Abstract
Nucleolus is the most conspicuous sub-nuclear compartment that is well known as the site of RNA polymerase I-mediated rDNA transcription and assembly of ribosome subunits in eukaryotes. Recent studies on mammalian cells suggest that functions of nucleolus are not limited to ribosome biogenesis, and that nucleolus is involved in a diverse array of nuclear and cellular processes such as DNA repair, stress responses, and protein sequestration. In fungi, knowledge of nucleolus and its functions was primarily gleaned from the budding yeast. However, little is known about nucleolus of the filamentous fungi. Considering that the filamentous fungi are multi-cellular eukaryotes and thus distinct from the yeast in many aspects, researches on nucleoli of filamentous fungi would have the potential to uncover the evolution of nucleolus and its roles in the diverse cellular processes. Here we provide a brief up-to-date overview of nucleolus in general, and evidence suggesting their roles in fungal physiology and development.
Keywords: Nucleolus, nucleolar protein, filamentous fungi, fungal physiology and development, rice blast fungus
1. Introduction
The nucleoli are membrane-less dense compartments central to rDNA transcription, pre-rRNA processing and ribosome assembly [1]. It was only in 1964 that a study with Xenopus laevis spurred interest in the function and structure of the nucleolus [2], although nucleoli themselves were first identified by bright-field microscopy as early as the 1800s [3]. Much work on the nucleolus has focused on RNA polymerase I-mediated transcription and ribosome biogenesis, establishing the nucleolus as a ribosome factory [1,4–7].
Animals and plants have a number of nucleoli in each cell. Based on studies using electron microscope, each nucleolus in their cells was initially described as tripartite structure that is organized into morphologically distinct sub-compartments at the ultrastructural level: fibrillar center (FC), dense fibrillar center (DFC) and granular component (GC) [8,9]. Such organization was shown to correspond to sequential processes in rDNA transcription, rRNA processing, and ribosome assembly [10]. The tripartite structure, however, is not universal, as many eukaryotes have bipartite nucleoli [9]. Phylogenetic pattern of tripartite and bipartite structures suggested emergence of a third nucleolar compartment during evolutionary transition between the anamniotes and the amniotes in animals [11].
A wealth of researches over the last few decades showed that the nucleolus is not just a ribosome factory but a multifunctional domain on which nucleus relies for many of its functions [12–14]. It has been also demonstrated that the nucleolus is a dynamic structure that changes in response to cell cycle, stresses, and metabolic activity of cells [8,12,15–17]. The list of roles that the nucleolus plays in regulating nuclear and cellular processes keeps growing to date.
Unlike animal and plant cells, the model yeast fungus, Saccharomyces cerevisiae, has only a single, crescent-shaped nucleolus that occupies up to one-third of the nucleoplasm [9] and organizes into bipartite structure. Though nucleoli of the budding yeast are quite distinct from animal and plant counterparts, most of our understanding of ribosome biogenesis at the molecular level in eukaryotes had been derived from studies in this fungus. Moreover, the nucleolar biology of the yeast does not seem to be a complete representation of the fungal nucleoli in general, as illustrated by the nucleolus of the fission yeast Schizosaccharomyces pombe that was reported to have tripartite structure [18].
Considering the hyper-diversity of the fungal kingdom, it is important to understand the variations present in nucleolar structures and functions of diverse fungal species and whether such variations are related to evolution, lifestyles and ecology of fungi. However, little is known about nucleolus and its functions in filamentous fungi to date. In this review, we first briefly summarize what has been known about functions of nucleolus and dynamics of nucleolar morphology, and then describe our recent observations on nucleolar dynamics in the rice blast fungus, Magnaporthe oryzae. Finally, we point out the knowledge gap that should be filled in future research endeavors regarding the fungal nucleoli.
2. Formation and functions of nucleolus
Nucleolus is a membrane-less organelle that is known to form as a new phase, separating from nucleoplasm [19,20]. However, it was shown that the initial assembly of nucleolus is guided by transcription of rRNA, conferring spatiotemporal precision in otherwise stochastic process by itself [21]. Nucleolar assembly involves the formation of pre-nucleolar bodies (PNB) and stabilization of the nucleolar machinery by precursor rRNA. As the initiation of nucleolus formation occurs around tandem arrays of ribosomal gene repeats, this chromosomal region is termed nucleolar organizer region (NOR). Details of NORs including genomic architecture are beyond the scope of this review, and thus readers are referred to the relevant review paper [22]. Cooperative interactions between chromosomal territories including NORs are known to complete nucleolar assembly process [23].
Functions of nucleoli including ribosome biogenesis hinge largely on the sequestration or release of specific proteins that enter the nucleoli (Figure 1A). It is well established that the import of proteins into nucleus is often mediated by a short stretch of basic amino acids termed as nuclear localization sequences (NLS). Unlike the nucleus separated from the cytoplasm by membrane and involving NLS-based active protein transport mechanism, the nucleolus does not have protein transport mechanisms [24]. Despite lack of active transport, it had been observed that nucleolar proteins are highly enriched in the nucleolus, while nuclear proteins without nucleolar functions are less concentrated in or completely excluded from this sub-nuclear compartment. Together with such observations, dynamic exchange of proteins between nucleolus and nucleoplasm/cytoplasm in response to different cellular conditions suggested that the specific signals or mechanisms dictate nucleolar localization and retention of proteins.
Figure 1.
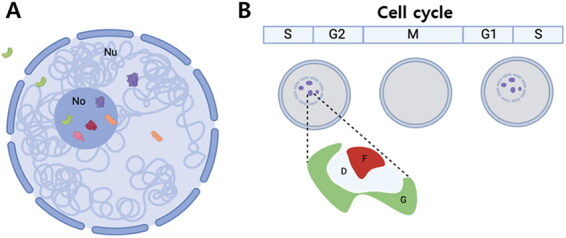
Schematic diagram showing dynamics of nucleolar proteins (A) and cell cycle-dependent changes in nucleoli (B). Some of the nucleolar proteins (red and pink) reside, functioning mainly in nucleolus, and others move between nucleoplasm and nucleolus (purple and orange) (a). They shuttle between nucleoplasm and nucleolus, either because they should be sequestered inside the nucleolus until they are required by the nucleus, or because they have major roles in the nucleolus and have moonlighting functions in the nucleoplasm. There are even proteins that go out to the cytoplasm (green). Nucleoli undergo disassembly and assembly during the cell cycle (B). The figures are created based on the mammalian nucleoli, which have tri-partite structure consisting of fibrillar center (F), dense fibrillar center (D), and granular component (G).
Based on the sequence composition and predicted secondary structure of known human nucleolar proteins, an artificial neural network (NoD server) was trained and used to systematically predict nucleolar localization sequences (NoLS) [25,26] (Note that interactive online version of NoD server is available at http://www.compbio.dundee.ac.uk/www-nod/). However, it was pointed out that there is a considerably high false positive rate in NoD prediction due to the subtle difference between NLS and NoLS with the latter being a few basic charged amino acids longer [24]. Although the defining properties of peptides and proteins that determine nucleolar localization have not been fully resolved, it seems clear that a positively charged peptide entity containing a patch of arginine is sufficient for accumulation of proteins in nucleoli [24].
The role of nucleolus is not limited to rRNA production and ribosome assembly, but includes RNA processing, assembly, and export of ribonucleoprotein (RNP) particles such as the signal recognition particle, and processing of precursor tRNA and U6 small nuclear RNA (snRNA). However, it has been revealed that functions of nucleolus are much more diverse and profound than previously recognized. For example, nucleolus is involved in DNA damage and repair, telomere maintenance and response to cellular stresses [13]. Many of these recently identified functions are related to the protein sequestration in the nucleolus or protein shuttling between the nucleolus and the nucleoplasm (Figure 1A). Some proteins are stored in the nucleolus, while others have main functions in the nucleolus and have moonlighting functions in the nucleoplasm. There are also proteins that are sequestered in the nucleolus until it should be released. Below we briefly describe discoveries made on nucleolar functions during the last decade. It should be noted that such discoveries have centered around mammalian cells.
Based on dynamics of nucleolar proteins, the nucleolus plays a role in DNA damage repair by participating in the recruitment and assembly of repair factors at sites of DNA damage. The nucleolus regulates availability and activity of repair factors by sequestering or release them in response to DNA damage [27,28]. The nucleolus is also known to be involved in telomere maintenance, which is crucial for preserving genomic stability and cell viability. It can control the availability of telomerases by sequestering them, and can play a role in the recruitment of telomeric proteins to telomeres by interacting with telomeric proteins and telomerase, ensuring proper telomere structure and function [29–32]. In addition, the nucleolus is known to play a role in the 3D organization of the genome by facilitating the spatial arrangement of chromatin and gene expression [33,34]. This is mediated by its interactions with specific genomic loci, recruitment of transcription factors, chromatin remodeling factor and histone modifiers.
In animal cells, the nucleolus and the Cajal body are two nuclear organelles that respond to stress by undergoing specific changes in morphology and composition. Although underlying mechanisms of nucleolar stress responses are complex and interconnected, they can be grouped into p53-dependent and -independent pathways in animal cells [16]. The tumor suppressor, p53, which is known as the “guardian of the genome” and kept low under normal physiological conditions due to its interaction with E3 ubiquitin ligases, appears to be the key in many pathways that translate stress signals into a cellular response [35,36]. Stresses such as genotoxic and ribosomal stress can disrupt ribosome subunit biogenesis and lead to an increase in free RPL11 and RPL26 (ribosomal proteins), which up-regulates p53 translation and inhibits Hdm2 (E3 ubiquitin ligase)-mediated p53 degradation [37,38]. This increases the opportunity for p53 to activate transcription of its target genes in the nucleoplasm. Furthermore, disruption of ribosome biogenesis can increase the transport of p53 to the cytoplasm, resulting in activation of the mitochondrial apoptotic pathway. For more details of nucleolar-related mechanisms to activate p53, please refer to the Boulon et al. [16].
3. Dynamics and regulation of nucleolus
The nucleolus is dynamic not just in terms of exchange of its proteins with nucleoplasm but also in the sense of changes in number and size. There are several factors such as cell cycle, nutrient availability and cellular stresses that influence the dynamics of nucleoli in the cell. In this section, we describe how nucleoli respond to those factors and what molecular mechanisms underlie the changes in nucleoli.
3.1. Cell cycle
During the cell cycle, ribosome production should be tightly regulated in coordination with the progression of cell cycle (Figure 1B). In higher eukaryotes, ribosome production starts to cease at prophase (at the beginning of mitosis), and then re-initiates at the end of mitosis. It increases during G1 through S, and then peaks in G2 phase [17,39]. At the beginning of mitosis, nucleolar disassembly occurs in a sequential process. First, nucleolar processing complexes, which consist of DFC and GC proteins such as fibrillarin and Nop52, respectively, is released from the nucleolus at the end of prophase, during which nuclear envelope breaks down and chromosomes are condensed, forming a structure called peri-chromosomal compartment [40,41]. Such release of proteins changes the shape of the nucleolus, and eventually makes the nucleoli invisible. Following the disassociation of the nucleolar processing complexes from the nucleolus, transcription of rDNA is arrested at the end of prophase, while the DNA polymerase I (pol I) transcription machinery remains associated with the rDNAs.
Although there are pol I machineries associated with rDNAs, some of them (pol I-specific transcription factors) are phosphorylated by the Cdk1-cyclin B kinase, resulting in the suppression of rDNA transcription [42]. The Cdk1-cyclin B kinase phosphorylates the nucleolar processing proteins as well. This phosphorylation decreases the RNA-binding affinity of the target proteins, possibly contributing to the dissociation of the proteins from the nucleolus during prophase [43].
The assembly of nucleolus, which starts at telophase, requires sophisticated coordination of rDNA transcription and activation of the RNA processing complexes [17]. Activation of rDNA transcription depends on the pol I machineries associated with NORs [44]. Repression of pol I transcription activity by Cdk1-cyclin B kinase during mitosis is derepressed by the phosphatases PP1 and PP2A [45]. It should be noted that control of phosphorylation/dephosphorylation is dependent upon the local concentration and distribution of kinases and phosphatases involved in cell cycle regulation. The resumption of rDNA transcription is necessary but not enough for the nucleolar assembly. Complete organization of the nucleolus also requires the snoRNAs, ribosomal proteins, rRNAs and other proteins that form the nucleolar processing complexes in foci designated PNBs. In human HeLa cells, the PNBs are known to be observed in telophase and present during early G1 period [46,47].
3.2. Nutrient availability and other stresses
It has long been known that the size and shape of nucleoli are intimately related to the growth and proliferation status of eukaryotic cell [48]. Nutrient availability is a crucial factor for the control of nucleolar activity, as the rDNA transcription and ribosome biogenesis are highly costly and demanding cellular processes in terms of energy and resource usage. For example, RNA pol I-mediated transcriptional activity accounts for 60% of the total transcription in case of the growing yeast. Transcription of ribosomal genes takes up approximately 50% of the total RNA pol II activity. The total mass of resulting ribosomal proteins accounts for almost half the total cellular protein mass [49]. These facts about rDNA transcription and ribosome production indicate that nucleolar activities should be adjusted and tightly controlled based on available resources.
In general, stresses including heat stress, UV radiation, oxidative stress, nitrogen starvation, glucose starvation, and calorie restriction can cause a shrinkage and reorganization of the nucleolus into a smaller and more compact structure (Figure 2). Such nucleolar shrinkage is concomitant with inhibition of rDNA transcription and reduction in ribosome biogenesis. On the contrary, nutrients and growth signals can induce the nucleolus to enlarge. As a result, rDNA transcription and subsequent ribosome biogenesis increase. This impinges on speed and accuracy of translation, resulting in loss of cellular protein homeostasis (proteostasis) [48].
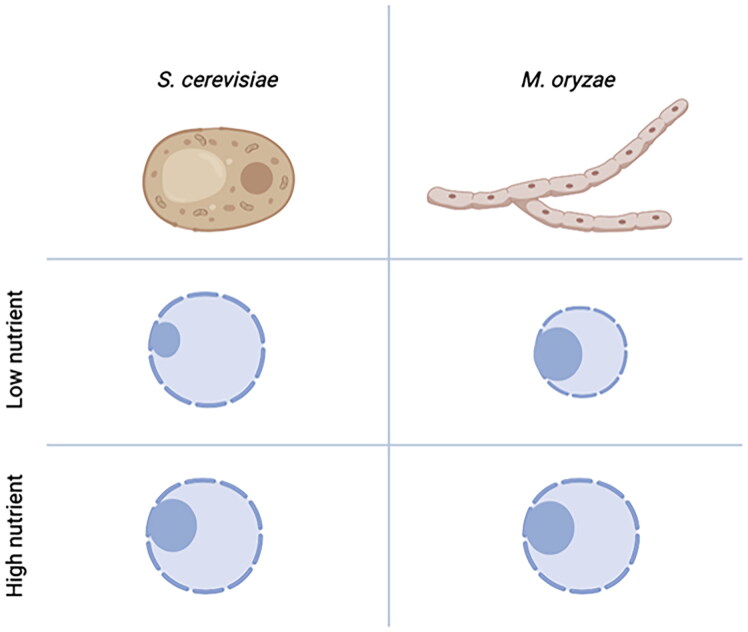
Figure 2.
Differences between the nucleoli of the budding yeast and Magnaporthe oryzae. At low concentration of nutrient, the budding yeast decreases the size of nucleolus, which correlates with reduced rDNA transcription activity. In the filamentous fungus, M. oryzae, however, the size of nucleolus remains the same, while the size of nucleus decreases.
The target of rapamycin (TOR) protein (serine/threonine) kinase is a evolutionarily conserved central regulator that links the quality and quantity of nutrients to developmental and metabolic processes [50]. In the budding yeast, there are two TOR kinases, TOR1 and TOR2, each of which forms TORC1 and TORC2 complexes in the budding yeast. In mammals, there is a single TOR kinase (mTOR) that exists in two functionally distinct mTORC1 and mTORC2 complexes. The TOR1 complex is rapamycin sensitive and involved in anabolic processes (cell growth), while the TOR2 complex is rapamycin insensitive and is involved in cell proliferation and survival [51]. Above mentioned stresses including nutrient availability and rapamycin can inhibit the TOR kinase pathway. In the yeast and mammals, such suppression/inhibition of TOR results in the reduced size of nucleolus in the cell. Detailed molecular mechanisms underlying the control of nucleolar size and its activity involve diverse transcription factors and epigenetic regulators downstream of TOR kinase, and are beyond the scope of this review (for more information, refer to the Srivastava et al. [52] and Gonzale and Rallis [53].
4. Nucleoli in filamentous fungi
In filamentous fungi, little is known about the nucleolar dynamics, its regulation and implications. Description of nucleolus in filamentous fungi dates as far back as the late 1960s, and its ultrastructural characteristics was first reported in Penicillium hyphae at 1972 [54]. Most of knowledge on fungal nucleoli are thus based on the studies of the nucleoli in the budding yeast, and work on nucleoli of filamentous fungi are scarce. However, there a few studies that demonstrate the importance of nucleoli in the biology of filamentous fungi.
For example, in a human opportunistic pathogen, Aspergillus fumigatus, the ability to grow rapidly at 37 °C is considered as a potential virulence factor [55]. Works on CgrA in A. fumigatus showed that CgrA is predominantly localized to nucleolus in a temperature-dependent manner [56], and is necessary for pre-rRNA processing and 60S ribosomal subunit assembly [57]. This established a link between thermotolerance and ribosome biogenesis, emphasizing the role of nucleolar functions in fungal pathogenesis, although how CgrA-mediated ribosome biogenesis contributes to the thermotolerance is not clear.
In a model plant pathogenic fungus, Magnaporthe oryzae, it was shown that unlike the yeast and mammalian counterparts, there is no direct relationship between the activity and size of nucleoli in this filamentous fungus [58]. Under the low nutrient concentration, nucleolar size appeared to be decoupled from the nucleolar activity in M. oryzae (Figure 2). Observation of nucleoli during germination of conidia and appressorium formation suggested the role of fungal nucleoli in the course of infection-specific development in this fungus. This study highlights the differences in the yeast and filamentous fungi in terms of regulation of nucleoli.
There are also a number of studies showing the importance of TOR kinase pathway in development and pathogenesis of filamentous fungi [59–63]. Although these studies did not directly analyze the nucleoli in the fungal species of interest, the fact that TOR is a conserved regulator of nucleolar functions strongly corroborates the importance of nucleolar function and its regulation in the biology of filamentous fungi.
Recently, there has been a report about fungal effector proteins being translocated into the nucleoli of host plant [64]. This is in parallel to the oomycete effectors being targeted to the host nucleoli and regulating nucleolar processes including ribosome biogenesis [65,66]. Such observations bring a few interesting questions into our attention: 1) do those effectors go into the fungal nucleolus as well? And is it a matter of when and where those effectors are expressed? 2) If they don’t localize to the fungal nucleolus when expressed in fungal vegetative hyphae, what would be the mechanism(s) that enables regulation of effector localization only to the host plant nucleolus? In line with the second question, 3) do those effectors have distinct (for example, plant-specific) nucleolar localization sequences? Attempts to answer the questions above would shed light on novel aspect of fungal nucleolus itself and spatial regulation of nucleolar protein.
5. Conclusion
Nucleoli is such an important organelle in terms of its cellular energy consumption and diverse functions including ribosome biogenesis. In this review, we argue that despite its implication in eukaryotic cells, organization and functions of nucleoli in filamentous fungi have been under-appreciated, in contrast to the nucleus within which the nucleolus is located and mRNA production is regulated. We propose that future studies addressing the following questions should be forthcoming in order to bridge this knowledge gap and better understand the nucleoli in the filamentous fungi. First, do nucleoli in filamentous fungi have bi-partite or tri-partite structural organization? This is related to the evolution of nucleoli and whether there is any correlation between the type of organization and biology of filamentous fungi such as lifestyle. Second, what are the nucleolar proteins other than the rRNA transcription and processing factors? Functions of particular organelles are largely dependent upon the proteins that reside in them. In mammalian cells, over 4,500 nucleolus-associated proteins were identified through proteomic experiments. However, except the yeast, no systematic approach has been made to identify the nucleolar proteins in filamentous fungi [67]. Identification of nucleolar protein repertoire in the filamentous fungi would not only shed light on the nucleolar functions that might have contributed to the evolutionary success of this immensely diverse group of eukaryotic microbes but also provide novel targets for therapeutics and agrochemicals.
Funding Statement
This work was supported by the 2022 Yeungnam University Research Grant.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- 1.Mélèse T, Xue Z.. The nucleolus: an organelle formed by the act of building a ribosome. Curr Opin Cell Biol. 1995;7(3):319–324. doi: 10.1016/0955-0674(95)80085-9. [DOI] [PubMed] [Google Scholar]
- 2.Brown DD, Gurdon JB.. Absence of ribosomal RNA synthesis in the anucleolate mutant of Xenopus laevis. Proc Natl Acad Sci U S A. 1964;51(1):139–146. doi: 10.1073/pnas.51.1.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pederson T. The nucleolus. Cold Spring Harb Perspect Biol. 2011;3(3):a000638–a000638. a000638. doi: 10.1101/cshperspect.a000638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Birnstiel ML, Wallace H, Sirlin JL, et al. . Localization of the ribosomal DNA complements in the nucleolar organizer region of Xenopus laevis. Natl Cancer Inst Monogr. 1966;23:431–447. [PubMed] [Google Scholar]
- 5.Wallace H, Birnstiel ML.. Ribosomal cistrons and the nucleolar organizer. Biochim Biophys Acta. 1966;114(2):296–310. doi: 10.1016/0005-2787(66)90311-x. [DOI] [PubMed] [Google Scholar]
- 6.Nissan TA, Bassler J, Petfalski E, et al. . 60S pre-ribosome formation viewed from assembly in the nucleolus until export to the cytoplasm. Embo J. 2002;21(20):5539–5547. doi: 10.1093/emboj/cdf547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Warner JR. The nucleolus and ribosome formation. Curr Opin Cell Biol. 1990;2(3):521–527. doi: 10.1016/0955-0674(90)90137-4. [DOI] [PubMed] [Google Scholar]
- 8.Shaw P, Brown J.. Nucleoli: composition, function, and dynamics. Plant Physiol. 2012;158(1):44–51. doi: 10.1104/pp.111.188052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Thiry M, Lafontaine DL.. Birth of a nucleolus: the evolution of nucleolar compartments. Trends Cell Biol. 2005;15(4):194–199. doi: 10.1016/j.tcb.2005.02.007. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Verdun D, Roussel P, Thiry M, et al. . The nucleolus: structure/function relationship in RNA metabolism. Wiley Interdiscip Rev RNA. 2010;1(3):415–431. doi: 10.1002/wrna.39. [DOI] [PubMed] [Google Scholar]
- 11.Thiry M, Lamaye F, Lafontaine DL.. The nucleolus: when 2 became 3. Nucleus. 2011;2(4):289–293. doi: 10.4161/nucl.2.4.16806. [DOI] [PubMed] [Google Scholar]
- 12.Boisvert FM, van Koningsbruggen S, Navascues J, et al. . The multifunctional nucleolus. Nat Rev Mol Cell Biol. 2007;8(7):574–585. doi: 10.1038/nrm2184. [DOI] [PubMed] [Google Scholar]
- 13.Iarovaia OV, Minina EP, Sheval EV, et al. . Nucleolus: a central hub for nuclear functions. Trends Cell Biol. 2019;29(8):647–659. doi: 10.1016/j.tcb.2019.04.003. [DOI] [PubMed] [Google Scholar]
- 14.Kalinina NO, Makarova S, Makhotenko A, et al. . The multiple functions of the nucleolus in plant development, disease and stress responses. Front Plant Sci. 2018;9:132. doi: 10.3389/fpls.2018.00132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leung AK, Lamond AI.. The dynamics of the nucleolus. Crit Rev Eukaryot Gene Expr. 2003;13(1):39–54. doi: 10.1615/critreveukaryotgeneexpr.v13.i1.40. [DOI] [PubMed] [Google Scholar]
- 16.Boulon S, Westman BJ, Hutten S, et al. . The nucleolus under stress. Mol Cell. 2010;40(2):216–227. doi: 10.1016/j.molcel.2010.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hernandez-Verdun D. Assembly and disassembly of the nucleolus during the cell cycle. Nucleus. 2011;2(3):189–194. doi: 10.4161/nucl.2.3.16246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Léger-Silvestre I, Noaillac-Depeyre J, Faubladier M, et al. . Structural and functional analysis of the nucleolus of the fission yeast Schizosaccharomyces pombe. Eur J Cell Biol. 1997;72(1):13–23. [PubMed] [Google Scholar]
- 19.Lafontaine DLJ, Riback JA, Bascetin R, et al. . The nucleolus as a multiphase liquid condensate. Nat Rev Mol Cell Biol. 2021;22(3):165–182. doi: 10.1038/s41580-020-0272-6. [DOI] [PubMed] [Google Scholar]
- 20.Staples MI, Frazer C, Fawzi NL, et al. . Phase separation in fungi. Nat Microbiol. 2023;8(3):375–386. doi: 10.1038/s41564-022-01314-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Falahati H, Pelham-Webb B, Blythe S, et al. . Nucleation by rRNA dictates the precision of nucleolus assembly. Curr Biol. 2016;26(3):277–285. doi: 10.1016/j.cub.2015.11.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McStay B. Nucleolar organizer regions: genomic ‘dark matter’ requiring illumination. Genes Dev. 2016;30(14):1598–1610. doi: 10.1101/gad.283838.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hernandez-Verdun D, Roussel P, Gébrane-Younès J.. Emerging concepts of nucleolar assembly. J Cell Sci. 2002;115(Pt 11):2265–2270. doi: 10.1242/jcs.115.11.2265. [DOI] [PubMed] [Google Scholar]
- 24.Martin RM, Ter-Avetisyan G, Herce HD, et al. . Principles of protein targeting to the nucleolus. Nucleus. 2015;6(4):314–325. doi: 10.1080/19491034.2015.1079680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott MS, Boisvert FM, McDowall MD, et al. . Characterization and prediction of protein nucleolar localization sequences. Nucleic Acids Res. 2010;38(21):7388–7399. doi: 10.1093/nar/gkq653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Scott MS, Troshin PV, Barton GJ.. NoD: a nucleolar localization sequence detector for eukaryotic and viral proteins. BMC Bioinformatics. 2011;12(1):317. doi: 10.1186/1471-2105-12-317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lirussi L, Antoniali G, Vascotto C, et al. . Nucleolar accumulation of APE1 depends on charged lysine residues that undergo acetylation upon genotoxic stress and modulate its BER activity in cells. Mol Biol Cell. 2012;23(20):4079–4096. doi: 10.1091/mbc.E12-04-0299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vascotto C, Fantini D, Romanello M, et al. . APE1/Ref-1 interacts with NPM1 within nucleoli and plays a role in the rRNA quality control process. Mol Cell Biol. 2009;29(7):1834–1854. doi: 10.1128/MCB.01337-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fu D, Collins K.. Purification of human telomerase complexes identifies factors involved in telomerase biogenesis and telomere length regulation. Mol Cell. 2007;28(5):773–785. doi: 10.1016/j.molcel.2007.09.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lee JH, Lee YS, Jeong SA, et al. . Catalytically active telomerase holoenzyme is assembled in the dense fibrillar component of the nucleolus during S phase. Histochem Cell Biol. 2014;141(2):137–152. doi: 10.1007/s00418-013-1166-x. [DOI] [PubMed] [Google Scholar]
- 31.Wong JM, Kusdra L, Collins K.. Subnuclear shuttling of human telomerase induced by transformation and DNA damage. Nat Cell Biol. 2002;4(9):731–736. doi: 10.1038/ncb846. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez OG, Assfalg R, Koch S, et al. . Telomerase stimulates ribosomal DNA transcription under hyperproliferative conditions. Nat Commun. 2014;5(1):4599. doi: 10.1038/ncomms5599. [DOI] [PubMed] [Google Scholar]
- 33.Van Koningsbruggen S, Gierlinski M, Schofield P, et al. . High-resolution whole-genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol Biol Cell. 2010;21(21):3735–3748. doi: 10.1091/mbc.E10-06-0508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Schöfer C, Weipoltshammer K.. Nucleolus and chromatin. Histochem Cell Biol. 2018;150(3):209–225. doi: 10.1007/s00418-018-1696-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Olson MO. Sensing cellular stress: another new function for the nucleolus? Sci STKE. 2004;2004(224):pe10. doi: 10.1126/stke.2242004pe10. [DOI] [PubMed] [Google Scholar]
- 36.Pederson T, Tsai RY.. In search of nonribosomal nucleolar protein function and regulation. J Cell Biol. 2009;184(6):771–776. doi: 10.1083/jcb.200812014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kruse JP, Gu W.. SnapShot: p53 posttranslational modifications. Cell. 2008;133(5):930–930.e1. doi: 10.1016/j.cell.2008.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee JT, Gu W.. The multiple levels of regulation by p53 ubiquitination. Cell Death Differ. 2010;17(1):86–92. doi: 10.1038/cdd.2009.77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sirri V, Roussel P, Hernandez-Verdun D.. The AgNOR proteins: qualitative and quantitative changes during the cell cycle. Micron. 2000;31(2):121–126. doi: 10.1016/s0968-4328(99)00068-2. [DOI] [PubMed] [Google Scholar]
- 40.Gautier T, Robert-Nicoud M, Guilly MN, et al. . Relocation of nucleolar proteins around chromosomes at mitosis. A study by confocal laser scanning microscopy. J Cell Sci. 1992;102 (4):729–737 doi: 10.1242/jcs.102.4.729. [DOI] [PubMed] [Google Scholar]
- 41.Van Hooser AA, Yuh P, Heald R.. The perichromosomal layer. Chromosoma. 2005;114(6):377–388. doi: 10.1007/s00412-005-0021-9. [DOI] [PubMed] [Google Scholar]
- 42.Heix J, Vente A, Voit R, et al. . Mitotic silencing of human rRNA synthesis: inactivation of the promoter selectivity factor SL1 by cdc2/cyclin B-mediated phosphorylation. Embo J. 1998;17(24):7373–7381. doi: 10.1093/emboj/17.24.7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Negi SS, Olson MO.. Effects of interphase and mitotic phosphorylation on the mobility and location of nucleolar protein B23. J Cell Sci. 2006;119(Pt 17):3676–3685. doi: 10.1242/jcs.03090. [DOI] [PubMed] [Google Scholar]
- 44.Roussel P, André C, Comai L, et al. . The rDNA transcription machinery is assembled during mitosis in active NORs and absent in inactive NORs. J Cell Biol. 1996;133(2):235–246. doi: 10.1083/jcb.133.2.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Trinkle-Mulcahy L, Lamond AI.. Mitotic phosphatases: no longer silent partners. Curr Opin Cell Biol. 2006;18(6):623–631. doi: 10.1016/j.ceb.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 46.Savino TM, Gébrane-Younès J, De Mey J, et al. . Nucleolar assembly of the rRNA processing machinery in living cells. J Cell Biol. 2001;153(5):1097–1110. doi: 10.1083/jcb.153.5.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Muro E, Gébrane-Younès J, Jobart-Malfait A, et al. . The traffic of proteins between nucleolar organizer regions and prenucleolar bodies governs the assembly of the nucleolus at exit of mitosis. Nucleus. 2010;1(2):202–211. doi: 10.4161/nucl.1.2.11334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Matos-Perdomo E, Machin F.. Nucleolar and ribosomal DNA structure under stress: yeast lessons for aging and cancer. Cells. 2019;8(8):779. doi: 10.3390/cells8080779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warner JR. The economics of ribosome biosynthesis in yeast. Trends Biochem Sci. 1999;24(11):437–440. doi: 10.1016/s0968-0004(99)01460-7. [DOI] [PubMed] [Google Scholar]
- 50.Dobrenel T, Caldana C, Hanson J, et al. . TOR signaling and nutrient sensing. Annu Rev Plant Biol. 2016;67(1):261–285. doi: 10.1146/annurev-arplant-043014-114648. [DOI] [PubMed] [Google Scholar]
- 51.González A, Hall MN, Lin S-C, et al. . AMPK and TOR: the yin and yang of cellular nutrient sensing and growth control. Cell Metab. 2020;31(3):472–492. doi: 10.1016/j.cmet.2020.01.015. [DOI] [PubMed] [Google Scholar]
- 52.Srivastava R, Srivastava R, Ahn SH.. The epigenetic pathways to ribosomal DNA silencing. Microbiol Mol Biol Rev. 2016;80(3):545–563. doi: 10.1128/MMBR.00005-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Gonzalez S, Rallis C.. The TOR signaling pathway in spatial and temporal control of cell size and growth. Front Cell Dev Biol. 2017;5:61. doi: 10.3389/fcell.2017.00061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Colman OD, Stockert JC.. The nucleolus in the vegetative cells of Penicillium. Caryologia. 1972;25(3):253–258. doi: 10.1080/00087114.1972.10796480. [DOI] [Google Scholar]
- 55.Bhabhra R, Askew DS.. Thermotolerance and virulence of Aspergillus fumigatus: role of the fungal nucleolus. Med Mycol. 2005;43 Suppl 1: s 87–S93. doi: 10.1080/13693780400029486. [DOI] [PubMed] [Google Scholar]
- 56.Bhabhra R, Zhao W, Rhodes JC, et al. . Nucleolar localization of Aspergillus fumigatus CgrA is temperature-dependent. Fungal Genet Biol. 2006;43(1):1–7. doi: 10.1016/j.fgb.2005.07.005. [DOI] [PubMed] [Google Scholar]
- 57.Bhabhra R, Miley MD, Mylonakis E, et al. . Disruption of the Aspergillus fumigatus gene encoding nucleolar protein CgrA impairs thermotolerant growth and reduces virulence. Infect Immun. 2004;72(8):4731–4740. doi: 10.1128/IAI.72.8.4731-4740.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cho E, Lee SH, Dean RA, et al. . Distinct dynamics of the nucleolus in response to nutrient availability and during development in the rice blast fungus. mBio. 2023;14(5):e0184423. doi: 10.1128/mbio.01844-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Sun G, Qi X, Wilson RA.. A feed-forward subnetwork emerging from integrated TOR- and cAMP/PKA-signaling architecture reinforces Magnaporthe oryzae appressorium morphogenesis. Mol Plant Microbe Interact. 2019;32(5):593–607. doi: 10.1094/MPMI-10-18-0287-R. [DOI] [PubMed] [Google Scholar]
- 60.Li G, Gong Z, Dulal N, et al. . A protein kinase coordinates cycles of autophagy and glutaminolysis in invasive hyphae of the fungus Magnaporthe oryzae within rice cells. Nat Commun. 2023;14(1):4146. doi: 10.1038/s41467-023-39880-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vangalis V, Markakis EA, Knop M, et al. . Components of TOR and MAP kinase signaling control chemotropism and pathogenicity in the fungal pathogen Verticillium dahliae. Microbiol Res. 2023;271:127361. doi: 10.1016/j.micres.2023.127361. [DOI] [PubMed] [Google Scholar]
- 62.Jiao W, Ding W, Rollins JA, et al. . Cross-talk and multiple control of target of rapamycin (TOR) in Sclerotinia sclerotiorum. Microbiol Spectr. 2023;11(2):e0001323. doi: 10.1128/spectrum.00013-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yu F, Gu Q, Yun Y, et al. . The TOR signaling pathway regulates vegetative development and virulence in Fusarium graminearum. New Phytol. 2014;203(1):219–232. doi: 10.1111/nph.12776. [DOI] [PubMed] [Google Scholar]
- 64.Petre B, Saunders DG, Sklenar J, et al. . Candidate effector proteins of the rust pathogen Melampsora larici-populina target diverse plant cell compartments. Mol Plant Microbe Interact. 2015;28(6):689–700. doi: 10.1094/MPMI-01-15-0003-R. [DOI] [PubMed] [Google Scholar]
- 65.Lee S, Kim J, Kim M-S, et al. . The Phytophthora nucleolar effector Pi23226 targets host ribosome biogenesis to induce necrotrophic cell death. Plant Commun. 2023;4(5):100606. doi: 10.1101/2022.05.02.490323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Pecrix Y, Buendia L, Penouilh‐Suzette C, et al. . Sunflower resistance to multiple downy mildew pathotypes revealed by recognition of conserved effectors of the oomycete Plasmopara halstedii. Plant J. 2019;97(4):730–748. doi: 10.1111/tpj.14157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ahmad Y, Boisvert F-M, Gregor P, et al. . NOPdb: nucleolar proteome database–2008 update. Nucleic Acids Res. 2009;37:D181–D184. doi: 10.1093/nar/gkn804. [DOI] [PMC free article] [PubMed] [Google Scholar]