Abstract
Human influenza A viruses replicate in the upper respiratory tract at a temperature of about 33°C, whereas avian viruses replicate in the intestinal tract at a temperature close to 41°C. In the present study, we analyzed the influence of low temperature (33°C) on RNA replication of avian and human viruses in cultured cells. The kinetics of replication of the NP segment were similar at 33 and 37°C for the human A/Puerto-Rico/8/34 and A/Sydney/5/97 viruses, whereas replication was delayed at 33°C compared to 37°C for the avian A/FPV/Rostock/34 and A/Mallard/NY/6750/78 viruses. Making use of a genetic system for the in vivo reconstitution of functional ribonucleoproteins, we observed that the polymerase complexes derived from avian viruses but not human viruses exhibited cold sensitivity in mammalian cells, which was determined mostly by residue 627 of PB2. Our results suggest that a reduced ability of the polymerase complex of avian viruses to ensure replication of the viral genome at 33°C could contribute to their inability to grow efficiently in humans.
Influenza A viruses have been isolated from a wide range of species and appear to cross the species barrier more or less easily, depending on the species involved (33). Human viruses do not spread in birds, and avian influenza viruses generally do not replicate efficiently or cause disease in humans (1). However, adaptation of an avian virus to the human host may occur either through genetic reassortment or following direct transmission and may result in influenza pandemics, as was the case in 1957 and 1968 (26). The molecular bases for the host specificity of human and avian influenza A viruses are not fully understood. The hemagglutinin (HA) and neuraminidase (NA) are considered possible determinants of host restriction because of different receptor specificities between avian and human viruses (5, 10, 13, 24, 25, 32). In addition, genetic studies have indicated that gene segments encoding internal proteins, especially the PB2 segment, carried determinants for host range (4, 17, 27, 29, 31).
The temperature at the site of infection of human and avian viruses differs. In humans, influenza viruses initiate replication in the upper respiratory tract at a temperature of about 33°C and induce an acute respiratory illness, whereas in aquatic birds, influenza viruses primarily infect the intestinal tract at a temperature close to 41°C and cause no clinical symptoms. Naturally occurring temperature-sensitive human influenza A viruses showing restricted growth at the elevated temperatures of 38 to 41°C have been described previously (3, 8, 11, 20). Moreover, it has been established that 33°C was more efficient than 37°C for the isolation and expansion of human influenza A viruses (28).
In the present study, we addressed the question of whether the natural temperature dependence of influenza A viruses could contribute to their host specificity by comparing the abilities of human and avian influenza A viruses to replicate at the temperature of the upper respiratory tract (33°C).
Temperature dependence of multiplication of avian and human influenza viruses.
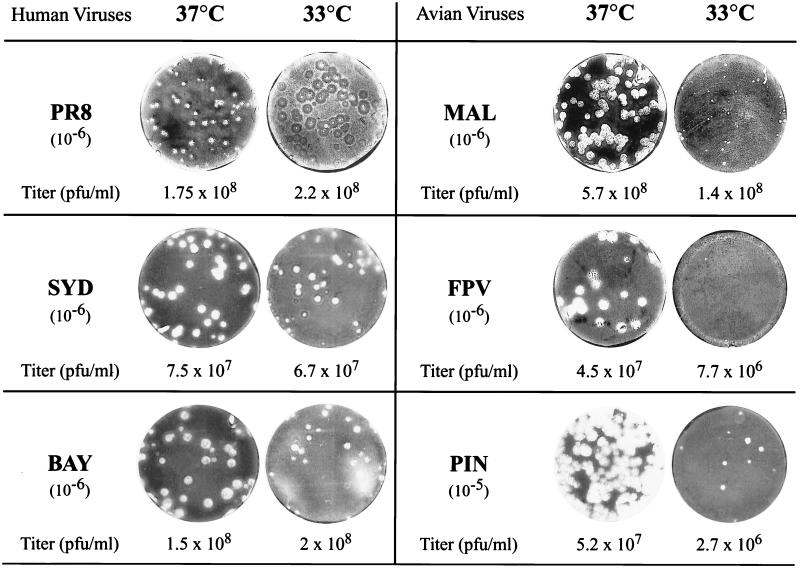
The avian viruses A/Mallard/NY/6750/78 (MAL, H2N2), A/FPV/Rostock/34 (FPV, H7N1), and A/Pintail/Alberta/79 (PIN, H4N6) and the human viruses A/Puerto-Rico/8/34 (PR8, H1N1), A/Bayern/7/95 (BAY, H1N1), and A/Sydney/5/97 (SYD, H3N2) were grown at 35°C in 11-day-old embryonated chicken eggs. Plaque assays were performed with the allantoic fluids on MDCK cells (106 cells in 35-mm dishes) at 37 and 33°C in parallel. Virus titers and plaque size were examined at 72 h postinfection (p.i.) for the MAL, PIN, BAY, and SYD viruses and at 96 h p.i. for the FPV and PR8 viruses. For the human viruses, titers were similar whether measured at 37 or 33°C. In contrast, for the avian viruses MAL, FPV, and PIN, the titers appeared to be lower (4-, 6-, and 20-fold, respectively) at 33°C than at 37°C (Fig. 1). Plaque size was only slightly reduced at 33°C compared to 37°C for the human viruses, but was strongly reduced for the avian viruses. Therefore, it cannot be excluded that MAL, FPV, and PIN titers at 33°C were underestimated, as the corresponding plaques became hardly visible. To our knowledge, the only report about the effect of low temperature on the growth of an avian influenza virus is the observation by Breuning and Scholtissek (2) that FPV titers on primary chicken embryo cells are reduced 10-fold when incubated at 33°C compared to 37°C, in accordance with our own results.
FIG. 1.
Comparison of virus titers and plaque size for human and avian influenza viruses at 37 and 33°C. MDCK cells were infected in duplicate with various dilutions of human PR8, SYD, or BAY or avian MAL, FPV, or PIN influenza virus. For each virus, MDCK cells which were infected with the same dilution (10−6 or 10−5) at either 37 or 33°C are shown. Titers were determined from dilutions that gave a minimum of 10 plaques.
In order to determine whether the efficiency of multiplication of avian viruses was impaired at 33°C, we examined the kinetics of RNA replication of PR8, SYD, BAY, MAL, FPV, and PIN viruses at 37 and 33°C. MDCK and COS-1 cells were infected at a multiplicity of infection (MOI) of 20 and 10 PFU/cell, respectively, or mock infected. After adsorption for 1 h at 33°C, cells were washed to remove unbound viruses and incubated with medium warmed to 37 or 33°C. At different times postinfection, total RNAs were extracted using the Trizol reagent (Gibco-BRL), slot blotted onto a nylon membrane (Hy bond), and probed with a 32P-radiolabeled riboprobe directed against the nucleoprotein (NP) segment of FPV.
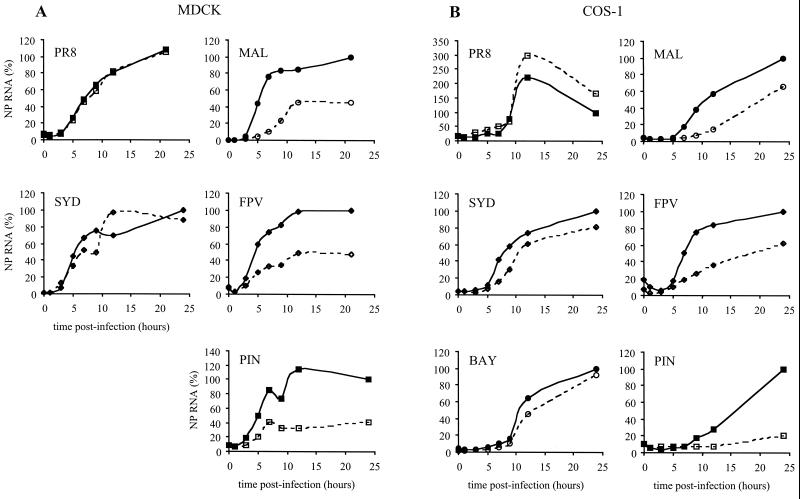
Quantitative analysis of the membranes was performed using a STORM820 optical scanner and Image Quant software (Molecular Dynamics). As shown in Fig. 2A, in MDCK cells the kinetics of replication of the human viruses PR8 and SYD were similar at 37 and 33°C. In contrast, replication of the MAL, FPV, and PIN viruses appeared delayed at 33°C compared to 37°C. The rate of accumulation of NP RNA was lower at 33°C than at 37°C at early times postinfection, and the overall levels of NP RNA synthesized in the infected cells at 33°C represented less than 50% of those produced at 37°C (Fig. 2A). Similar results were obtained in COS-1 cells (Fig. 2B). For the avian viruses MAL, FPV, and PIN but not for the human viruses PR8, SYD, and BAY, the accumulation of NP RNA was greatly delayed at 33°C. The overall levels of NP RNA synthesized at 21 or 24 h p.i. at 33°C represented only 66% (MAL), 62% (FPV), and 20% (PIN) of those produced at 37°C.
FIG. 2.
Kinetics of replication of the NP RNA segment from human and avian influenza viruses at 37 and 33°C. (A) MDCK cells were infected at an MOI of 20 PFU/cell with either PR8, SYD, MAL, FPV, or PIN virus or mock infected. (B) COS-1 cells were infected at an MOI of 10 PFU/cell with either PR8, SYD, BAY, MAL, FPV, or PIN virus or mock infected. Infected cells were incubated at either 37°C (solid symbols, solid lines) or 33°C (open symbols, dashed lines). The amount of NP vRNA was evaluated at 0, 1, 3, 5, 7, 9, 12, and 21 or 24 h p.i. The signal measured for a given virus at a given time was corrected by subtracting the signal measured at the same time point in mock-infected cells. For each virus, the corrected values were then expressed as a percentage of the value obtained at 21 or 24 h p.i. and at 37°C. The results are from one experiment representative of two independent experiments (A, PR8, MAL, and FPV) or of one experiment (A, SYD and PIN, and B).
Temperature dependence of polymerase complex of avian and human influenza viruses.
We investigated whether the lower efficiency of viral multiplication observed at 33°C for MAL and FPV was linked to a defect in transcription and/or replication of the viral RNA segments using a previously described plasmid-based genetic system that allows the in vivo reconstitution of functional ribonucleoproteins (RNPs) (23). As we described previously (17), subconfluent monolayers of COS-1 cells were transfected with four plasmids encoding the PB1, PB2, PA, and NP proteins (1, 1, 1, and 2 μg, respectively) derived from the PR8, MAL, or FPV strain or from the A/Victoria/3/75 (VIC) human strain, together with 1 μg of the pPolI-CAT-RT plasmid, which drives the expression of a virus-like chloramphenicol acetyltransferase (CAT) reporter RNA. Transfected cells were incubated for 48 h at 37 or 33°C. The efficiency of transcription-replication of the reconstituted RNPs was then evaluated by measuring the levels of CAT in cell extracts using the CAT enzyme-linked immunosorbent assay (ELISA) kit (Roche). As shown in Fig. 3, the CAT levels measured at 33°C in cells expressing the PB1, PB2, PA, and NP proteins derived from the human PR8 or VIC strain represented 162% ± 1% and 60% ± 15%, respectively, of the levels measured at 37°C. In contrast, the CAT levels measured at 33°C in cells expressing the FPV- or MAL-derived proteins represented 12% ± 0.2% and less than 2%, respectively, of the levels measured at 37°C. This indicated that the efficiencies with which the polymerase complexes ensured transcription-replication of the virus-like reporter RNA were similar at both temperatures, for the human viruses, while they were reduced by nearly 10- to 100-fold at 33°C for the avian viruses.
FIG. 3.
Transcription-replication of a virus-like CAT reporter RNA with homospecific polymerase complexes at 37 and 33°C in COS-1 cells. COS-1 cells were cotransfected in duplicate with pPolI-CAT-RT and four plasmids expressing the PB1, PB2, PA, and NP proteins derived from PR8, VIC, FPV, or MAL. Cell extracts were prepared and tested for CAT expression following 48 h of incubation at 37 or 33°C. For a given complex, the CAT levels measured at 33°C (grey bars) were compared to the CAT levels measured at 37°C (black bars). The results are expressed as concentration values and as the mean ± standard deviation (SD) of duplicate samples from one experiment representative of two independent experiments (A) or as percentages and as the mean ± SD of two independent experiments (B).
Identification of residue 627 of PB2 as a major determinant for cold sensitivity of the polymerase complex in COS-1 cells.
To determine whether cold sensitivity could be attributed to one or another of the proteins involved in the transcription and replication of the viral RNA, the PB1, PB2, PA, and NP proteins derived from FPV were each tested in combination with the three other proteins derived from PR8 at 37 and 33°C. In these experiments we used FPV- rather than MAL-derived proteins because of the very low levels of CAT measured in cells expressing heterospecific MAL/PR8-derived complexes (17). As shown in Fig. 4A, in cells expressing the FPV PB1 or NP protein in association with PR8 proteins, the CAT levels measured at 33°C (4,702 ± 482 and 1,081 ± 124 ng/ml, respectively) were slightly higher than those measured at 37°C. In contrast, in cells expressing the FPV PB2 or PA protein in association with PR8 proteins, the CAT levels measured at 33°C (2 ± 0.05 and 1,366 ± 20 ng/ml, respectively) represented about 6 and 60%, respectively, of those measured at 37°C. These observations suggested that the proteins responsible for the cold sensitivity of the transcription-replication complexes derived from avian viruses were mainly PB2 and to a lesser extent PA. Residue 627 of PB2 (Glu in PB2 proteins of avian origin, Lys in PB2 proteins of human origin) had previously been identified as an important determinant of the species specificity of influenza viruses (17, 29). In order to determine whether residue 627 of PB2 was also involved in cold sensitivity of the polymerase complex, we made use of plasmids encoding mutant FPV or MAL PB2 protein with a Lys instead of a Glu at position 627 (E627K-PB2 proteins). As shown in Fig. 4B, in cells expressing the FPV or MAL complexes with an E627K-PB2 protein, the CAT levels measured at 33°C (2,503 ± 174 and 1,421 ± 456 ng/ml, respectively) represented about 55 and 32%, respectively, of those measured at 37°C. In contrast, in cells expressing the FPV or MAL complexes with a wild-type PB2 protein, the CAT levels measured at 33°C (409 ± 52 and 24 ± 4 ng/ml, respectively) represented only 14 and 3%, respectively, of those measured at 37°C. This identified residue 627 of PB2 as a major determinant of cold sensitivity of the transcription-replication activity of the polymerase complex. Using Western blot analysis, we established that the steady-state levels of expression of PB2 proteins of human or avian origin were similar at 33 and 37°C (data not shown). This suggested that the function of PB2 proteins was impaired at the low temperature, rather than their expression or stability.
FIG. 4.
Transcription-replication of a virus-like CAT reporter RNA with heterospecific polymerase complexes at 37 and 33°C in COS-1 cells. COS-1 cells were cotransfected with pPolI-CAT-RT and four plasmids encoding the PB1, PB2, PA, and NP proteins derived from PR8, FPV, or MAL viruses, including, in the case of FPV and MAL, either the wild-type PB2 (A and B) or mutant E627K-PB2 (B), as indicated. Cell extracts were prepared and tested for CAT expression following 48 h of incubation at 37 or 33°C. For a given combination of PB1, PB2, PA, and NP, the CAT levels measured at 33°C were compared to the CAT levels measured at 37°C (100%, solid line). The results are expressed as percentages and as the mean ± SD of duplicate samples from one experiment representative of two independent experiments (A) or as the mean ± SD of two independent experiments (B).
Influence of residue 627 of PB2 on activity of polymerase complex in avian cells.
Because it has been shown to be a major determinant of host range in influenza viruses (29), we examined the influence of residue 627 of PB2 on the activity of polymerase complexes reconstituted in avian QT6 cells. A plasmid-based genetic system analogous to the one described above was used. Since the human polymerase I (PolI) promoter is functional exclusively in primate cells, the pPolI-CAT-RT plasmid could not be used in QT6 cells. Thus, a plasmid (pT7-CAT-RT) similar to plasmid pT7NSCAT-RT, described by Ortega et al. (19), was constructed so that sequences corresponding to the virus-like reporter RNA were driven by the T7 promoter and followed by a hepatitis delta virus ribozyme and the T7 terminator. Subconfluent monolayers of COS-1 or QT6 cells were infected for 1 h at an MOI of 5 PFU/cell with a recombinant vaccinia virus encoding the T7 RNA polymerase and then transfected with pT7-CAT-RT (12 μg) together with four plasmids encoding, under the control of the T7 promoter, the PB1, PB2, PA, and NP proteins of VIC or MAL (3, 3, 1, and 12 μg, respectively) according to Ortega et al. (19). Transfected cells were incubated for 18 h at 37 or 33°C. The efficiency of transcription-replication of the reconstituted RNPs was then evaluated as described previously. As shown in Fig. 5A, at 37°C, the CAT levels measured in COS-1 cells expressing the VIC-derived complex were about 3-fold lower than those measured when using the pPolI-CAT-RT plasmid, whereas the CAT levels in cells expressing the MAL-derived complex were undetectable. This reflected a lower sensitivity of the system with pT7-CAT-RT than with pPolI-CAT-RT, which might be related to a lower level of synthesis of the virus-like RNA from the T7 promoter than from the PolI promoter. In COS-1 cells expressing the MAL-derived complex, the CAT levels measured at 37°C were at least 10-fold higher with E627K-PB2 than with the wild-type PB2 protein (Fig. 5A), confirming that the nature of residue 627 of PB2 was a determinant for the efficiency of transcription-replication activity in COS-1 cells (Fig. 4B) (17). The levels of CAT measured in cells expressing either the VIC or MAL (including MAL-E627K-PB2) proteins and incubated at 33°C represented 78% ± 14% and 32% ± 2%, respectively, of those measured at 37°C (Fig. 5A), which was in agreement with the results obtained using pPolI-CAT-RT (Fig. 3 and 4B). In QT6 cells expressing the VIC proteins, the CAT levels measured at 37°C were lower than those measured in COS-1 cells (Fig. 5B), which may in part be related to a lower efficiency of the T7 expression system in QT6 cells, as documented using a reporter pT7 plasmid (data not shown). However, one cannot exclude that this may additionally reflect less efficient interactions of the VIC complex with avian cellular proteins. In QT6 cells expressing the VIC complex at 33°C, the CAT levels represented 49% ± 5% of those measured at 37°C (Fig. 5B), while in COS-1 cells expressing the VIC complex, the CAT levels were similar whether the cells were incubated at 33 or 37°C (Fig. 5A). Interestingly, the CAT levels measured at 37°C in QT6 cells expressing the MAL complex with either wild-type PB2 or E627K-PB2 were in the same range (Fig. 5B). The CAT levels measured at 33°C represented 17% ± 6% and 31% ± 10% of those measured at 37°C in cells expressing MAL PB2 and MAL E627K-PB2, respectively (Fig. 5B). These data indicated that in QT6 cells, contrary to what was observed in COS-1 cells, residue 627 of PB2 was not a determinant for the transcription-replication activity of the MAL complex at either 37 or 33°C. Additional experiments would be needed in order to determine whether the reduction of activity at 33°C observed for VIC and MAL complexes in QT6 cells is related to the cellular environment and/or to the viral proteins.
FIG. 5.
Transcription-replication of a virus-like CAT reporter RNA with homospecific polymerase complexes at 37 and 33°C in QT6 cells and COS-1 cells. COS-1 cells (A) or QT6 cells (B) were infected for 1 h at an MOI of 5 PFU/cell with a recombinant vaccinia virus expressing the T7 RNA polymerase and then transfected with pT7-CAT-RT and four plasmids encoding the PB1, PB2, PA, and NP proteins derived from VIC or MAL viruses, including in the latter case either the wild-type MAL PB2 or the mutant MAL E627K-PB2, as indicated. Cell extracts were prepared and tested for CAT expression following 18 h of incubation at 37 or 33°C. For a given complex, the CAT levels measured at 33°C (grey bars) were compared to the CAT levels measured at 37°C (black bars). The mean ± SD values are from duplicate samples from one experiment representative of two independent experiments.
Temperature dependence of transcription and/or replication of viral RNA of avian and human influenza viruses.
In order to determine whether the cold sensitivity linked to PB2 proteins of avian origin was related to transcription and/or replication, we analyzed the various RNA species involved in these processes. COS-1 cells were transfected with plasmids encoding either PR8- or MAL-derived core proteins together with the pPolI-CAT-RT plasmid as indicated above and incubated at either 33 or 37°C. Total RNA was prepared at 48 h posttransfection using the Trizol reagent (Gibco-BRL) and analyzed using an RNase protection assay. Total RNA (5 μg) was hybridized overnight at 42°C with an excess of a negative-sense CAT-specific 32P-radiolabeled riboprobe (105 cpm). Single-stranded RNAs were digested with a mixture of RNase A (2.5 U/ml) and RNase T1 (100 U/ml). Double-stranded RNAs were precipitated, diluted in loading buffer, and analyzed on a 6% polyacrylamide–8 M urea denaturating gel. The amounts of CAT-specific RNAs were determined using the STORM820 optical scanner and the Image Quant program (Molecular Dynamics). In cells expressing the PR8-derived proteins, the amounts of CAT-specific mRNA were similar whether the incubation temperature was 37 or 33°C, while the amount of cRNA at 33°C was twice the amount at 37°C (Fig. 6, lanes 4 and 10). This confirmed that neither the replication nor the transcription activity of the PR8-derived complex was impaired at 33°C. In cells expressing the MAL-derived proteins, the overall levels of cRNA and mRNA were dramatically reduced compared to PR8 at 37°C (Fig. 6, lane 5) and were not detectable at 33°C (Fig. 6, lane 11), consistent with the previous observation of reduced CAT levels (Fig. 3). These observations suggested that either the replication and transcription activities of the MAL complex together or the replication activity alone (as the reduction of mRNA levels might be just a consequence of the reduction of vRNA levels) was impaired in COS-1 cells. However, the very low levels of cRNA in cells expressing the MAL-derived proteins at 37°C as well as 33°C did not allow reliable quantification, and thus the question of the involvement of transcription and/or replication in the cold sensitivity of the avian-derived complex could not be addressed.
FIG. 6.
Synthesis of cRNA and mRNA at 37 and 33°C with avian and human virus-derived polymerase complexes. COS-1 cells were cotransfected with pPolI-CAT-RT and the PB1, PB2, PA, and NP expression plasmids derived from PR8 (lanes 4 and 10) or from MAL, including in the latter case either the wild-type MAL PB2 (lanes 5 and 11) or the mutant MAL E627K-PB2 (lanes 6 and 12) expression plasmid. Controls included COS-1 cells transfected with pPolI-CAT-RT without protein expression plasmids (lanes 1 and 7), PR8 protein expression plasmids without pPolI-CAT-RT (lanes 2 and 8), and MAL protein expression plasmids without pPolI-CAT-RT (lanes 3 and 9). Following 48 h of incubation at 37°C (lanes 1 to 6) or 33°C (lanes 7 to 12), total RNAs were extracted, and 5 μg was analyzed by RNase protection assay as described in the text. The image obtained by scanning the gel with a STORM820 optical scanner is shown. The length expected for the undigested riboprobe was 190 nucleotides (nt) (lane C+). No signal was expected when 5 μg of yeast tRNA was analyzed (lane C−). The expected lengths for the riboprobe fragments protected by cRNA or mRNA were 175 and 159 nt, respectively. MW, molecular size markers.
In cells expressing the MAL complex with the E627K-PB2 protein, the amounts of CAT-specific cRNA and mRNA were significantly higher than in cells expressing the wild-type MAL complex (Fig. 6, lanes 6 and 12) and were only slightly reduced (2-fold at 37°C, 4-fold at 33°C) compared to PR8, which was consistent with previous measurement of the CAT levels (Fig. 4B). These data suggested that either the replication and transcription activities of the MAL complex together or the replication activity alone was enhanced with E627K-PB2 compared with the wild-type PB2 protein. Interestingly, while the question of whether PB2 is required for replication of the viral genome appears to be controversial in the literature (18, 22), these observations favor an involvement of PB2 in replication.
There are numerous reports of PB2 being involved in sensitivity to high temperatures (37 to 40°C) or adaptation to low temperatures (25 to 33°C) of influenza A viruses. More than 20 different PB2 mutations responsible for such phenotypes have been identified using genomic sequencing (6, 9, 12, 14, 34) or reverse genetics methods (16, 21, 30). Here we showed that residue 627 of PB2 (a Lys in all human-derived proteins and a Glu in all avian-derived proteins) is a determinant of the natural cold sensitivity of avian-derived polymerase complexes when reconstituted in COS-1 cells. Remarkably, this same residue of PB2 has already been identified as a major determinant of the ability of reassortant influenza A viruses to replicate in mammalian (MDCK) cells (29) and of the activity of polymerase complexes reconstituted into mammalian (COS-1) cells (17). One hypothesis is that, in mammalian cells, the determination of host range and cold sensitivity could both rely on the same molecular interactions between PB2 and cellular proteins. These interactions would be strong at 37°C as well as at 33°C when PB2 is derived from a human virus, while they would be weaker (at 37°C and even more so at 33°C) when PB2 is derived from an avian virus. In contrast to what was observed in COS-1 cells, the nature of residue 627 of PB2 did not appear to be critical with respect to the activity of polymerase complexes reconstituted into QT6 cells. Hence, in avian cells, unlike in mammalian cells, the molecular interactions involving residue 627 of PB2 could be equally efficient whether it is a Lys or a Glu and whether the temperature is 37 or 33°C.
Interestingly, residue 627 of PB2 differed among the avian H5N1 viruses that were responsible for respiratory disease in humans in Hong Kong in 1997 (7). Using BALB/c mice as a mammalian model for evaluation of the pathogenesis of human H5N1 viruses, two groups of viruses of distinct virulence were clearly identified (7, 15). Among the viruses with high virulence, two had a Lys (typical of human strains), two had a Glu (typical of avian strains), and one was found to be a mixed population of viruses having a Lys and a Glu. Among the viruses with low virulence, four viruses out of four showed a Glu at residue 627. These findings suggested that the Glu627Lys substitution in PB2, in association with other molecular determinants, could have contributed to an increased replicative efficiency and pathogenicity of the H5N1 viruses in humans.
Accordingly, our results suggest that a reduced ability of the polymerase complex of avian viruses to ensure replication of the viral genome at 33°C, mostly determined by residue 627 of PB2, could contribute to their inability to grow efficiently in humans. The use of reassortant viruses or plasmid-based reverse genetics techniques will be needed to confirm this hypothesis and will probably help to elucidate the molecular mechanisms involved.
Acknowledgments
We are very grateful to J. Pavlovic (Institut für Medizinische Virologie, Zurich, Switzerland) for providing the expression plasmids for PR8 proteins, to P. Palese (Mount Sinai Medical Center, New York, N.Y.) for providing plasmid pPolI-CAT-RT, to A. Portela (Instituto Carlos III, Madrid, Spain) for providing the pGEM recombinant plasmids for VIC proteins, and to J. Ortin (Universidad Autonoma de Madrid, Madrid, Spain) for providing a polyclonal serum specific for PB2 protein. The technical assistance of Ida Rijks and Maryse Tardy-Panit for the production of influenza viruses is gratefully acknowledged. We thank Marco Vignuzzi for critical reading of the manuscript.
This work was supported in part by the Ministère de l'Education Nationale, de la Recherche et de la Technologie (EA 302).
REFERENCES
- 1.Beare A S, Webster R G. Replication of avian influenza viruses in humans. Arch Virol. 1991;119:37–42. doi: 10.1007/BF01314321. [DOI] [PubMed] [Google Scholar]
- 2.Breuning A, Scholtissek C. A reassortant between influenza A viruses (H7N2) synthesizing an enzymatically inactive neuraminidase at 40°C which is not incorporated into infectious particles. Virology. 1986;150:65–74. doi: 10.1016/0042-6822(86)90266-7. [DOI] [PubMed] [Google Scholar]
- 3.Chu C M, Tian S F, Ren G F, Zhang Y M, Zhang L X, Liu G Q. Occurrence of temperature-sensitive influenza A viruses in nature. J Virol. 1982;41:353–359. doi: 10.1128/jvi.41.2.353-359.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clements M L, Snyder M H, Buckler-White A J, Tierney E L, London W T, Murphy B R. Evaluation of avian-human reassortant influenza A/Washington/897/80 × A/Pintail/119/79 virus in monkeys and adult volunteers. J Clin Microbiol. 1986;24:47–51. doi: 10.1128/jcm.24.1.47-51.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Connor R J, Kawaoka Y, Webster R G, Paulson J C. Receptor specificity in human, avian, and equine H2 and H3 influenza virus isolates. Virology. 1994;205:17–23. doi: 10.1006/viro.1994.1615. [DOI] [PubMed] [Google Scholar]
- 6.Cox N J, Kitame F, Kendal A P, Maassab H F, Naeve C. Identification of sequence changes in the cold-adapted, live attenuated influenza vaccine strain. A/Ann Arbor/6/60 (H2N2) Virology. 1988;167:554–567. [PubMed] [Google Scholar]
- 7.Gao P, Watanabe S, Ito T, Goto H, Wells K, McGregor M, Cooley A J, Kawaoka Y. Biological heterogeneity, including systemic replication in mice, of H5N1 influenza A virus isolates from humans in Hong Kong. J Virol. 1999;73:3184–3189. doi: 10.1128/jvi.73.4.3184-3189.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Giesendorf B, Bosch F X, Wahn K, Rott R. Temperature sensitivity in maturation of mammalian influenza A viruses. Virus Res. 1984;1:655–667. [Google Scholar]
- 9.Herlocher M L, Clavo A C, Maassab H F. Sequence comparisons of A/AA/6/60 influenza viruses: mutations which may contribute to attenuation. Virus Res. 1996;42:11–25. doi: 10.1016/0168-1702(96)01292-0. [DOI] [PubMed] [Google Scholar]
- 10.Ito T, Suzuki Y, Suzuki T, Takada A, Horimoto T, Wells K, Kida H, Otsuki K, Kiso M, Ishida H, Kawaoka Y. Recognition of N-glycolylneuraminic acid linked to galactose by the α2,3 linkage is associated with intestinal replication of influenza A virus in ducks. J Virol. 2000;74:9300–9305. doi: 10.1128/jvi.74.19.9300-9305.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kendal A P, Galphin J C, Palmer E L. Replication of influenza virus at elevated temperatures: production of virus-like particles with reduced matrix protein content. Virology. 1977;76:186–195. doi: 10.1016/0042-6822(77)90295-1. [DOI] [PubMed] [Google Scholar]
- 12.Klimov A I, Cox N J, Wagner W V, Rocha E, Alexandrova G I, Kendal A P. Sequences changes in the live attenuated, cold-adapted variants of influenza A/Leningrad/134/57 (H2N2) virus. Virology. 1992;186:795–797. doi: 10.1016/0042-6822(92)90050-y. [DOI] [PubMed] [Google Scholar]
- 13.Kobasa D, Kodihalli S, Luo M, Castrucci M R, Donatelli I, Suzuki Y, Suzuki T, Kawaoka Y. Amino acid residues contributing to the substrate specificity of the influenza A virus neuraminidase. J Virol. 1999;73:6743–6751. doi: 10.1128/jvi.73.8.6743-6751.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lawson C M, Subbarao E K, Murphy B R. Nucleotide sequence changes in the polymerase basic protein 2 gene of temperature-sensitive mutants of influenza A virus. Virology. 1992;191:506–510. doi: 10.1016/0042-6822(92)90221-a. [DOI] [PubMed] [Google Scholar]
- 15.Lu X, Tumpey T M, Morken T, Zaki S R, Cox N J, Katz J M. A mouse model for the evaluation of pathogenesis and immunity to influenza A (H5N1) viruses isolated from humans. J Virol. 1999;73:5903–5911. doi: 10.1128/jvi.73.7.5903-5911.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murphy B R, Park E J, Gottlieb P, Subbarao K. An influenza A live attenuated reassortant virus possessing three temperature-sensitive mutations in the PB2 polymerase gene rapidly loses temperature sensitivity following replication in hamsters. Vaccine. 1997;15:1372–1378. doi: 10.1016/s0264-410x(97)00031-5. [DOI] [PubMed] [Google Scholar]
- 17.Naffakh N, Massin P, Escriou N, Crescenzo-Chaigne B, van der Werf S. Genetic analysis of the compatibility between polymerase proteins from human and avian strains of influenza A viruses. J Gen Virol. 2000;81:1283–1291. doi: 10.1099/0022-1317-81-5-1283. [DOI] [PubMed] [Google Scholar]
- 18.Nakagawa Y, Oda K, Nakada S. The PB1 subunit alone can catalyze cRNA synthesis, and the PA subunit in addition to the PB1 subunit is required for viral RNA synthesis in replication of the influenza virus genome. J Virol. 1996;70:6390–6394. doi: 10.1128/jvi.70.9.6390-6394.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ortega J, Martin-Benito J, Zurcher T, Valpuesta J M, Carrascosa J L, Ortin J. Ultrastructural and functional analyses of recombinant influenza virus ribonucleoproteins suggest dimerization of nucleoprotein during virus amplification. J Virol. 2000;74:156–163. doi: 10.1128/jvi.74.1.156-163.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Oxford J S, Corcoran T, Schild G C. Naturally occurring temperature-sensitive influenza A viruses of the H1N1 and H3N2 subtypes. J Gen Virol. 1980;48:383–389. doi: 10.1099/0022-1317-48-2-383. [DOI] [PubMed] [Google Scholar]
- 21.Parkin N T, Chiu P, Coelingh K. Genetically engineered live attenuated influenza A virus vaccine candidates. J Virol. 1997;71:2772–2778. doi: 10.1128/jvi.71.4.2772-2778.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Perales B, Ortin J. The influenza A virus PB2 polymerase subunit is required for the replication of viral RNA. J Virol. 1997;71:1381–1385. doi: 10.1128/jvi.71.2.1381-1385.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pleschka S, Jaskunas R, Engelhardt O G, Zurcher T, Palese P, Garcia-Sastre A. A plasmid-based reverse genetics system for influenza A virus. J Virol. 1996;70:4188–4192. doi: 10.1128/jvi.70.6.4188-4192.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers G N, D'Souza B L. Receptor binding properties of human and animal H1 influenza virus isolates. Virology. 1989;173:317–322. doi: 10.1016/0042-6822(89)90249-3. [DOI] [PubMed] [Google Scholar]
- 25.Rogers G N, Paulson J C. Receptor determinants of human and animal influenza virus isolates: differences in receptor specificity of the H3 hemagglutinin based on species of origin. Virology. 1983;127:361–373. doi: 10.1016/0042-6822(83)90150-2. [DOI] [PubMed] [Google Scholar]
- 26.Scholtissek C, Rohde W, von Hoyningen V, Rott R. On the origin of the human influenza subtype H2N2 and H3N2. Virology. 1978;87:13–20. doi: 10.1016/0042-6822(78)90153-8. [DOI] [PubMed] [Google Scholar]
- 27.Snyder M H, Buckler-White A J, London W T, Tierney E L, Murphy B R. The avian influenza virus nucleoprotein gene and a specific constellation of avian and human virus polymerase genes each specify attenuation of avian-human influenza A/Pintail/79 reassortant viruses for monkeys. J Virol. 1987;61:2857–2863. doi: 10.1128/jvi.61.9.2857-2863.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Stern H, Tippett K C. Primary isolation of influenza viruses at 33°C. Lancet. 1963;i:1301–1302. doi: 10.1016/s0140-6736(63)91992-5. [DOI] [PubMed] [Google Scholar]
- 29.Subbarao E K, London W, Murphy B R. A single amino acid in the PB2 gene of influenza A virus is a determinant of host range. J Virol. 1993;67:1761–1764. doi: 10.1128/jvi.67.4.1761-1764.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Subbarao E K, Park E J, Lawson C M, Chen A Y, Murphy B R. Sequential addition of temperature-sensitive missense mutations into the PB2 gene of influenza A transfectant viruses can effect an increase in temperature sensitivity and attenuation and permits the rational design of a genetically engineered live influenza A virus vaccine. J Virol. 1995;69:5969–5977. doi: 10.1128/jvi.69.10.5969-5977.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tian S F, Buckler-White A J, London W T, Reck L J, Chanock R M, Murphy B R. Nucleoprotein and membrane protein genes are associated with restriction of replication of influenza A/Mallard/NY/78 virus and its reassortants in squirrel monkey respiratory tract. J Virol. 1985;53:771–775. doi: 10.1128/jvi.53.3.771-775.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vines A, Wells K, Matrosovich M, Castrucci M R, Ito T, Kawaoka Y. The role of influenza A virus hemagglutinin residues 226 and 228 in receptor specificity and host-range restriction. J Virol. 1998;72:7626–7631. doi: 10.1128/jvi.72.9.7626-7631.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Webster R G, Bean W J., Jr . Evolution and ecology of Influenza viruses: interspecies transmission, P. 109–119. In: Nicholson K G, Webster R G, Hay A J, editors. Textbook of influenza. Oxford, England: Blackwell Science, Ltd.; 1998. [Google Scholar]
- 34.Yamanaka K, Ogasawara N, Ueda M, Yoshikawa H, Ishihama A, Nagata K. Characterisation of a temperature-sensitive mutant in the RNA polymerase PB2 subunit gene of influenza A/WSN/33 virus. Arch Virol. 1990;114:65–73. doi: 10.1007/BF01311012. [DOI] [PubMed] [Google Scholar]