Abstract
Promyelocytic leukemia (PML) oncogenic domains (PODs) accumulate the transcriptional cofactor named CREB binding protein (CBP) and have been suggested to function as centers of transcription. Transcriptional activation by nuclear hormones, such as glucocorticoids, is augmented by the key constituent of PODs, the PML protein, and decreased by the POD-associated Tax protein of human T-cell leukemia virus type 1 (HTLV-1). This led to the hypothesis that intact PODs might play a positive role in the activation of these promoters. We report here that transiently expressed E4orf3 protein of adenovirus type 5, immediate-early protein 1 of human cytomegalovirus, and the PML-retinoic acid receptor fusion protein from leukemia cells each redistribute CBP within the nucleus. However, unlike the Tax protein of HTLV-1, these factors did not inhibit a glucocorticoid-inducible promoter but strongly enhanced its activity. Thus, at least glucocorticoid-induced transcription does not depend on POD integrity.
A number of viral and cellular oncoproteins act to modify dot-like intranuclear structures termed promyelocytic leukemia (PML) oncogenic domains (PODs), nuclear bodies, or nuclear domain 10. The functions of PODs are not fully understood but appear to involve the regulation of virus replication (reviewed in reference 29), transcription (13, 18, 38), apoptosis (25, 34), DNA repair (13, 18, 38), cell proliferation (21, 33), and oncogenesis (33). The PML protein is consistently found in this structure, and the integrity of PODs depends on PML (15). In addition, numerous cellular proteins were found to accumulate in the PODs (29, 38), including a transcriptional cofactor termed cyclic AMP-responsive element binding protein (CREB) binding protein (CBP) (18). It was suggested that PODs might represent centers of RNA synthesis (18). In parallel, promoters that are induced by nuclear hormone receptors were found to be activated by PML overexpression (13). In contrast, the Tax protein from human T-cell leukemia virus type 1 (HTLV-1) partially localizes to PODs (11), redistributes at least one component of PODs (the Int-6 protein) within the nucleus (10), and downregulates the activity of nuclear-hormone-inducible promoters, an effect that can be reversed by PML overexpression (11). Thus, it seemed conceivable that intact PODs might activate such promoters, perhaps by forming a suitable environment for their transcription (13, 18).
A tool with which to test this hypothesis is provided by protein factors that disrupt the integrity of PODs. One of these POD-disrupting factors is encoded by the third open reading frame (orf3) within the E4 region of adenovirus (Ad) type 5. E4orf3 colocalizes with PML in infected cells, and it changes the PODs from a spherical to a rod-like shape (9, 12) termed “tracks” (24). Further, the E4orf3 protein has been shown to facilitate the malignant transformation of rodent cells (23). POD disruption and cotransformation are activities shared with a different viral protein, the IE1 protein of human cytomegalovirus (CMV) (1, 2, 28). Finally, in promyelocytic leukemia, the PML gene is frequently fused to the gene encoding retinoic acid receptor (RAR) alpha to express a fusion protein termed PML-RAR (16). The expression of this fusion protein is known to disrupt the PODs and to redistribute their components in the nucleus (35). When these leukemic cells are treated with retinoic acid, the PODs are reestablished. In parallel, the cells differentiate and ultimately undergo apoptosis. Treatment of patients with retinoic acid successfully takes advantage of this phenomenon to eliminate leukemic cells (see references 20 and 29 for reviews).
If PODs contribute directly to nuclear-hormone-induced promoter activation by providing an environment for transcription, then one would expect that POD-disrupting factors should downregulate these promoters, like the Tax protein of HTLV-1. Surprisingly, we found that Ad E4orf3, CMV IE1, and PML-RAR each strongly activate a glucocorticoid-responsive promoter. Thus, POD integrity is not a requirement for promoter activation, suggesting that, at least in the case of glucocorticoid-induced transcription, PODs and their components act as indirect transcriptional modulators rather than physically constitute centers of mRNA synthesis.
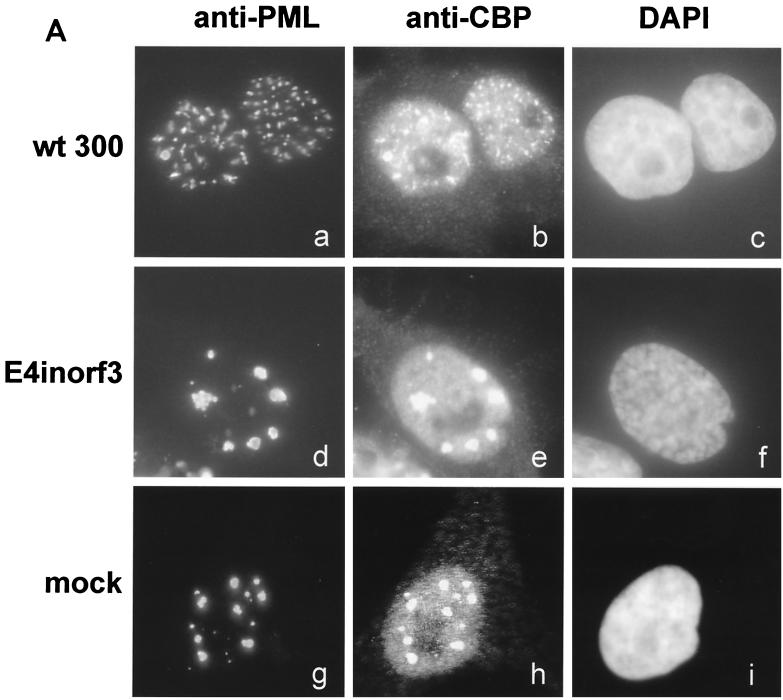
Previous reports have suggested that PODs might be involved in the regulation of transcription, possibly through the transcriptional cofactor CBP (18). Therefore, we tested whether CBP might be relocalized along with PML in the presence of a POD-disrupting factor, such as Ad E4orf3. PML++ HeLa cells (a generous gift from H. Will and T. Sternsdorf) were used in this experiment to facilitate the visualization of PODs. These cells allow the inducible overexpression of PML after the removal of tetracycline (30). Fourteen hours after infection with wild-type Ad type 5 (wt300) or an otherwise identical virus lacking E4orf3 (E4inorf3 [14]; both viruses were kindly provided by P. Hearing), PML++ HeLa cells were fixed and simultaneously stained for the PML protein and CBP (antibodies [from Santa Cruz] PG-M3 [sc-966] and A-22 [sc-369], respectively), as described previously (17). The typical POD morphology (Fig. 1A, g) was largely unaffected in E4inorf3-infected cells (Fig. 1A, d), and CBP localized within the PODs (Fig. 1A, e and h). In contrast, when cells were infected with wild-type Ad, PML was relocalized to numerous track-like structures (Fig. 1A, a), confirming previous results (9, 12). When these cells were stained with antibodies against CBP, this protein was found in the same tracks as PML (Fig. 1A, b) and was no longer associated with spherical structures. These results document that CBP, like PML, is relocalized in Ad-infected cells and that the expression of E4orf3 at least contributes to this relocalization.
FIG. 1.
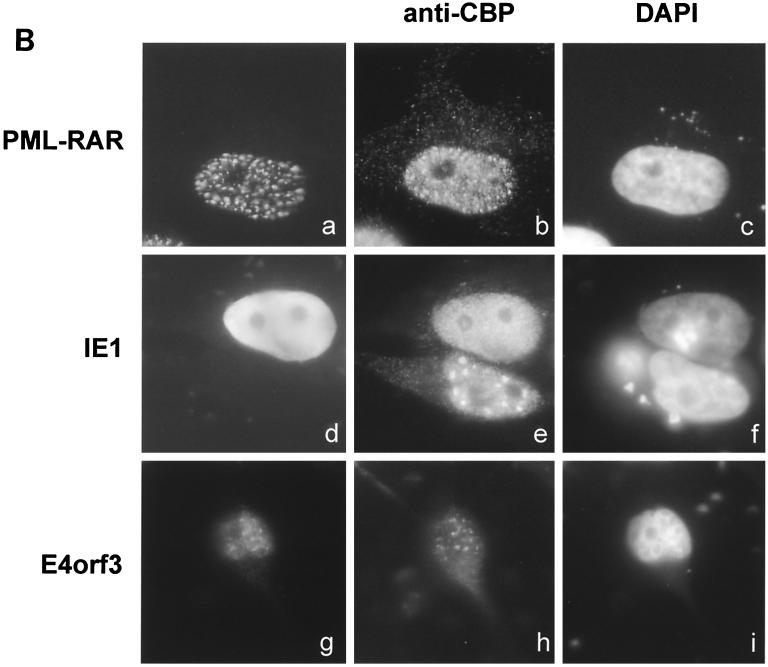
Relocalization of the CREB binding protein by E4orf3, CMV IE1, and PML-RAR. (A) PML++ HeLa cells (30) were induced to express PML by removal of tetracycline and infected with Ad (multiplicity of infection [MOI] of 5) 24 h later or mock infected (g to i). The virus employed was either the wild type (termed wt300) (a to c) or a mutant lacking E4orf3 expression (E4inorf3) (d to f). Fourteen hours after infection, PML (a, d, and g) and CBP (b, e, and h) were stained with antibodies to these proteins, followed by secondary antibodies coupled to fluorescent dyes. The nuclei were visualized using 4′,6′-diamidino-2-phenylindole (DAPI) stain (c, f, and i). (B) PML++ HeLa cells were induced to express PML and then transfected using Superfect (Qiagen) with expression plasmids for a PML-RAR fusion protein (a to c), CMV IE1 (d to f), or Ad E4orf3 (g to i). Twenty-four hours after transfection, the cells were fixed and stained with antibodies against PML (a), the hemagglutinin tag (d), or the FLAG tag (g). Simultaneously, CBP was stained in all cells (b, e, and h) and the nuclei were visualized with DAPI (c, f, and i). Note that in part e, the pattern of CBP distribution is shown in a nontransfected cell next to a transfected cell.
We then tested whether E4orf3, as well as other viral and cellular proteins known to interfere with POD organization, were sufficient to relocalize CBP. To this end, PML++ HeLa cells were transiently transfected with expression vectors for PML-RAR (Fig. 1B, a to c), CMV IE1 (Fig. 1B, d to f), and E4orf3 (Fig. 1B, g to i). Expression of these proteins was detected by immunostaining with corresponding antibodies (Fig. 1B, a, d, and g). In parallel, CBP was stained (Fig. 1B, b, e, and h) and found to be relocalized from the PODs in a pattern similar, if not identical, to the distribution of the POD-disrupting factors. CBP relocalization was observed in all cells (100%) that expressed detectable PML-RAR, CMV IE1, or E4orf3, respectively. Thus, all three of these factors are capable of relocalizing CBP.
A similar set of experiments was carried out with antibodies to the CBP homologue p300. However, we were unable to detect p300 within PODs (data not shown). Thus, despite the similarities between CBP and p300, the localization of CBP to PODs might constitute a difference between the two factors. However, it remains formally possible that p300 localizes to PODs but cannot be immunostained there because of steric hindrance, as has been found with CBP when using some antibody preparations (18; our unpublished observations).
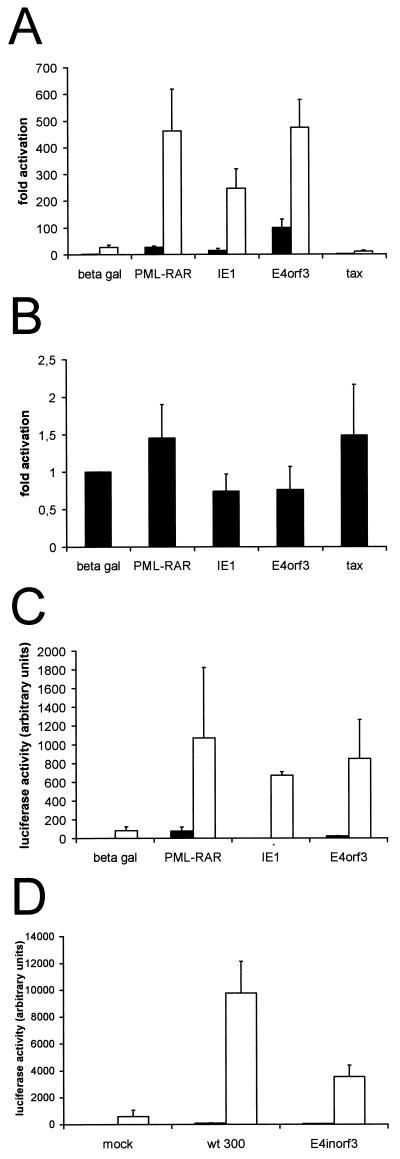
Given the relocalization of CBP by E4orf3, PML-RAR, and CMV IE1, we tested whether these three factors might also influence CBP-dependent transcription. The transcriptional activation by glucocorticoid receptors was previously shown to involve CBP (32). Overexpression of PML was found to enhance glucocorticoid-induced transcription. Further, HTLV-1 Tax protein was reported to downregulate this transcriptional activity while PML overexpression was found to compensate for the inhibitory effect of Tax (18). Therefore, we anticipated that disruption of PODs by E4orf3, PML-RAR, and CMV IE1 might have the opposite effect of PML overexpression and reduce glucocorticoid-induced transcription. To test this, HeLa cells were transfected with a luciferase reporter construct containing a glucocorticoid-responsive promoter (5′ long terminal repeat [LTR] from mouse mammary tumor virus [MMTV]; kindly provided by M. Beato), along with expression constructs for the three POD-disrupting factors and Tax (27). The cells were then either left untreated or incubated with dexamethasone. With or without the steroid, reporter expression was strongly increased for all three of the POD-disrupting factors tested (Fig. 2A) but moderately reduced when Tax was expressed (Fig. 2A) and largely unaffected when an NFκB-responsive reporter construct was used as a control (Fig. 2B). This was surprising, since PML overexpression alone led to a similar stimulation of glucocorticoid-responsive promoters in previous experiments (18; our unpublished observations). Thus, it seems that establishment of prominent PODs through overexpression of PML and disruption of the PODs through expression of PML-RAR, CMV IE1, or E4orf3 have parallel, not opposite, effects on glucocorticoid-mediated transcription.
FIG. 2.
Activation of a glucocorticoid-responsive promoter by E4orf3, CMV IE1, and PML-RAR. (A) HeLa cells were transfected with expression plasmids (2 μg) for β-galactosidase (beta gal), PML-RAR, CMV IE1, Ad5 E4orf3, and HTLV-1 Tax (0.5 μg), together with a reporter plasmid (1 μg) containing the MMTV 5′ LTR regulating the expression of luciferase. Twenty-four hours after transfection, the cells were treated with dexamethasone (1 μM) for another 24 h (open bars) or left without further treatment (filled bars). Subsequently, the cells were harvested and luciferase activity was determined from at least five independent experiments. The mean values are shown along with the standard error. (B) HeLa cells were transfected and processed as described for panel A, but instead of the MMTV LTR reporter construct, an NFκB-responsive reporter plasmid (Stratagene) was used and no dexamethasone was added. (C) HeLa cells were transfected with the same MMTV LTR-luciferase reporter construct as described for panel A (3 μg) along with an expression plasmid conferring neomycin resistance (pCIN4 [26]; 100 ng), and a pool of stably transfected cells was selected with G418 (0.4 μg/ml). These cells were then transiently transfected with expression plasmids for β-galactosidase, PML-RAR, CMV IE1, and Ad5 E4orf3, as indicated. Reporter activities in the absence (filled bars) or presence (open bars) of dexamethasone are shown as in panel A. (D) The same pool of stably transfected HeLa cells was infected (MOI = 1) with wild-type Ad or a mutant lacking E4orf3 or mock infected as indicated. Immediately after infection, dexamethasone was added in one set of experiments (open bars) or omitted (filled bars), and luciferase activity was determined 18 h after infection.
CBP is known to function as a histone acetyltransferase, modulating chromatin structure (6). Therefore, it was important to test the effects of POD-disrupting factors on a chromosomally integrated reporter rather than on a transiently transfected reporter plasmid. A pool of HeLa cells was created that was stably transfected with the same MMTV LTR reporter construct as in the previous experiment. When POD-disrupting factors were expressed in this pool of cells, the expression pattern of the chromosomally integrated reporter was comparable to that of the transiently transfected plasmid (Fig. 2C), arguing that the disruption of PODs by these factors occurs in parallel with the activation of a glucocorticoid-responsive promoter within the cellular genome.
To further ensure that the modulation of glucocorticoid-responsive transcription is a physiological function of E4orf3, the pool of stably transfected HeLa cells was infected with Ad. In parallel, cells were mock infected or infected with a recombinant virus lacking E4orf3 expression (E4inorf3). The cells were then treated with dexamethasone or left untreated. Twenty-four hours later, luciferase activity was measured. Again, infection with the wild-type virus augmented reporter expression whereas the recombinant virus lacking E4orf3 did so only to a lesser extent (Fig. 2D). Thus, while Ad infection as such seems to stimulate a glucocorticoid-responsive promoter under the conditions employed here, the expression of E4orf3 by Ad significantly enhances this effect. This suggests that the activation of glucocorticoid-responsive promoters can be considered a physiological effect of E4orf3 expression during infection with Ad.
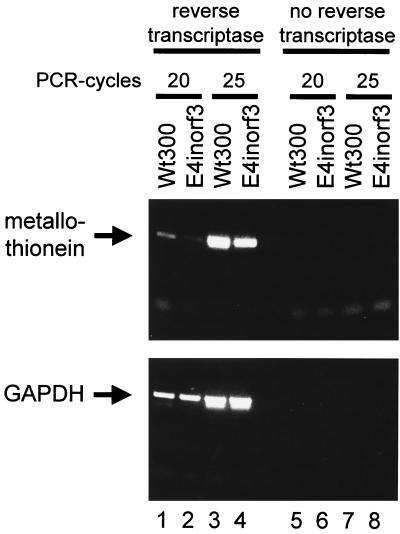
Finally, the metallothionein IIa mRNA gene, a cellular gene known to be activated by glucocorticoids (31), was analyzed during Ad infection. HeLa cells were infected with Ad expressing or lacking E4orf3. Twenty-four hours later, the cells were harvested and cellular RNA was prepared (Trizol; Life Technologies). The levels of metallothionein IIa mRNA were measured by quantitative reverse transcription-PCR. Reverse transcription was carried out using Superscript II (Life Technologies) and the primer AGCAAACGGTCACGGTCAGGGTTG, which corresponds to the reverse complement of metallothionein IIa mRNA. The cDNA obtained was amplified using the primers GCCGCCGGTGACTCCTGCACCTGC and TCCAGGTTTGTGGAAGTCGCGTTC and Pfu turbo DNA polymerase (Stratagene). In parallel reactions, the mRNA for glyceraldehyde phosphate dehydrogenase (GAPDH) was reverse transcribed with the primer GGTTCACACCCATGACGAACATG and amplified with the primers TGAAGGTCGGAGTCAACGGATTTGGT and GCAGAGATGATGACCCTTTTGGCTC. After 20 and 25 amplification cycles, the reaction was stopped and the PCR products were analyzed on an agarose gel (Fig. 3, lanes 1 to 4). Control reactions were carried out omitting the reverse transcription step (Fig. 3, lanes 5 to 8). Using this assay, the metallothionein IIa mRNA levels were found to be considerably enhanced when cells had been infected with wild-type Ad, compared to Ad lacking E4orf3 (Fig. 3, compare lanes 1 and 2, as well as lanes 3 and 4). In contrast, no difference was detected when assaying for GAPDH mRNA levels in parallel. Therefore, we concluded that E4orf3 enhances the amount of mRNA of a cellular glucocorticoid-responsive gene under the physiological conditions of virus infection.
FIG. 3.
Induction of the cellular glucocorticoid-responsive gene for metallothionein IIa by E4orf3 during Ad infection. HeLa cells were infected (MOI = 1) with Ad expressing (wt300, lanes 1, 3, 5, and 7) or lacking E4orf3 (E4inorf3, lanes 2, 4, 6, and 8). Twenty-four hours after infection, RNA was extracted from the cells and the metallothionein and GAPDH mRNA levels were determined by quantitative reverse transcription-PCR as described in the text. The reaction was stopped at 20 (lanes 1, 2, 5, and 6) or 25 (lanes 3, 4, 7, and 8) amplification cycles, and the PCR products were visualized by ethidium bromide staining of an agarose gel. In control reactions, the reverse transcription step was omitted (lanes 5 to 8).
In parallel with the disruption of PODs and the intracellular redistribution of CBP, transcription from glucocorticoid-responsive promoters is enhanced. This is in seeming contrast to previous reports that suggested that PODs might assist in CBP-mediated transcriptional activation by providing a suitable environment for transcription (13, 18). If CBP-mediated transcription actually had to occur within the PODs, one would expect downregulated, rather than enhanced, promoter activities after disruption of the PODs. Indeed, it was found that overexpression of PML upregulates CBP-mediated transcription, possibly through direct association of PML with CBP (13). Downregulation of some nuclear-hormone-responsive promoters has been observed upon expression of another POD-targeted protein, the tax gene product of HTLV-1 (11). We show here that other factors that target the PODs, namely, AdE4orf3, CMV IE1, and PML-RAR, all have the opposite effect of Tax on glucocorticoid-induced transcription. The reason for this seeming contradiction could be that transcription is indeed regulated by PODs and the factors contained therein but in a manner that does not require PODs as a scaffold for mRNA synthesis. This explanation is further supported by the fact that PML is required for POD formation (15), while mice lacking PML are viable and show only subtle differences from their PML+/+ littermates (33). These observations suggest that PODs do not represent “factories” needed to synthesize essential mRNA species. In addition, studies involving electron microscopy showed no evidence for RNA accumulation in PODs (7). Several models for POD-regulated transcription have been suggested (38). PODs might act as storage sites for transcription factors that can be released in a controlled fashion. Further, transcription factors may transiently localize to PODs in order to be modified posttranslationally, e.g., by SUMO (22, 30). We do not know if these putative functions are destroyed when PODs are disrupted by E4orf3, CMV IE1, or PML-RAR or whether the redistributed components of the PODs may still act as “mini-PODs” that retain at least some of the POD functions. Further, it remains formally possible that E4orf3, CMV IE1, or PML-RAR might affect posttranscriptional events such as RNA stability or translational efficiency, which would also result in enhanced reporter expression. However, given the specificity of the promoter, we favor a model that agrees with the notion that these proteins augment the efficiency of mRNA synthesis.
To further clarify the role of E4orf3, we performed mutagenesis on it. Thereby, we attempted to examine a possible correlation between the relocalization of PML and CBP and activation of the MMTV LTR promoter. Out of 116 amino acid residues of E4orf3, the following triplets of amino acids were mutated into alanine residues: 23 to 25, 34 to 36, 38 to 40, 52 to 54, 55 to 57, 66 to 68, 80 to 82, and 97 to 99. These residues were predicted to be exposed to the surface of the protein or were conserved between different Ad strains. However, all of these mutants still relocalized PML and were therefore not suitable for the study of the correlations outlined above (C. König, unpublished observations).
Does the activation of glucocorticoid-responsive promoters (and possibly other promoters that are responsive to nuclear hormones) constitute an advantage for virus propagation? An Ad lacking the coding sequence for it replicates nearly as well as wild-type Ad (14). Hence, the function of E4orf3 might be relevant for Ad spread in an infected organism but is not essential in cell culture. Therefore, it is hard to determine how E4orf3 and its ability to modulate transcription contribute to virus spread. However, it is tempting to speculate that E4orf3 and other POD-disrupting factors might influence the expression of major histocompatibility complex molecules, as does the PML protein in certain cells (37). Thereby, E4orf3 might act as a modulator of the immune response. It has been found by several groups that the expression of the E4orf3 gene by Ad vectors enhances transgene expression and prolongs vector persistence in transduced tissue (4, 5, 8, 19, 36). This might be due to modulated major histocompatibility complex expression. Alternatively, transcriptional activation by nuclear hormone receptors might constitute a mechanism by which to enhance transgene expression directly. Accordingly, the CMV major immediate-early promoter that is commonly used for transgene expression can be activated by nuclear hormones (3).
Despite the intensive study of PODs, their functions remain poorly understood. The hypothesis that they constitute centers of transcription is attractive, but the data presented here argue against this idea, at least in the case of glucocorticoid-induced transcription. Our results suggest that PODs do not directly serve as RNA factories. However, they do argue that PODs and their constituents regulate transcription in a more indirect way. The underlying mechanisms, as well as the full spectrum of target genes, remain subjects for future investigations.
Acknowledgments
We thank H.-D. Klenk for generous support; C. Lenz-Stöppler for excellent technical assistance; P. Hearing for Ad wt300 and E4inorf3 and helpful discussion; H. Will and T. Sternsdorf for PML++ HeLa cells and advice; M. Beato, P. Chambon, B. Cullen, A. Dejean, R. Evans, R. Grassmann, and T. Shenk for plasmids; J. Roth, C. König, and Y. Shen for many helpful suggestions; and U. Hobom for critically reading the manuscript.
This work was supported by the German Research Foundation and the P. E. Kempkes Foundation. S.W. received a fellowship from the Hoechst Foundation, and M.D. was a recipient of the Stipendium für Infektionsbiologie of the German Cancer Research Center.
REFERENCES
- 1.Ahn J H, Brignole III E J, Hayward G S. Disruption of PML subnuclear domains by the acidic IE1 protein of human cytomegalovirus is mediated through interaction with PML and may modulate a RING finger-dependent cryptic transactivator function of PML. Mol Cell Biol. 1998;18:4899–4913. doi: 10.1128/mcb.18.8.4899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ahn J H, Hayward G S. The major immediate-early proteins IE1 and IE2 of human cytomegalovirus colocalize with and disrupt PML-associated nuclear bodies at very early times in infected permissive cells. J Virol. 1997;71:4599–4613. doi: 10.1128/jvi.71.6.4599-4613.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Angulo A, Ghazal P. Regulation of human cytomegalovirus by retinoic acid. Scand J Infect Dis Suppl. 1995;99:113–115. [PubMed] [Google Scholar]
- 4.Armentano D, Smith M P, Sookdeo C C, Zabner J, Perricone M A, St. George J A, Wadsworth S C, Gregory R J. E4ORF3 requirement for achieving long-term transgene expression from the cytomegalovirus promoter in adenovirus vectors. J Virol. 1999;73:7031–7034. doi: 10.1128/jvi.73.8.7031-7034.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Armentano D, Zabner J, Sacks C, Sookdeo C C, Smith M P, St. George J A, Wadsworth S C, Smith A E, Gregory R J. Effect of the E4 region on the persistence of transgene expression from adenovirus vectors. J Virol. 1997;71:2408–2416. doi: 10.1128/jvi.71.3.2408-2416.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bannister A J, Kouzarides T. The CBP coactivator is a histone acetyltransferase. Nature. 1996;384:641–643. doi: 10.1038/384641a0. [DOI] [PubMed] [Google Scholar]
- 7.Boisvert F M, Hendzel M J, Bazett-Jones D P. Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol. 2000;148:283–292. doi: 10.1083/jcb.148.2.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brough D E, Hsu C, Kulesa V A, Lee G M, Cantolupo L J, Lizonova A, Kovesdi I. Activation of transgene expression by early region 4 is responsible for a high level of persistent transgene expression from adenovirus vectors in vivo. J Virol. 1997;71:9206–9213. doi: 10.1128/jvi.71.12.9206-9213.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carvalho T, Seeler J S, Ohman K, Jordan P, Petterson U, Akusjarvi G, Carmo-Fonseca M, Dejean A. Targeting of adenovirus E1A and E4-ORF3 proteins to nuclear matrix-associated PML bodies. J Cell Biol. 1995;131:45–56. doi: 10.1083/jcb.131.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Desbois C, Rousset R, Bantignies F, Jalinot P. Exclusion of Int-6 from PML nuclear bodies by binding to the HTLV-I Tax oncoprotein. Science. 1996;273:951–953. doi: 10.1126/science.273.5277.951. [DOI] [PubMed] [Google Scholar]
- 11.Doucas V, Evans R M. Human T-cell leukemia retrovirus-Tax protein is a repressor of nuclear receptor signaling. Proc Natl Acad Sci USA. 1999;96:2633–2638. doi: 10.1073/pnas.96.6.2633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Doucas V, Ishov A M, Romo A, Juguilon H, Weitzman M D, Evans R M, Maul G G. Adenovirus replication is coupled with the dynamic properties of the PML nuclear structure. Genes Dev. 1996;10:196–207. doi: 10.1101/gad.10.2.196. [DOI] [PubMed] [Google Scholar]
- 13.Doucas V, Tini M, Egan D A, Evans R M. Modulation of CREB binding protein function by the promyelocytic (PML) oncoprotein suggests a role for nuclear bodies in hormone signaling. Proc Natl Acad Sci USA. 1999;96:2627–2632. doi: 10.1073/pnas.96.6.2627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang M M, Hearing P. Adenovirus early region 4 encodes two gene products with redundant effects in lytic infection. J Virol. 1989;63:2605–2615. doi: 10.1128/jvi.63.6.2605-2615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ishov A M, Sotnikov A G, Negorev D, Vladimirova O V, Neff N, Kamitani T, Yeh E T, Strauss J F, 3rd, Maul G G. J. Cell Biol. 147:221–234. 1999. PML is critical for ND10 formation and recruits the PML-interacting protein daxx to this nuclear structure when modified by SUMO-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kakizuka A, Miller W H, Jr, Umesono K, Warrell R P, Jr, Frankel S R, Murty V V, Dmitrovsky E, Evans R M. Chromosomal translocation t(15;17) in human acute promyelocytic leukemia fuses RAR alpha with a novel putative transcription factor, PML. Cell. 1991;66:663–674. doi: 10.1016/0092-8674(91)90112-c. [DOI] [PubMed] [Google Scholar]
- 17.König C, Roth J, Dobbelstein M. Adenovirus type 5 E4orf3 protein relieves p53 inhibition by E1B-55-kilodalton protein. J Virol. 1999;73:2253–2262. doi: 10.1128/jvi.73.3.2253-2262.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.La Morte V J, Dyck J A, Ochs R L, Evans R M. Localization of nascent RNA and CREB binding protein with the PML-containing nuclear body. Proc Natl Acad Sci USA. 1998;95:4991–4996. doi: 10.1073/pnas.95.9.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lusky M, Grave L, Dieterle A, Dreyer D, Christ M, Ziller C, Furstenberger P, Kintz J, Hadji D A, Pavirani A, Mehtali M. Regulation of adenovirus-mediated transgene expression by the viral E4 gene products: requirement for E4 ORF3. J Virol. 1999;73:8308–8319. doi: 10.1128/jvi.73.10.8308-8319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mu Z M, Le X F, Glassman A B, Chang K S. The biologic function of PML and its role in acute promyelocytic leukemia. Leuk Lymphoma. 1996;23:277–285. doi: 10.3109/10428199609054830. [DOI] [PubMed] [Google Scholar]
- 21.Mu Z M, Le X F, Vallian S, Glassman A B, Chang K S. Stable overexpression of PML alters regulation of cell cycle progression in HeLa cells. Carcinogenesis. 1997;18:2063–2069. doi: 10.1093/carcin/18.11.2063. [DOI] [PubMed] [Google Scholar]
- 22.Muller S, Matunis M J, Dejean A. Conjugation with the ubiquitin-related modifier SUMO-1 regulates the partitioning of PML within the nucleus. EMBO J. 1998;17:61–70. doi: 10.1093/emboj/17.1.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nevels M, Tauber B, Kremmer E, Spruss T, Wolf H, Dobner T. Transforming potential of the adenovirus type 5 E4orf3 protein. J Virol. 1999;73:1591–1600. doi: 10.1128/jvi.73.2.1591-1600.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ornelles D A, Shenk T. Localization of the adenovirus early region 1B 55-kilodalton protein during lytic infection: association with nuclear viral inclusions requires the early region 4 34-kilodalton protein. J Virol. 1991;65:424–429. doi: 10.1128/jvi.65.1.424-429.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quignon F, De Bels F, Koken M, Feunteun J, Ameisen J C, de The H. PML induces a novel caspase-independent death process. Nat Genet. 1998;20:259–265. doi: 10.1038/3068. [DOI] [PubMed] [Google Scholar]
- 26.Rees S, Coote J, Stables J, Goodson S, Harris S, Lee M G. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. BioTechniques. 1996;20:102–104. doi: 10.2144/96201st05. , 106, 108–110. [DOI] [PubMed] [Google Scholar]
- 27.Rimsky L, Hauber J, Dukovich M, Malim M H, Langlois A, Cullen B R, Greene W C. Functional replacement of the HIV-1 rev protein by the HTLV-1 rex protein. Nature. 1988;335:738–740. doi: 10.1038/335738a0. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Zhu H, Shenk T. Human cytomegalovirus IE1 and IE2 proteins are mutagenic and mediate “hit-and-run” oncogenic transformation in cooperation with the adenovirus E1A proteins. Proc Natl Acad Sci USA. 1997;94:3341–3345. doi: 10.1073/pnas.94.7.3341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sternsdorf T, Grotzinger T, Jensen K, Will H. Nuclear dots: actors on many stages. Immunobiology. 1997;198:307–331. doi: 10.1016/S0171-2985(97)80051-4. [DOI] [PubMed] [Google Scholar]
- 30.Sternsdorf T, Jensen K, Will H. Evidence for covalent modification of the nuclear dot-associated proteins PML and Sp100 by PIC1/SUMO-1. J Cell Biol. 1997;139:1621–1634. doi: 10.1083/jcb.139.7.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strahle U, Boshart M, Klock G, Stewart F, Schutz G. Glucocorticoid- and progesterone-specific effects are determined by differential expression of the respective hormone receptors. Nature. 1989;339:629–632. doi: 10.1038/339629a0. [DOI] [PubMed] [Google Scholar]
- 32.Torchia J, Rose D W, Inostroza J, Kamei Y, Westin S, Glass C K, Rosenfeld M G. The transcriptional co-activator p/CIP binds CBP and mediates nuclear-receptor function. Nature. 1997;387:677–684. doi: 10.1038/42652. [DOI] [PubMed] [Google Scholar]
- 33.Wang Z G, Delva L, Gaboli M, Rivi R, Giorgio M, Cordon-Cardo C, Grosveld F, Pandolfi P P. Role of PML in cell growth and the retinoic acid pathway. Science. 1998;279:1547–1551. doi: 10.1126/science.279.5356.1547. [DOI] [PubMed] [Google Scholar]
- 34.Wang Z G, Ruggero D, Ronchetti S, Zhong S, Gaboli M, Rivi R, Pandolfi P P. PML is essential for multiple apoptotic pathways. Nat Genet. 1998;20:266–272. doi: 10.1038/3073. [DOI] [PubMed] [Google Scholar]
- 35.Weis K, Rambaud S, Lavau C, Jansen J, Carvalho T, Carmo-Fonseca M, Lamond A, Dejean A. Retinoic acid regulates aberrant nuclear localization of PML-RAR alpha in acute promyelocytic leukemia cells. Cell. 1994;76:345–356. doi: 10.1016/0092-8674(94)90341-7. [DOI] [PubMed] [Google Scholar]
- 36.Yew N S, Marshall J, Przybylska M, Wysokenski D M, Ziegler R J, Rafter P W, Li C, Armentano D, Cheng S H. Increased duration of transgene expression in the lung with plasmid DNA vectors harboring adenovirus E4 open reading frame 3. Hum Gene Ther. 1999;10:1833–1843. doi: 10.1089/10430349950017518. [DOI] [PubMed] [Google Scholar]
- 37.Zheng P, Guo Y, Niu Q, Levy D E, Dyck J A, Lu S, Sheiman L A, Liu Y. Proto-oncogene PML controls genes devoted to MHC class I antigen presentation. Nature. 1998;396:373–376. doi: 10.1038/24628. [DOI] [PubMed] [Google Scholar]
- 38.Zhong S, Salomoni P, Pandolfi P P. The transcriptional role of PML and the nuclear body. Nat Cell Biol. 2000;2:E85–E90. doi: 10.1038/35010583. [DOI] [PubMed] [Google Scholar]