, editor. PCR protocols. Totowa, N.J: Humana Press; 1993. pp. 277–286. [Google Scholar]
- 6.Bol J F. Alfalfa mosaic virus and ilarviruses: involvement of coat protein in multiple steps of the replication cycle. J Gen Virol. 1999;80:1089–1102. doi: 10.1099/0022-1317-80-5-1089. [DOI] [PubMed] [Google Scholar]
- 7.Carrington J C, Kasschau K D, Mahajan A K, Schaad M C. Cell-to-cell and long-distance transport of viruses in plants. Plant Cell. 1996;8:1669–1681. doi: 10.1105/tpc.8.10.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Choi Y G, Grantham G L, Rao A L N. Molecular studies on bromovirus capsid protein. VI. Contributions of the N-terminal arginine-rich motif of BMV capsid protein to virion stability and RNA packaging. Virology. 2000;270:377–385. doi: 10.1006/viro.2000.0312. [DOI] [PubMed] [Google Scholar]
- 9.Cunningham B C, Wells J A. High-resolution epitope mapping of hGH-receptor interactions by alanine-scanning mutagenesis. Science. 1989;244:1081–1085. doi: 10.1126/science.2471267. [DOI] [PubMed] [Google Scholar]
- 10.Damayanti T A, Nagano H, Mise K, Furusawa I, Okuno T. Brome mosaic virus defective RNAs generated during infection of barley plants. J Gen Virol. 1999;80:2511–2518. doi: 10.1099/0022-1317-80-9-2511. [DOI] [PubMed] [Google Scholar]
- 11.Dasgupta R, Kaesberg P. Complete nucleotide sequences of coat protein messenger RNAs of brome mosaic virus and cowpea chlorotic mottle virus. Nucleic Acids Res. 1982;10:703–713. doi: 10.1093/nar/10.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dawson W O. Tobamovirus-plant interactions. Virology. 1992;186:359–367. doi: 10.1016/0042-6822(92)90001-6. [DOI] [PubMed] [Google Scholar]
- 13.Dhingra O, Sinclair J. Basic plant pathology methods. Boca Raton, Fla: CRC Press; 1985. p. 317. [Google Scholar]
- 14.Emini E A, Hughes J V, Perlow D C, Boger J. Induction of hepatitis A virus-neutralizing antibody by a virus-specific synthetic peptide. J Virol. 1985;55:836–839. doi: 10.1128/jvi.55.3.836-839.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Flasinski S, Dzianott A, Pratt S, Bujarski J J. Mutational analysis of the coat protein gene of brome mosaic virus: effects on replication and movement in barley and in Chenopodium hybridum. Mol Plant-Microbe Interact. 1995;8:23–31. doi: 10.1094/mpmi-8-0023. [DOI] [PubMed] [Google Scholar]
- 16.Fujita M, Mise K, Kajiura Y, Dohi K, Furusawa I. Nucleic acid-binding properties and subcellular localization of the 3a protein of brome mosaic bromovirus. J Gen Virol. 1998;79:1273–1280. doi: 10.1099/0022-1317-79-5-1273. [DOI] [PubMed] [Google Scholar]
- 17.Fujita Y, Mise K, Okuno Y, Ahlquist P, Furusawa I. A single codon change in a conserved motif of a bromovirus movement protein gene confers compatibility with a new host. Virology. 1996;223:283–291. doi: 10.1006/viro.1996.0480. [DOI] [PubMed] [Google Scholar]
- 18.Fujita Y, Mise K, Furusawa I. Genotypic and phenotypic analysis of bromovirus adaptive mutants derived from a single plant. Microbiol Immunol. 1999;43:181–185. doi: 10.1111/j.1348-0421.1999.tb02391.x. [DOI] [PubMed] [Google Scholar]
- 19.Fujita Y, Fujita M, Mise K, Kobori T, Osaki T, Furusawa I. Bromovirus movement protein conditions for the host specificity of virus movement through the vascular system and affects pathogenicity in cowpea. Mol Plant-Microbe Interact. 2000;13:1195–1203. doi: 10.1094/MPMI.2000.13.11.1195. [DOI] [PubMed] [Google Scholar]
- 20.Horikoshi M, Nakayama M, Yamaoka N, Furusawa I, Shishiyama J. Brome mosaic virus coat protein inhibits viral RNA synthesis in vitro. Virology. 1987;158:15–19. doi: 10.1016/0042-6822(87)90232-7. [DOI] [PubMed] [Google Scholar]
- 21.Horikoshi M, Mise K, Furusawa I, Shishiyama J. Further studies on inhibitory effect of brome mosaic virus coat protein on the viral RNA synthesis in vitro. Ann Phytopath Soc Jpn. 1988;54:533–535. [Google Scholar]
- 22.Janda M, French R, Ahlquist P. High efficiency polymerase synthesis of infectious RNA from cloned brome mosaic virus cDNA and effects of 5′ extensions on transcript infectivity. Virology. 1987;158:259–262. doi: 10.1016/0042-6822(87)90265-0. [DOI] [PubMed] [Google Scholar]
- 23.Jansen K A, Wolfs C J A M, Lohuis H, Goldbach R W, Verduin B J M. Characterization of the brome mosaic virus movement protein expressed in E. coli. Virology. 1998;242:387–394. doi: 10.1006/viro.1997.9000. [DOI] [PubMed] [Google Scholar]
- 24.Kaido M, Mori M, Mise K, Okuno T, Furusawa I. Inhibition of brome mosaic virus (BMV) amplification in protoplasts from transgenic tobacco plants expressing replicable BMV RNAs. J Gen Virol. 1995;76:2827–2833. doi: 10.1099/0022-1317-76-11-2827. [DOI] [PubMed] [Google Scholar]
- 25.Kasteel D T J, van der Wel N N, Jansen K A J, Goldbach R W, van Lent J W M. Tubule-forming capacity of the movement proteins of alfalfa mosaic virus and brome mosaic virus. J Gen Virol. 1997;78:2089–2093. doi: 10.1099/0022-1317-78-8-2089. [DOI] [PubMed] [Google Scholar]
- 26.Kroner P, Ahlquist P. RNA based viruses. In: Gurr S J, McPherson M J, Bowles D J, editors. Molecular plant pathology: a practical approach. I. Oxford, United Kingdom: IRL Press; 1992. pp. 23–34. [Google Scholar]
- 27.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 28.Lane L C. Bromoviruses. In: Kurstak E, editor. Handbook of plant virus infections and comparative diagnosis. Amsterdam, The Netherlands: Elsevier Biomedical Press; 1981. pp. 333–376. [Google Scholar]
- 29.Lazarowitz S G, Beachy R N. Viral movement proteins as probes for intracellular and intercellular trafficking in plants. Plant Cell. 1999;11:535–548. doi: 10.1105/tpc.11.4.535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mise K, Tsuge S, Nagano K, Okuno T, Furusawa I. Nucleotide sequence responsible for the synthesis of a truncated coat protein of brome mosaic virus strain ATCC66. J Gen Virol. 1992;73:2543–2551. doi: 10.1099/0022-1317-73-10-2543. [DOI] [PubMed] [Google Scholar]
- 31.Mise K, Allison R F, Janda M, Ahlquist P. Bromovirus movement protein genes play a crucial role in host specificity. J Virol. 1993;67:2815–2823. doi: 10.1128/jvi.67.5.2815-2823.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mise K, Ahlquist P. Host-specificity restriction by bromovirus cell-to-cell movement protein occurs after initial cell-to-cell spread of infection in nonhost plants. Virology. 1995;206:276–286. doi: 10.1016/s0042-6822(95)80043-3. [DOI] [PubMed] [Google Scholar]
- 33.Nagano H, Okuno T, Mise K, Furusawa I. Deletion of the C-terminal 33 amino acids of cucumber mosaic virus movement protein enables a chimeric brome mosaic virus to move from cell to cell. J Virol. 1997;71:2270–2276. doi: 10.1128/jvi.71.3.2270-2276.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okuno T, Nakayama M, Yoshida S, Furusawa I, Komiya T. Comparative susceptibility of transgenic tobacco plants and protoplasts expressing the coat protein gene of cucumber mosaic virus to infection with virions and RNA. Phytopathology. 1993;83:542–547. [Google Scholar]
- 35.Petty I T D, Jackson A O. Mutational analysis of barley stripe mosaic virus RNA β. Virology. 1990;179:712–718. doi: 10.1016/0042-6822(90)90138-h. [DOI] [PubMed] [Google Scholar]
- 36.Rao A L N, Grantham G L. Biological significance of the seven amino-terminal basic residues of brome mosaic virus coat protein. Virology. 1995;211:42–52. doi: 10.1006/viro.1995.1377. [DOI] [PubMed] [Google Scholar]
- 37.Rao A L N, Grantham G L. Molecular studies on bromovirus capsid protein. II. Functional analysis of the amino-terminal arginine-rich motif and its role in encapsidation, movement and pathology. Virology. 1996;226:294–305. doi: 10.1006/viro.1996.0657. [DOI] [PubMed] [Google Scholar]
- 38.Romero J, Dzianott A M, Bujarski J J. The nucleotide sequence and genome organization of the RNA2 and RNA3 segments of broad bean mottle virus. Virology. 1992;187:671–681. doi: 10.1016/0042-6822(92)90470-a. [DOI] [PubMed] [Google Scholar]
- 39.Sacher R, Ahlquist P. Effects of deletions in the N-terminal basic arm of brome mosaic virus coat protein on RNA packaging and systemic infection. J Virol. 1989;63:4545–4552. doi: 10.1128/jvi.63.11.4545-4552.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 41.Sasaki N, Fujita Y, Mise K, Furusawa I. Site-specific single amino acid changes to Lys or Arg in the central region of the movement protein of a hybrid bromovirus are required for adaptation to a nonhost. Virology. 2001;279:47–57. doi: 10.1006/viro.2000.0518. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz I, Rao A L N. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology. 1996;226:281–293. doi: 10.1006/viro.1996.0656. [DOI] [PubMed] [Google Scholar]
- 43.Shintaku H M, Zhang L, Palukaitis P. A single amino acid substitution in the coat protein of cucumber mosaic virus induces chlorosis in tobacco. Plant Cell. 1992;4:751–757. doi: 10.1105/tpc.4.7.751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sholthof H B, Morris T J, Jackson A O. The capsid protein gene of tomato bushy stunt virus is dispensable for systemic movement and can be replaced for localized expression of foreign genes. Mol Plant-Microbe Interact. 1993;6:309–322. [Google Scholar]
- 45.Speir J A, Munshi S, Wang G, Baker T S, Johnson J E. Structures of the native and swollen forms of cowpea chlorotic mottle virus determined by X-ray crystallography and cryo-electron microscopy. Structure. 1995;3:63–78. doi: 10.1016/s0969-2126(01)00135-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sullivan L M, Ahlquist P. cis-Acting signals in bromovirus RNA replication and gene expression: networking with viral proteins and host factors. Semin Virol. 1997;8:221–230. [Google Scholar]
- 47.Taraporewala Z F, Culver J N. Identification of an elicitor active site within the three-dimensional structure of the tobacco mosaic tobamovirus coat protein. Plant Cell. 1996;8:169–178. doi: 10.1105/tpc.8.2.169. [DOI] [PMC free article] [PubMed] [Google Scholar]