ABSTRACT
Prior to 2017, the family Bunyaviridae included five genera of arthropod and rodent viruses with tri-segmented negative-sense RNA genomes related to the Bunyamwera virus. In 2017, the International Committee on Taxonomy of Viruses (ICTV) promoted the family to order Bunyavirales and subsequently greatly expanded its composition by adding multiple families for non-segmented to polysegmented viruses of animals, fungi, plants, and protists. The continued and accelerated discovery of bunyavirals highlighted that an order would not suffice to depict the evolutionary relationships of these viruses. Thus, in April 2024, the order was promoted to class Bunyaviricetes. This class currently includes two major orders, Elliovirales (Cruliviridae, Fimoviridae, Hantaviridae, Peribunyaviridae, Phasmaviridae, Tospoviridae, and Tulasviridae) and Hareavirales (Arenaviridae, Discoviridae, Konkoviridae, Leishbuviridae, Mypoviridae, Nairoviridae, Phenuiviridae, and Wupedeviridae), for hundreds of viruses, many of which are pathogenic for humans and other animals, plants, and fungi.
KEYWORDS: arenavirid, Arenaviridae, arenavirus, bunyaviral, Bunyavirales, bunyaviricete, Bunyaviricetes, bunyavirus, crulivirid, Cruliviridae, discovirid, Discoviridae, fimovirid, Fimoviridae, fimovirus, ellioviral, Elliovirales, Ellioviricetes, hantavirid, Hantaviridae, hantavirus, hareaviral, Hareavirales, ICTV, International Committee on Taxonomy of Viruses, konkovirid, Konkoviridae, leishbuvirid, Leishbuviridae, megaclassification, megataxonomy, mypovirid, Mypoviridae, nairovirid, Nairoviridae, nairovirus, negarnaviricot, Negarnaviricota, Orthornavirae, peribunyavirid, Peribunyaviridae, peribunyavirus, phasmavirid, Phasmaviridae, phenuivirid, Phenuiviridae, phenuivirus, polyploviricotine, Polyploviricotina, RdRp, Riboviria, ribovirian, RNA-dependent RNA polymerase, RNA-directed RNA polymerase, tospovirid, Tospoviridae, tospovirus, tulasvirid, Tulasviridae, virus classification, virus nomenclature, virus taxonomy, wupedevirid, Wupedeviridae
COMMENTARY
In 1973, a viral family, Bunyaviridae, including one genus, Bunyavirus, was proposed to be established for numerous insect viruses based on serological cross-reactivities and similarities and differences in physical properties and virion morphologies (1). The names of these taxa were derived from the “type” virus used for all analyses, Bunyamwera virus, which was first isolated in 1946 from Aedes mosquitoes in Semliki Forest of Uganda Protectorate (2). In 1975, both family and genus were accepted by the International Committee on Taxonomy of Viruses (ICTV) (3). At the time of the family/genus proposal (1), it was already obvious that there were numerous ‘bunyavirus-like viruses’, many of them pathogenic for humans and other animals, that should be family members but should not be assigned to genus Bunyavirus. In subsequent years, additional genera were established for these ‘bunyavirus-like viruses’ and newly discovered viruses. By 1981, Bunyavirus (by then all taxon names were italicized) was joined by genera Nairovirus, Phlebovirus, and Uukuvirus to accommodate insect and tick viruses serologically related to Nairobi sheep disease virus/Crimean-Congo hemorrhagic fever virus, sandfly fever Sicilian virus, and Uukuniemi virus, respectively (4, 5). The family experienced its next major expansion when the genus Hantavirus was included for a variety of human-pathogenic rodent-borne viruses (Hantaan virus and relatives) (6). Then, in 1990, the genus Tospovirus was added for pathogenic plant viruses transmitted by thrips (tomato spotted wilt virus and relatives) (7), and the genus Uukuvirus was absorbed in the genus Phlebovirus (8).
Promotion of family Bunyaviridae to order Bunyavirales
Since 1990, improved genome sequencing methodologies, followed by increasingly efficient metagenomic sequencing and improved tools to depict phylogenies, revealed a plethora of viruses closely related to the viruses classified in the family Bunyaviridae (9–14). Yet, many of these viruses, which were found in animals, fungi, plants, and protists, could not be assigned to the established genera and those that could represent divergent clades; i.e., the three-taxon (family, genus, and species) hierarchy did not suffice to represent the diversity of the family. Consequently, in 2017, the ICTV promoted the family to order Bunyavirales and the five genera to families (Bunyavirus→Peribunyaviridae; Hantavirus→Hantaviridae; Nairovirus→Nairoviridae; Phlebovirus→Phenuiviridae; and Tospovirus→Tospoviridae) (15). In subsequent years, the order underwent repeated and substantial revisions (through the addition of numerous novel families, genera, and species) (16–22), was included in a megataxonomic framework (realm Riboviria, kingdom Orthornavirae, phylum Negarnaviricota, subphylum Polyploviricotina, class Ellioviricetes) (23–25), and was updated via the consistent application of a Latinized binomial species nomenclature (21, 22).
Establishment of class Bunyaviricetes
The continued and accelerated discovery of bunyavirals (26–36) and their RNA-directed RNA polymerase (RdRp) phylogeny revealed that all of these viruses cluster in two readily distinguishable groups (37–39), i.e., clade 1 (Cruliviridae, Fimoviridae, Hantaviridae, Peribunyaviridae, Phasmaviridae, Tospoviridae, and Tulasviridae) and clade 2 (Arenaviridae, Discoviridae, Leishbuviridae, Mypoviridae, Nairoviridae, Phenuiviridae, and Wupedeviridae). In April 2024, the ICTV approved taxonomic proposal (TaxoProp) 2023.024M.Bunyaviricetes (40), resulting in class Ellioviricetes being renamed Bunyaviricetes (Fig. 1); order Bunyavirales being renamed Elliovirales and all clade 2 taxa removed from this order; and establishment of a new order, Hareavirales (from Basque harea, meaning sand; a reference to Arenaviridae and sandfly-borne viruses of Phenuiviridae), for all clade 2 taxa. In addition, a new family, Konkoviridae, was established and included in order Hareavirales [TaxoProps 2023.006M.Bunyavirales_1nfam_1ngen_1nsp and 2023.006MX.Bunyavirales_1nfam_1ngen_1nsp_Error_Correction (40)] (Fig. 2). The decision to change the name of class Ellioviricetes to Bunyaviricetes was made to ensure that the word stem “bunya” remains applicable to all viruses of the original five genera established in former family Bunyaviridae.
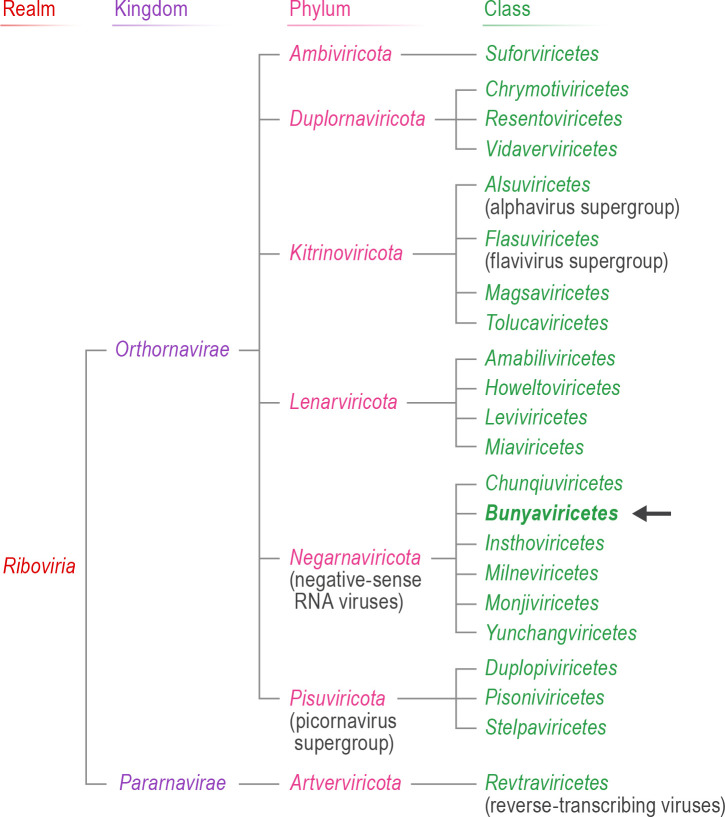
Fig 1.
Megataxonomic position of class Bunyaviricetes. Schematic of the megataxonomic organization of realm Riboviria down to the class rank, indicating the relative position of class Bunyaviricetes (bold, arrow). Adapted from Fig. 11A in reference (23) and Fig. 1A in reference (41).
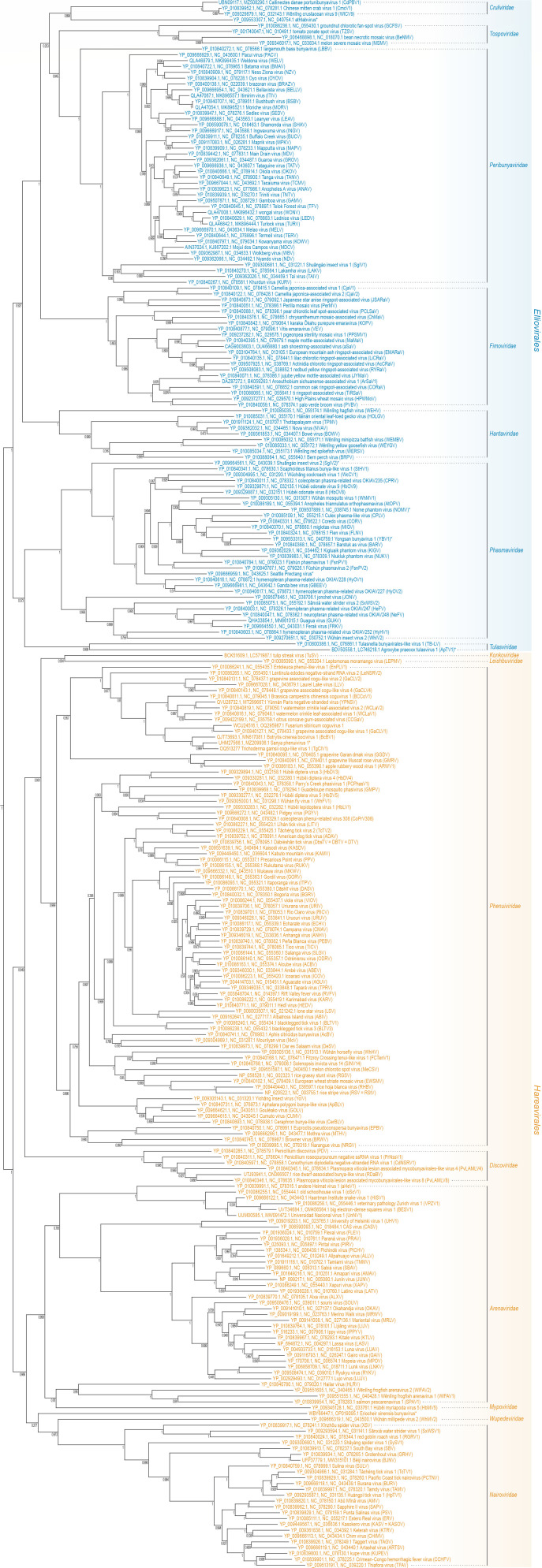
Fig 2.
Class Bunyaviricetes as of 2024. Phylogenetic tree using FastTree after MUSCLE alignment (with maxiters 100) of full-length GenBank large (L) protein (containing RNA-directed RNA polymerase domain) sequences. Highlighted are the now-established orders Elliovirales (blue) and Hareavirales (orange) and their families. Asterisks indicate unclassified viruses. Adapted from Fig. 1 in TaxoProp 2023.024M.Bunyaviricetes (40).
Bunyaviricete diversity
Classification of viruses by the ICTV is dependent on the submission of TaxoProps by the virology community and succeeds only by meeting specific rigorous standards, including the now-mandatory requirement of coding-complete virus genome sequences (42). Alas, many viruses that appear to be obvious bunyaviricetes remain unclassified because suitable proposals have not been prepared and submitted. The number of classified bunyaviricetes pales in comparison to the number of known related RdRp sequences in GenBank and sequence read archives; these RdRp sequences represent viruses but by themselves do not suffice for official classification. However, they can be used to establish phylogenetic trees, thereby providing a glimpse into the future of bunyaviricete classification. A cursory examination of such trees reveals that, for instance, fungal, invertebrate, plant, and stramenopile bunyaviricete diversity is likely enormous, but many of these viruses could likely be accommodated within the established two orders. Indeed, the Halophytophthora RNA virus 8 RdRp and its relatives form an obvious ellioviral family ‘Epsilonmycobunyaviridae’ (29), and the recently suggested ‘Rhizoctobunyaviridae’ (43) should be absorbed in ellioviral family Tulasviridae adjacent to a new tentative family ‘Sclerobunyaviridae’ (44). Order Hareavirales may be expanded in the future by tentative families ‘Gammaymycobunyaviridae’ (45) and ‘Zetamycobunyaviridae’ (46–49).
Many deposited RdRp sequences already portend challenges to the current classification. For instance, the Botrytis cinerea negative-stranded RNA virus 1 and Macrophomina phaseolina negative-stranded RNA virus 1 RdRps and their numerous immediate relatives (29, 50) together with a sister clade of lower plant virus RdRps (33) could represent tentatively named ellioviral families ‘Deltamycobunyaviridae’ and ‘Viridisbunyaviridae’ but may also be considered a third bunyaviricete order to include many families. However, coding-complete genome sequences are not available for most of these viruses and hence they may not be classified (42, 51). In fact, in many cases, not even nucleocapsid-encoding sequences have been uncovered in conjunction with the RdRp-encoding sequences. These nucleocapsid-encoding sequences may be too divergent from known sequences to be readily identified. Alternatively, some of these viruses may have acquired alternative structural protein genes and/or evolved capsidless lifestyles. In the latter case, deep phylogenetic analysis will have to be performed to determine whether these entities can still be considered (derived) viruses as per the current ICTV-adopted virus definition (51, 52) or whether they constitute non-viral mobile genetic elements related to viruses.
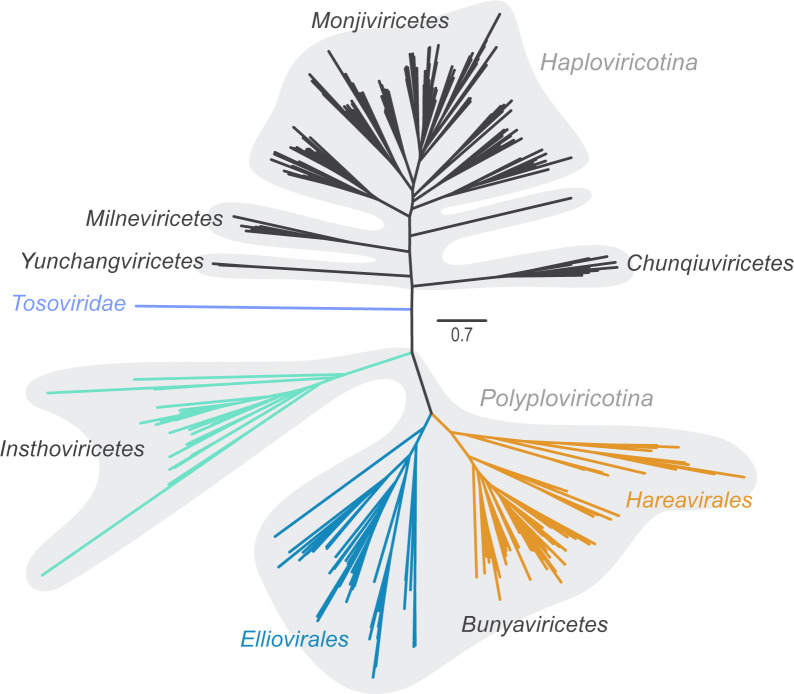
Other imminent challenges include determining the taxonomic position of the family Tosoviridae and class Insthoviricetes (families Amnoonviridae and Orthomyxoviridae). Family Tosoviridae was established in 2023 (22, 53) for a single virus, turtle fraservirus 1 (TFV1; species Fraservirus testudinis), isolated from diseased freshwater turtles (54). At the time of discovery, the TFV1 RdRp was not assignable to either of the two negarnaviricot subphyla (Haploviricotina or Polyploviricotina) and hence this family is currently “free-floating” in the phylum (22). In the same analysis, class Insthoviricetes clustered separately from all bunya-like viruses but within Polyploviricotina (54) (Fig. 3). Analyses performed with the now newly increased RdRp sequence data set indicate that both Tosoviridae and Insthoviricetes may have to be absorbed by Bunyaviricetes (Y. I. Wolf., unpublished data). However, whereas TFV1 at least resembles hareaviral arenavirids in the genomic organization, insthoviricetes appear to be highly dissimilar from classified bunyaviricetes. Additional phylogenetic and structural analyses of non-RdRp proteins encoded by bunyaviricetes and insthoviricetes may clarify this issue and should be encouraged (along with submission of TaxoProps) to assist in further improving polyploviricotine taxonomy.
Fig 3.
Negarnaviricot RdRp diversity. Unrooted maximum-likelihood tree (inferred with IQ-tree and the rtREV+F+R9 model) constructed with negarnaviricot RdRps available in 2021. The scale bar indicates average amino acid substitutions per site. Adapted from Fig. 10 in reference (54).
ACKNOWLEDGMENTS
The authors thank Anya Crane and Jiro Wada (Integrated Research Facility at Fort Detrick, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Fort Detrick, Frederick, MD, USA) for critically editing the text and creating figures, respectively.
This manuscript was prepared whilst K.E. held a National Research Council (NRC) Research Associateship Award at the Walter Reed Biosystematics Unit, through the Walter Reed Army Institute of Research, Silver Spring, MD, USA.
This work was supported in part through the Laulima Government Solutions, LLC, prime contract with the U.S. National Institute of Allergy and Infectious Diseases (NIAID) under Contract No. HHSN272201800013C. J.H.K. performed this work as an employee of Tunnell Government Services (TGS), a subcontractor of Laulima Government Solutions, LLC, under Contract No. HHSN272201800013C. The views and conclusions contained in this document are those of the authors and should not be interpreted as necessarily representing the official policies, either expressed or implied, of the U.S. Departments of the Army, Defense, Navy, or Health and Human Services or of the institutions and companies affiliated with the authors, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. Material contained within this publication has been reviewed by the Walter Reed Army Institute of Research. There is no objection to its presentation and/or publication. The findings and conclusions in this report are the opinions of the authors and do not necessarily represent the official position of the Centers for Disease Control and Prevention.
J.H.K., S.A., J.C.D.L.T., M.D., H.R.H., S.J., A.J.L., P.M., M.M., G.P., T.S., M.S., Y.-Z.Z., Y.I.W., and M.T. are members of the 2020–2023 International Committee on Taxonomy of Viruses (ICTV) Ellioviricetes Study Group.
The views expressed in this article do not necessarily reflect the views of the journal or of ASM.
Contributor Information
Jens H. Kuhn, Email: kuhnjens@mail.nih.gov.
Massimo Turina, Email: massimo.turina@ipsp.cnr.it.
Felicia Goodrum, The University of Arizona, Tucson, Arizona, USA.
ETHICS APPROVAL
This work did not include any work with humans or animals.
REFERENCES
- 1. Murphy FA, Harrison AK, Whitfield SG. 1973. Bunyaviridae: morphologic and morphogenetic similarities of Bunyamwera serologic supergroup viruses and several other arthropod-borne viruses. Intervirology 1:297–316. doi: 10.1159/000148858 [DOI] [PubMed] [Google Scholar]
- 2. Mahaffy AF, Haddow AJ, Smithburn KC. 1946. A neurotropic virus isolated from Aedes mosquitoes caught in the Semliki Forest. Am J Trop Med Hyg s1-26:189–208. doi: 10.4269/ajtmh.1946.s1-26.189 [DOI] [PubMed] [Google Scholar]
- 3. International Committee on Taxonomy of Viruses . 1975. Minutes of the third meeting of ICTV held in Madrid, 12 and 16 September 1975. Available from: https://ictv.global/ictv/proposals/Ratification_1975.pdf
- 4. Bishop DHL, Calisher CH, Casals J, Chumakov MP, Ya Gaidamovich S, Hannoun C, Lvov DK, Marshall ID, Oker-Blom N, Pettersson RF, Porterfield JS, Russell PK, Shope RE, Westaway EG. 1980. Bunyaviridae. Intervirology 14:125–143. doi: 10.1159/000149174 [DOI] [PubMed] [Google Scholar]
- 5. International Committee on Taxonomy of Viruses . 1981. Minutes of the 5th meeting of the ICTV. Strasbourg, 4 August 1981. Available from: https://ictv.global/ictv/proposals/Ratification_1981.pdf
- 6. International Committee on Taxonomy of Viruses . 1987. Minutes of the 7th meeting of the ICTV Edmonton, Canada, 12th August 1987. Available from: https://ictv.global/ictv/proposals/Ratification_1987.pdf
- 7. International Committee on Taxonomy of Viruses . 1987. Minutes of the 8th plenary meeting of the ICTV, Berlin, 29 August 1990. Available from: https://ictv.global/ictv/proposals/Ratification_1990.pdf
- 8. Francki RIB, Fauquet CM, Knudson DL, Brown F. 1991. Fifth report of the international committee on taxonomy of viruses. Archives of virology supplementum. In Classification and nomenclature of viruses. Vol. 2. Springer-Verlag, Vienna, Austria. [Google Scholar]
- 9. Marklewitz M, Handrick S, Grasse W, Kurth A, Lukashev A, Drosten C, Ellerbrok H, Leendertz FH, Pauli G, Junglen S. 2011. Gouleako virus isolated from West African mosquitoes constitutes a proposed novel genus in the family Bunyaviridae. J Virol 85:9227–9234. doi: 10.1128/JVI.00230-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Stenglein MD, Sanders C, Kistler AL, Ruby JG, Franco JY, Reavill DR, Dunker F, Derisi JL. 2012. Identification, characterization, and in vitro culture of highly divergent arenaviruses from boa constrictors and annulated tree boas: candidate etiological agents for snake inclusion body disease. mBio 3:e00180-12. doi: 10.1128/mBio.00180-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Marklewitz M, Zirkel F, Rwego IB, Heidemann H, Trippner P, Kurth A, Kallies R, Briese T, Lipkin WI, Drosten C, Gillespie TR, Junglen S. 2013. Discovery of a unique novel clade of mosquito-associated bunyaviruses. J Virol 87:12850–12865. doi: 10.1128/JVI.01862-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ballinger MJ, Bruenn JA, Hay J, Czechowski D, Taylor DJ. 2014. Discovery and evolution of bunyavirids in arctic phantom midges and ancient bunyavirid-like sequences in insect genomes. J Virol 88:8783–8794. doi: 10.1128/JVI.00531-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Marklewitz M, Zirkel F, Kurth A, Drosten C, Junglen S. 2015. Evolutionary and phenotypic analysis of live virus isolates suggests arthropod origin of a pathogenic RNA virus family. Proc Natl Acad Sci U S A 112:7536–7541. doi: 10.1073/pnas.1502036112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. doi: 10.7554/eLife.05378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Adams MJ, Lefkowitz EJ, King AMQ, Harrach B, Harrison RL, Knowles NJ, Kropinski AM, Krupovic M, Kuhn JH, Mushegian AR, Nibert M, Sabanadzovic S, Sanfaçon H, Siddell SG, Simmonds P, Varsani A, Zerbini FM, Gorbalenya AE, Davison AJ. 2017. Changes to taxonomy and the international code of virus classification and nomenclature ratified by the international committee on taxonomy of viruses (2017). Arch Virol 162:2505–2538. doi: 10.1007/s00705-017-3358-5 [DOI] [PubMed] [Google Scholar]
- 16. Maes P, Alkhovsky SV, Bào Y, Beer M, Birkhead M, Briese T, Buchmeier MJ, Calisher CH, Charrel RN, Choi IR, et al. 2018. Taxonomy of the family Arenaviridae and the order Bunyavirales: update 2018. Arch Virol 163:2295–2310. doi: 10.1007/s00705-018-3843-5 [DOI] [PubMed] [Google Scholar]
- 17. Abudurexiti A, Adkins S, Alioto D, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, et al. 2019. Taxonomy of the order Bunyavirales: update 2019. Arch Virol 164:1949–1965. doi: 10.1007/s00705-019-04253-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maes P, Adkins S, Alkhovsky SV, Avšič-Županc T, Ballinger MJ, Bente DA, Beer M, Bergeron É, Blair CD, Briese T, et al. 2019. Taxonomy of the order Bunyavirales: second update 2018. Arch Virol 164:927–941. doi: 10.1007/s00705-018-04127-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kuhn JH, Adkins S, Alioto D, Alkhovsky SV, Amarasinghe GK, Anthony SJ, Avšič-Županc T, Ayllón MA, Bahl J, Balkema-Buschmann A, et al. 2020. 2020 taxonomic update for phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch Virol 165:3023–3072. doi: 10.1007/s00705-020-04731-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Kuhn JH, Adkins S, Agwanda BR, Al Kubrusli R, Alkhovsky SV, Amarasinghe GK, Avšič-Županc T, Ayllón MA, Bahl J, Balkema-Buschmann A, et al. 2021. 2021 taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch Virol 166:3513–3566. doi: 10.1007/s00705-021-05143-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kuhn JH, Adkins S, Alkhovsky SV, Avšič-Županc T, Ayllón MA, Bahl J, Balkema-Buschmann A, Ballinger MJ, Bandte M, Beer M, et al. 2022. 2022 taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch Virol 167:2857–2906. doi: 10.1007/s00705-022-05546-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kuhn JH, Abe J, Adkins S, Alkhovsky SV, Avšič-Županc T, Ayllón MA, Bahl J, Balkema-Buschmann A, Ballinger MJ, Kumar Baranwal V, et al. 2023. Annual (2023) taxonomic update of RNA-directed RNA polymerase-encoding negative-sense RNA viruses (realm Riboviria: kingdom Orthornavirae: phylum Negarnaviricota). J Gen Virol 104:001864. doi: 10.1099/jgv.0.001864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH. 2020. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19. doi: 10.1128/MMBR.00061-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA virome. mBio 9:e02329-18. doi: 10.1128/mBio.02329-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. International Committee on Taxonomy of Viruses Executive Committee . 2020. The new scope of virus taxonomy: partitioning the virosphere into 15 hierarchical ranks. Nat Microbiol 5:668–674. doi: 10.1038/s41564-020-0709-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Grybchuk D, Akopyants NS, Kostygov AY, Konovalovas A, Lye L-F, Dobson DE, Zangger H, Fasel N, Butenko A, Frolov AO, Votýpka J, d’Avila-Levy CM, Kulich P, Moravcová J, Plevka P, Rogozin IB, Serva S, Lukeš J, Beverley SM, Yurchenko V. 2018. Viral discovery and diversity in trypanosomatid protozoa with a focus on relatives of the human parasite Leishmania. Proc Natl Acad Sci U S A 115:E506–E515. doi: 10.1073/pnas.1717806115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Käfer S, Paraskevopoulou S, Zirkel F, Wieseke N, Donath A, Petersen M, Jones TC, Liu S, Zhou X, Middendorf M, Junglen S, Misof B, Drosten C. 2019. Re-assessing the diversity of negative strand RNA viruses in insects. PLoS Pathog 15:e1008224. doi: 10.1371/journal.ppat.1008224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Wu H, Pang R, Cheng T, Xue L, Zeng H, Lei T, Chen M, Wu S, Ding Y, Zhang J, Shi M, Wu Q. 2020. Abundant and diverse RNA viruses in insects revealed by RNA-Seq analysis: ecological and evolutionary implications. mSystems 5:e00039-20. doi: 10.1128/mSystems.00039-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Botella L, Janoušek J, Maia C, Jung MH, Raco M, Jung T. 2020. Marine oomycetes of the genus Halophytophthora harbor viruses related to Bunyaviruses. Front Microbiol 11:1467. doi: 10.3389/fmicb.2020.01467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, Martino E, Girlanda M, Perotto S, Turina M. 2020. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol 6:veaa076. doi: 10.1093/ve/veaa076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Botella L, Jung T. 2021. Multiple viral infections detected in Phytophthora condilina by total and small RNA sequencing. Viruses 13:620. doi: 10.3390/v13040620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Zhang YY, Chen Y, Wei X, Cui J. 2022. Viromes in marine ecosystems reveal remarkable invertebrate RNA virus diversity. Sci China Life Sci 65:426–437. doi: 10.1007/s11427-020-1936-2 [DOI] [PubMed] [Google Scholar]
- 33. Mifsud JCO, Gallagher RV, Holmes EC, Geoghegan JL. 2022. Transcriptome mining expands knowledge of RNA viruses across the plant kingdom. J Virol 96:e0026022. doi: 10.1128/jvi.00260-22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Raco M, Vainio EJ, Sutela S, Eichmeier A, Hakalová E, Jung T, Botella L. 2022. High diversity of novel viruses in the tree pathogen Phytophthora castaneae revealed by high-throughput sequencing of total and small RNA. Front Microbiol 13:911474. doi: 10.3389/fmicb.2022.911474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Botella L, Jung MH, Rost M, Jung T. 2022. Natural populations from the Phytophthora palustris complex show a high diversity and abundance of ssRNA and dsRNA viruses. J Fungi (Basel) 8:1118. doi: 10.3390/jof8111118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Chen Z, Zhao H, Li Z, Huang M, Si N, Zhao H, Wei X, Sun B, Gao GF, Xu Z, Liu WJ. 2023. First discovery of phenuiviruses within diverse RNA viromes of asiatic toad (Bufo gargarizans) by metagenomics sequencing. Viruses 15:750. doi: 10.3390/v15030750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Herath V, Romay G, Urrutia CD, Verchot J. 2020. Family level phylogenies reveal relationships of plant viruses within the order bunyavirales. Viruses 12:1010. doi: 10.3390/v12091010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Olendraite I, Brown K, Firth AE. 2023. Identification of RNA virus-derived RdRp sequences in publicly available transcriptomic data sets. Mol Biol Evol 40:msad060. doi: 10.1093/molbev/msad060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Orba Y, Abu YE, Chambaro HM, Lundu T, Muleya W, Eshita Y, Qiu Y, Harima H, Kajihara M, Mori-Kajihara A, Matsuno K, Sasaki M, Hall WW, Hang’ombe BM, Sawa H. 2023. Expanding diversity of bunyaviruses identified in mosquitoes. Sci Rep 13:18165. doi: 10.1038/s41598-023-45443-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. International Committee on Taxonomy of Viruses Executive Committee . 2024. Approved proposals. Animal_dsRNA_and_ssRNA-_viruses. 2023. Available from: https://ictv.global/files/proposals/approved?fid=15260#block-teamplus-page-title
- 41. Koonin EV, Kuhn JH, Dolja VV, Krupovic M. 2024. Megataxonomy and global ecology of the virosphere. ISME J 18:1–16. doi: 10.1093/ismejo/wrad042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Simmonds P, Adams MJ, Benkő M, Breitbart M, Brister JR, Carstens EB, Davison AJ, Delwart E, Gorbalenya AE, Harrach B, et al. 2017. Consensus statement: virus taxonomy in the age of metagenomics. Nat Rev Microbiol 15:161–168. doi: 10.1038/nrmicro.2016.177 [DOI] [PubMed] [Google Scholar]
- 43. Li W, Sun H, Cao S, Zhang A, Zhang H, Shu Y, Chen H. 2023. Extreme diversity of mycoviruses present in single strains of Rhizoctonia cerealis, the pathogen of wheat sharp eyespot. Microbiol Spectr 11:e0052223. doi: 10.1128/spectrum.00522-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Jia J, Fu Y, Jiang D, Mu F, Cheng J, Lin Y, Li B, Marzano S-YL, Xie J. 2021. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol 7:veab032. doi: 10.1093/ve/veab032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Marzano S-Y, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL. 2016. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863. doi: 10.1128/JVI.00357-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Chiapello M, Rodríguez-Romero J, Ayllón MA, Turina M. 2020. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol 6:veaa058. doi: 10.1093/ve/veaa058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Picarelli M, Forgia M, Rivas EB, Nerva L, Chiapello M, Turina M, Colariccio A. 2019. Extreme diversity of mycoviruses present in isolates of Rhizoctonia solani AG2-2 LP from Zoysia japonica from Brazil. Front Cell Infect Microbiol 9:244. doi: 10.3389/fcimb.2019.00244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ruiz-Padilla A, Rodríguez-Romero J, Gómez-Cid I, Pacifico D, Ayllón MA. 2021. Novel mycoviruses discovered in the mycovirome of a necrotrophic fungus. mBio 12:e03705-20. doi: 10.1128/mBio.03705-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pagnoni S, Oufensou S, Balmas V, Bulgari D, Gobbi E, Forgia M, Migheli Q, Turina M. 2023. A collection of Trichoderma isolates from natural environments in Sardinia reveals a complex virome that includes negative-sense fungal viruses with unprecedented genome organizations. Virus Evol 9:vead042. doi: 10.1093/ve/vead042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Donaire L, Pagán I, Ayllón MA. 2016. Characterization of Botrytis cinerea negative-stranded RNA virus 1, a new mycovirus related to plant viruses, and a reconstruction of host pattern evolution in negative-sense ssRNA viruses. Virology 499:212–218. doi: 10.1016/j.virol.2016.09.017 [DOI] [PubMed] [Google Scholar]
- 51. International Committee on Taxonomy of Viruses Executive Committee . 2021. The international code of virus classification and nomenclature (ICVCN). Available from: https://ictv.global/about/code
- 52. Koonin EV, Dolja VV, Krupovic M, Kuhn JH. 2021. Viruses defined by the position of the virosphere within the replicator space. Microbiol Mol Biol Rev 85:e0019320. doi: 10.1128/MMBR.00193-20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Zerbini FM, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Dempsey DM, Dutilh BE, García ML, Hendrickson RC, Junglen S, Krupovic M, Kuhn JH, Lambert AJ, Łobocka M, Oksanen HM, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Doorslaer K, Vandamme A-M, Varsani A. 2023. Changes to virus taxonomy and the ICTV statutes ratified by the international committee on taxonomy of viruses (2023). Arch Virol 168:175. doi: 10.1007/s00705-023-05797-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Waltzek TB, Stacy BA, Ossiboff RJ, Stacy NI, Fraser WA, Yan A, Mohan S, Koonin EV, Wolf YI, Rodrigues TCS, Viadanna PHO, Subramaniam K, Popov VL, Guzman-Vargas V, Shender LA. 2022. A novel group of negative-sense RNA viruses associated with epizootics in managed and free-ranging freshwater turtles in Florida, USA. PLoS Pathog 18:e1010258. doi: 10.1371/journal.ppat.1010258 [DOI] [PMC free article] [PubMed] [Google Scholar]