Abstract
The success of structural studies performed on an individual target in small scale or on many targets in the systemwide scale of structural genomics depends critically on three parameters: (i) obtaining an expression system capable of producing large quantities of the macromolecule(s) of interest, (ii) purifying this material in soluble form, and (iii) obtaining diffraction-quality crystals suitable for x-ray analysis. The attrition rate caused by these constraints is often quite high. Here, we present a strategy that addresses each of these three parameters simultaneously. Using DNA shuffling to introduce functional sequence variability into a protein of interest, we screened crude lysate supernatants for soluble variants that retain enzymatic activity. Crystallization trials performed on three WT and eight shuffled enzymes revealed two variants that crystallized readily. One of these was used to determine the high-resolution structure of the enzyme by x-ray analysis. The sequence diversity introduced through shuffling efficiently samples crystal packing space by modifying the surface properties of the enzyme. The approach demonstrated here does not require guidance as to the type of mutation necessary for improvements in expression, solubility, or crystallization. The method is scaleable and can be applied in situations where a single protein is being studied or in high-throughput structural genomics programs. Furthermore, it should be readily applied to structural studies of soluble proteins, membrane proteins, and macromolecular complexes.
Keywords: directed evolution, protein crystallography
Structural biology has been transformed over the past decade in large part because of the efforts of structural genomics initiatives established around the world. Many technological advances have been made to support these programs, including automated methods for crystallization, x-ray data collection, and structure determination (1–3). By some estimates, however, structural genomics programs have succeeded in obtaining diffraction-quality crystals for <10% of the expressed proteins tested (4–6). These programs have run into many of the same technical difficulties that have traditionally presented barriers to structural analysis (7). Foremost among these are: (i) obtaining high-level expression, (ii) obtaining soluble and monodisperse purified protein, and (iii) obtaining diffraction-quality crystals for x-ray analysis.
A number of approaches have been used in an attempt to overcome these barriers (8). For proteins that are difficult to express in Escherichia coli, there are several options, including the use of different promoters, affinity tags (e.g., maltose-binding protein, 6×His), protein fragments (e.g., domain mapping), and naturally occurring homologs. In cases where the protein under study is of eukaryotic origin, bacterial expression is often unsuccessful. Directed evolution can be used to generate sequence variants that express in E. coli (9); more commonly, expression efforts turn to less convenient organisms such as yeast, baculovirus, and other eukaryotic expression systems.
Oftentimes proteins that express to high levels are insoluble because of aggregation or misfolding. In addition to the strategies described above, directed evolution methods offer a potential solution to this problem (10). For example, libraries of sequence variants fused to GFP may be screened for fluorescence to identify soluble, properly folded protein (11, 12).
Alternatively, systematic approaches to refolding may be used to identify conditions under which inclusion bodies are solubilized and refolded into functional protein. Replacing hydrophobic residues with polar and charged residues has been used successfully to improve protein solubility (13, 14). This approach benefits from a priori structural information to target sites that preserve the structure and activity of the protein.
Even when sufficient quantities of soluble protein are obtained, crystallization is not guaranteed. The availability of commercial screens combined with recent advances in automation make it possible to screen tens of thousands of variables (pH, ionic strength, precipitant, etc.) in a single day (1). In cases where the protein of interest fails to yield diffraction-quality crystals from this type of screening, the intrinsic properties of the protein may be varied (15). Mapping stable fragments and/or domains by partial proteolysis or high-resolution deuterium exchange MS (16) can be used to define constructs that are more likely to crystallize. Altering surface properties by chemical modification, e.g., reductive methylation and carboxymethylation, is an approach that has been used successfully (17). Similarly, site-directed mutagenesis may be used to rationally change the surface properties of a protein (18–21). Finally, a time-tested approach is to screen naturally occurring homologs of the protein under study (22).
Work in our laboratory is focused on developing a glyphosate resistance strategy in plants. Previously we described the discovery of three closely related Bacillus licheniformis enzymes possessing weak N-acetylation activity against glyphosate (23). These genes provided the starting point in a directed evolution program designed to obtain an enzyme with a high enough level of glyphosate N-acetyltransferase (GAT) activity to confer resistance to glyphosate when expressed in transgenic plants. GAT variants with a broad range of activities were identified after multiple rounds of DNA shuffling (24, 25) and high-throughput screening. These include enzymes with nearly 10,000-fold improvements in kcat/KM for glyphosate, relative to the starting genes (23).
Here, we report on the application of DNA shuffling and screening to the crystallization of GAT. In this instance, the main obstacle to structural analysis was obtaining diffraction-quality crystals. Whereas crystallization trials on the WT enzymes failed to yield diffraction-quality crystals, trials carried out on eight shuffled GAT variants identified two that yield well formed crystals. One of these variants was used to solve the structure of GAT. Analysis of the structure with respect to the crystal packing reveals that the sequence variation introduced through shuffling efficiently samples crystal packing space by modifying the surface properties of the enzyme. The approach described here is broadly applicable. In its most general form, it combines directed evolution with high-throughput crystallization to simultaneously address each of the main barriers to macromolecular structure determination.
Materials and Methods
DNA Shuffling and High-Throughput Screening. Multigene DNA shuffling of three WT gat genes was carried out as described (23). For screening, GAT variants in E. coli were grown in 100 μl of LB containing 50 μg/ml carbenicillin and 1 mM isopropyl β-d-thiogalactoside in V-bottom 96-well polystyrene plates. Cells were harvested by centrifugation and resuspended in 20 μl of B-PER lysis solution (Pierce). After shaking the cells for 10 min, 80 μl of assay buffer (25 mM Hepes, pH 6.8/10% ethylene glycol) was added, and debris was removed by centrifugation. Reactions were performed by adding 290 μl of assay buffer containing 150 μM AcCoA and 0.3 mM ammonium glyphosate to 10 μl of crude lysate supernatant in a 96-well UV assay plate (Corning). Initial rates of hydrolysis of the thioester bond of AcCoA were monitored at 235 nm with a SPECTRAmax PLUS384 (Molecular Devices).
Enzyme Purification. GAT was purified from E. coli cell lysates by using CoA-agarose affinity chromatography and gel filtration. Typically, a 100-ml culture of E. coli with a gat gene in expression vector pQE80 was grown overnight in LB containing 50 μg/ml carbenicillin. This culture was used to inoculate 1 liter of LB containing 50 μg/ml carbenicillin. After 1 h, isopropyl β-d-thiogalactoside was added to 1 mM, and the culture was grown for an additional 6 h. Cells were harvested by centrifugation and lysed in 25 mM Hepes (pH 7.2), 100 mM KCl, 10% methanol (termed HKM), 1 mM DTT, 2 mg/ml of protease inhibitor mixture (Sigma-Aldrich), and 1 mg/ml of chicken egg lysozyme. After 30 min at room temperature, the lysate was sonicated, centrifuged to remove cell debris, and desalted by passage through Sephadex G25 equilibrated with HKM. The extract was passed through a bed of CoA-Agarose, washed with several bed volumes of HKM, and eluted in 1.5 bed volumes of HKM containing 1 mM AcCoA. Further purification was obtained by passage through a Superdex 75 column equilibrated with HKM. Selenomethionine-containing GAT protein was prepared following the method of Van Duyne et al. (26) and purified as described above.
Crystallization. Purified WT and variant GAT enzymes used in crystallization trials were buffer-exchanged and concentrated to 5–10 mg/ml in 5 mM Hepes, pH 7.5. The proteins were incubated in each of three conditions: (i) enzyme alone, (ii) 1 mM AcCoA, and (iii)1mMCoA + 20 mM glyphosate. Enzymes were screened by vapor diffusion in sitting drops by using the Hampton Research (Riverside, CA) HT Crystal Screen kit. Optimization was carried out by using grid screens in hanging drop format. Crystals of selenomethionine-containing GAT were grown in the presence of 1 mM CoA from a crystallization solution containing 100 mM NaOAc (pH 4.6), 250 mM ammonium sulfate (AS), and 25% polyethylene glycol (PEG) 4000.
Structure Determination. Data collection and refinement statistics are summarized in Table 1. For data collection, the crystals were mounted in nylon loops, after first transferring them to cryoprotectant comprising 100 mM NaOAc (pH 4.6), 250 mM AS, 25% PEG 4000 supplemented with 20% glycerol, and 2 mM CoA. Diffraction experiments were carried out at Advanced Light Source Beamline 5.0.2 (Berkeley, CA) with an Area Detector Systems (Poway, CA) Quantum-4 charge-coupled device detector. The crystals were maintained at 100 K by using an Oxford cryostream. The selenomethionine crystals diffracted to beyond 1.63 Å (see Fig. 2), and a single-wavelength anomalous data set was collected at 0.9794 Å (peak wavelength). The data were processed by using mosflm (27) and programs from the CCP4 suite (28), yielding unit cell parameters of a = 69.3, b = 49.4, c = 46.5 Å, α = γ = 90, and β = 103.4° for space group C2. Initial phases were based on the selenium anomalous signal, and single-wavelength anomalous dispersion electron density maps solvent-flattened by solomon (29) were of excellent quality, allowing us to build >98% of the protein structure. Refinement was carried out in refmac5 (30).
Table 1. Data collection and refinement statistics.
| Data collection | |
| Resolution range, Å | 50.0-1.63 |
| Total/unique reflections | 55,614/18,939 |
| Completeness, % | 97.4 (94.1) |
| 〈I/sigI〉 | 20.2 (8.0) |
| Rsym, % | 7.2 (16.2) |
| Refinement | |
| Rcryst, % | 17.9 |
| Rfree, % | 21.3 |
| No. of protein atoms | 1,192 |
| No. of water atoms | 161 |
| Average B factor, Å2 | 18.6 |
| rms deviation bond lengths, Å | 0.010 |
| rms deviation bond angles, ° | 1.4 |
Values in parentheses are for the high-resolution shell.
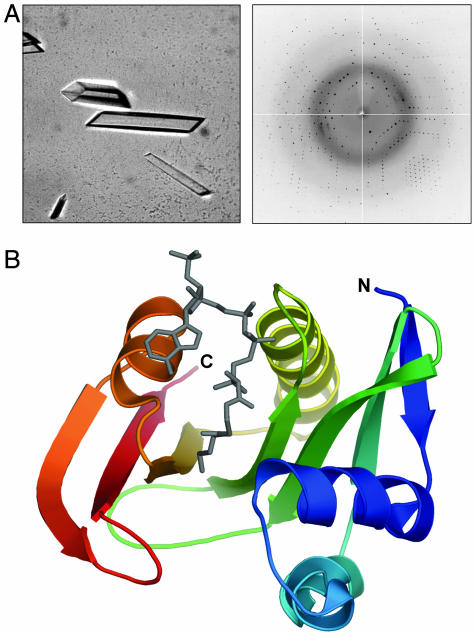
Fig. 2.
Crystallographic analysis of GAT. (A) Crystals of selenomethionine-containing variant 5 GAT after optimization diffract beyond 1.6-Å resolution. (B) Ribbon representation of GAT bound to oxidized CoA (gray), colored from the N terminus (blue) to C terminus (red). All structure figures were prepared with pymol (33).
Results
We initiated structural studies on three WT GAT enzymes to learn more about the mechanism of glyphosate N-acetylation. Sparse matrix screens, set up in the presence or absence of ligands (CoA, AcCoA, and glyphosate), were carried out on each of the three WT enzymes. These initial crystallization trials typically yielded thin plates or clusters of needles under a variety of different PEG and AS conditions. However, extensive attempts to optimize the crystallization conditions by varying the parameters of salt, pH, precipitant, temperature, etc., failed to produce crystals suitable for structure determination. Thus, we pursued an alternative strategy, varying the protein sequence to obtain diffraction-quality crystals.
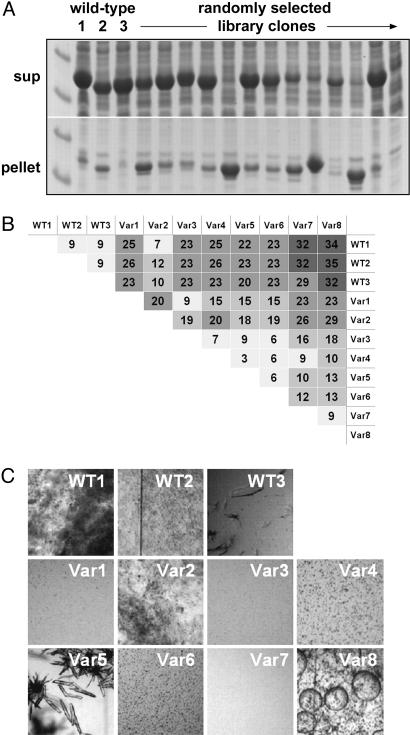
To test the idea that variations in protein sequence could facilitate the growth of diffraction-quality crystals, we identified shuffled gat genes that encode enzymes satisfying the following criteria: (i) contain a diverse set of amino acid changes, (ii) are expressed to high levels in E. coli, (iii) are soluble, and (iv) are functional. In a set of experiments carried out in parallel with this work, we used multiple rounds of DNA shuffling coupled with high-throughput screening to identify GAT variants with improved activity against glyphosate (23). In addition to containing variants with diverse functional properties, libraries of shuffled gat genes encode enzymes that express to different levels and with varying solubility. To illustrate this, we analyzed randomly selected clones from a library generated by a single round of shuffling of three WT gat genes (sharing between 93% and 95% homology at the nucleotide level). After expressing the library in E. coli, we examined the supernatant and pellet fractions of crude lysate supernatants for the presence of GAT. In this case, ≈85% of the shuffled clones expressed to high levels, while ≈60% produced predominantly soluble enzyme (Fig. 1A). For structural characterization we chose eight shuffled GAT variants containing between 7- and 35-aa changes relative to the WT enzymes (Fig. 1B). These eight were purified and subjected to crystallization trials in the presence and absence of ligands, as described above.
Fig. 1.
Sequence variation affects enzyme properties. (A) SDS/PAGE comparing the expression and solubility of randomly selected, shuffled GAT variants with those of the three WT enzymes. (B) Pairwise amino acid identity expressed as the number of differences for 11 enzymes that were selected for crystallization trials. Shading is from fewest (light gray) to most (dark gray) differences. (C) Side-by-side comparison of the behavior of the three WT and eight variant enzymes in the HT crystal screen condition yielding the best unoptimized crystals for variant 5.
Many of the variant GAT enzymes yielded the same needles and plates seen with the WT enzymes. However, as expected for a set of enzymes with a range of sequence homology, clear differences in crystallization behavior were also observed between them (Fig. 1C). In particular, variant 5 gave rise to well formed crystals in a number of PEG/AS and AS-only conditions directly from the crystal screen. Variant 2 gave rise to a well formed crystal from a single, no-salt PEG condition (data not shown).
Based on the robustness with which it crystallized from a variety of related conditions in the initial screen and the fact that it possesses a higher level of enzymatic activity, we focused our optimization efforts on variant 5. By varying PEG and AS concentrations we rapidly established conditions from which large, well formed rods could be grown overnight at room temperature (Fig. 2A). Because GAT possesses low sequence homology to proteins of known structure, selenomethionine-labeled protein was prepared and crystallized under similar conditions to the native enzyme. A high-resolution data set was collected to 1.6 Å (see Fig. 2 A and Table 1), and the structure was solved by a combination of single-wavelength anomalous dispersion and solvent flattening. GAT belongs to the GCN5-related superfamily of N-acetyltransferases (Fig. 2B). To understand how sequence variations introduced through shuffling alter the crystallization properties of the enzyme, we analyzed the location and type of these variations with respect to the observed crystal packing.
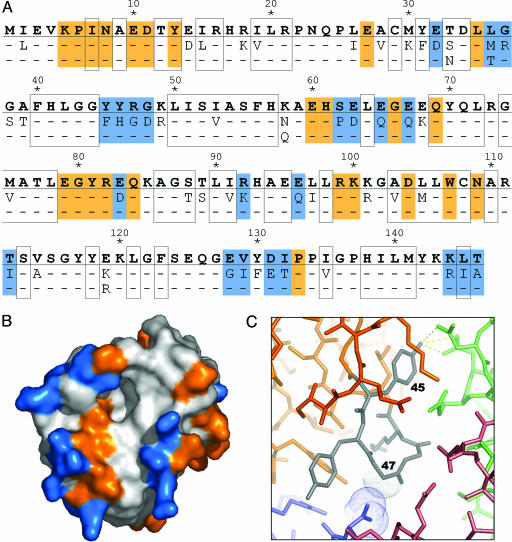
There are a total of 50 positions in the GAT sequence where the amino acid identity differs in at least one of the 11 enzymes tested (Fig. 3A). Of these 50 “variable” positions, more than three-quarters (76%) are located on the enzyme surface where they are positioned to affect the crystal packing (Fig. 3B). Of the 46 GAT residues mediating protein–protein interactions in this crystal form, 22 are located at variable positions (Fig. 3A, blue), whereas the remaining 24 are located at fixed positions (Fig. 3A, orange). Thus, residues located at variable positions mediate almost half (48%) of the protein–protein contacts observed in the crystal.
Fig. 3.
Sequence variation efficiently samples crystal-packing space by modifying the surface properties of the enzyme. (A) The amino acid sequence corresponding to one of the WT gat genes (GenBank accession no. AX543338) is shown in bold, and the 50 positions that vary among at least one of the 11 enzymes tested for crystallization are indicated below. Buried residues are boxed. Residues mediating protein–protein contacts in the crystal are shaded orange (constant positions) or blue (variable positions). (B) Solvent-accessible surface representation of GAT, colored as above. (C) Close-up showing a surface loop (residues 45–49; gray) of one GAT molecule (orange) that mediates protein–protein contacts with three symmetry-related GAT molecules (colored blue, red, and green) in the crystal. The sequence variation at position 45 (Phe vs. Tyr) and 47 (Arg vs. Gly) illustrates two different types of changes that contribute to the formation of a well ordered crystal lattice (see text for details).
Many factors (hydrogen bonding, salt bridges, hydrophobic surface area, rigidity of loops, and side-chain entropy) influence the ability of a protein to crystallize in a particular packing arrangement. Given the complexity of the GAT system described here, it is difficult to systematically analyze the relationship between sequence, structure, and the ability to crystallize. For example, an amino acid substitution that is favorable in one sequence context may be offset by variations at other positions. Similarly, an amino acid substitution that is favorable in one crystal form may negatively affect packing in a different one. Clearly, the effect of a particular sequence variation on crystallization is context-dependent.
Nevertheless, examination of the types of sequence variation present in this set of shuffled GAT variants provides insight into the altered crystallization properties of the enzymes. In particular, almost one-third (32%) of the variable positions encode nonconservative sequence diversity. These changes, including hydrophobic-to-polar/charged variation (10 instances), positive-to-negative charge variation (2 instances), and large steric changes (4 instances), are almost exclusively found at the surface of the enzyme and are representative of the types of sequence variation that are known to influence crystallization (15, 20).
Two such examples are found within a surface loop (located at position 45–49) that mediates an extensive crystal contact between four symmetry-related GAT monomers (Fig. 3C). This loop is comprised entirely of variable position residues (Fig. 3A), and its sequence is unique to variant 5. The entire contact region is highly ordered; residues within the loop possess an average B factor of 14.6 Å2 versus an average B factor of 18.6 Å2 for all protein atoms in the crystal. One of the residues within the loop, located at position 45, is either a Phe or a Tyr. In variant 5, this position is a Tyr, and it makes a series of well ordered hydrogen bonds to two different side chains from a symmetry-related GAT monomer (Fig. 3C, dashed yellow lines). A second residue in the loop, located at position 47, is either an Arg or a Gly. In variant 5, this position is a Gly, and it packs tightly against a second symmetry-related GAT monomer (Fig. 3C; close packing indicated by van der Waals dots). By providing additional hydrogen bonding potential (F45Y) and reducing steric hindrance and conformational entropy (R47G), these sequence variations contribute to an energetically favorable protein–protein interface within the crystal.
Discussion
The initial goal of this work was to solve the structure of GAT, a newly discovered enzyme that detoxifies the herbicide glyphosate through acetyl-CoA-dependent N-acetylation (23). Extensive crystallization trials with three WT homologs of GAT failed to produce crystals that could be used to solve the structure. As an alternative strategy, we tested the idea that by modifying the sequence of an enzyme (in this case, GAT), without prior structural information, we could obtain variants that form well ordered crystals.
Multigene DNA shuffling was used in combination with a high-throughput functional screen to isolate GAT variants with improved enzymatic properties that express to high levels in soluble form (23, 31). Eight of these, with between 7 and 35 amino acid changes relative to the WT proteins, were screened in crystallization experiments. As predicted, the enzymes behaved differently in crystallization trials, and one of the variants yielded crystals that were used to solve the high-resolution structure of GAT. Analysis of the crystal packing with respect to the locations of the sequence variation indicates that most of the variable positions are found on the surface of the enzyme. In addition, many of these surface changes are nonconservative in nature. Thus, the diversity introduced by DNA shuffling and screened for by activity is effectively sampling crystallization space by providing types of sequence variation (nonconservative) in regions of the enzyme (especially the surface) that directly affect the crystallization properties of the shuffled variants.
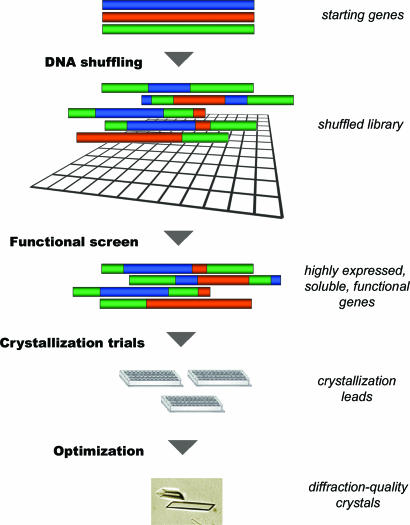
The combination of diversity generation and functional screening can be used to simultaneously address issues of protein expression, solubility, and crystallization. This strategy is outlined in Fig. 4. The initial step involves creating new sequence diversity by DNA shuffling. Once a high-quality library is created, it is interrogated to identify individual proteins with the desired set of properties, namely: (i) high levels of protein expression, (ii) production of soluble protein, and (iii) retention of biological activity. With a robust activity assay such as the one described here, these properties are simultaneously screened for with a high rate of success. Once suitable variants are obtained, a few or many of these are expressed, purified, and tested in standard crystallization screens.
Fig. 4.
Generalization of the crystallization strategy. See text for details.
As a means of generating sequence diversity for crystallization of difficult proteins, DNA shuffling has a number of key benefits. First, because multigene DNA shuffling results in libraries that are enriched in conservative changes at sites that are critical to maintaining structure and/or biological activity and nonconservative changes at the protein surface, it allows for surface modification without prior structural information. Second, DNA shuffling may be performed in a variety of different formats (24, 25, 31, 32), allowing the user to control the number and type of mutations introduced into the gene of interest. Third, the process of constructing shuffled libraries lends itself to high-throughput approaches, allowing for the rapid preparation of variants for crystal trials.
The question of how much diversity to introduce is a complex function of a number of parameters including: (i) the nature of the property one is trying to improve, (ii) the sensitivity of the protein structure to amino acid substitutions, and (iii) the ability to measure activity by using a functional assay. In general, we expect that screening relatively few variants from small libraries created from highly homologous genes will be sufficient to determine whether surface modification can overcome the limiting parameter(s) for a given system. For example, shuffling two genes encoding proteins of 200 aa and sharing 90% identity results in a library of ≈106 variants, with diversity spread over 20 variable positions. After screening a subset (100s-1,000s) for expression, solubility and activity, the most promising variants (10s-100s) may be subjected to crystallization trials. In the event that only distantly related sequence homologs are available (e.g., <60% nucleotide homology), then oliogonucleotide-based synthetic shuffling strategies (32) may be used to introduce a subset of diversity into the gene of interest.
The general nature of this approach is limited in certain instances. For example, in the case of ≈50% of proteins in sequenced genomes that have no known function, screening for activity is not possible. However, by designing libraries with modest levels of sequence diversity and screening for high-level expression of soluble protein, it should be possible to identify useful variants that retain WT properties. In the case of proteins containing large unstructured regions or multiple domains, the use of deuterium-exchange MS to identify crystallizable fragments is likely to be a more robust approach (16). And in the case of drug design, where there is a need to determine the structure of a specific protein sequence, the relevance of a variant structure must be carefully evaluated. However, so long as the crystallized variant retains its activity, the structural details in the biologically relevant regions of the protein should be largely intact. In this sense shuffled variants are similar to any other “naturally occurring” homolog. The variant structures can be used to guide additional experiments, and in some cases, may be used to facilitate the structure determination of any other sequence variant, including the WT sequence.
The strategy outlined here establishes an alternative approach to the structural characterization of macromolecules that are difficult to analyze with traditional methods. One of the important features of this strategy is that it does not require guidance as to the type of mutation that is needed for improvements in expression, solubility, or crystallization. The method is scaleable and can be incorporated into existing high-throughput approaches to macromolecular crystallization. Furthermore, it should be readily applied to a variety of systems, including membrane proteins, as well as protein–protein and protein–nucleic acid complexes.
Acknowledgments
We thank Michael Lassner and John Bedbrook for their support of this project, the University of California, San Francisco, Macromolecular Structure Group and the Advanced Light Source Beamline 5.0.2 staff for support, and Doug Freymann and Andrew Shiau for constructive comments on the manuscript.
Author contributions: R.J.K., D.L.S., and L.A.C. designed research; R.J.K., D.L.S., R.G., and L.A.C. performed research; R.J.K. analyzed data; and R.J.K. wrote the paper.
Abbreviations: GAT, glyphosate N-acetyltransferase; HKM, Hepes/KCl/methanol; AS, ammonium sulfate; PEG, polyethylene glycol.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2bsw).
References
- 1.Stevens, R. C. (2000) Curr. Opin. Struct. Biol. 10, 558–563. [DOI] [PubMed] [Google Scholar]
- 2.Abola, E., Kuhn, P., Earnest, T. & Stevens, R. C. (2000) Nat. Struct. Biol. 7, Suppl., 973–977. [DOI] [PubMed] [Google Scholar]
- 3.Lamzin, V. S. & Perrakis, A. (2000) Nat. Struct. Biol. 7, Suppl., 978–981. [DOI] [PubMed] [Google Scholar]
- 4.Claverie, J. M., Monchois, V., Audic, S., Poirot, O. & Abergel, C. (2002) Comb. Chem. High Throughput Screen 5, 511–522. [DOI] [PubMed] [Google Scholar]
- 5.Ding, H. T., Ren, H., Chen, Q., Fang, G., Li, L. F., Li, R., Wang, Z., Jia, X. Y., Liang, Y. H., Hu, M. H., et al. (2002) Acta Crystallogr. D 58, 2102–2108. [DOI] [PubMed] [Google Scholar]
- 6.Sulzenbacher, G., Gruez, A., Roig-Zamboni, V., Spinelli, S., Valencia, C., Pagot, F., Vincentelli, R., Bignon, C., Salomoni, A., Grisel, S., et al. (2002) Acta Crystallogr. D 58, 2109–2115. [DOI] [PubMed] [Google Scholar]
- 7.Service, R. F. (2002) Science 298, 948–950. [DOI] [PubMed] [Google Scholar]
- 8.Derewenda, Z. S. (2004) Methods 34, 354–363. [DOI] [PubMed] [Google Scholar]
- 9.Aharoni, A., Gaidukov, L., Yagur, S., Toker, L., Silman, I. & Tawfik, D. S. (2004) Proc. Natl. Acad. Sci. USA 101, 482–487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Waldo, G. S. (2003) Curr. Opin. Chem. Biol. 7, 33–38. [DOI] [PubMed] [Google Scholar]
- 11.Yang, J. K., Park, M. S., Waldo, G. S. & Suh, S. W. (2003) Proc. Natl. Acad. Sci. USA 100, 455–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pedelacq, J. D., Piltch, E., Liong, E. C., Berendzen, J., Kim, C. Y., Rho, B. S., Park, M. S., Terwilliger, T. C. & Waldo, G. S. (2002) Nat. Biotechnol. 20, 927–932. [DOI] [PubMed] [Google Scholar]
- 13.Dyda, F., Hickman, A. B., Jenkins, T. M., Engelman, A., Craigie, R. & Davies, D. R. (1994) Science 266, 1981–1986. [DOI] [PubMed] [Google Scholar]
- 14.Jenkins, T. M., Hickman, A. B., Dyda, F., Ghirlando, R., Davies, D. R. & Craigie, R. (1995) Proc. Natl. Acad. Sci. USA 92, 6057–6061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dale, G. E., Oefner, C. & D'Arcy, A. (2003) J. Struct. Biol. 142, 88–97. [DOI] [PubMed] [Google Scholar]
- 16.Pantazatos, D., Kim, J. S., Klock, H. E., Stevens, R. C., Wilson, I. A., Lesley, S. A. & Woods, V. L., Jr. (2004) Proc. Natl. Acad. Sci. USA 101, 751–756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rypniewski, W. R., Holden, H. M. & Rayment, I. (1993) Biochemistry 32, 9851–9858. [DOI] [PubMed] [Google Scholar]
- 18.Longenecker, K. L., Garrard, S. M., Sheffield, P. J. & Derewenda, Z. S. (2001) Acta Crystallogr. D 57, 679–688. [DOI] [PubMed] [Google Scholar]
- 19.Mateja, A., Devedjiev, Y., Krowarsch, D., Longenecker, K., Dauter, Z., Otlewski, J. & Derewenda, Z. S. (2002) Acta Crystallogr. D 58, 1983–1991. [DOI] [PubMed] [Google Scholar]
- 20.Derewenda, Z. S. (2004) Structure (London) 12, 529–535. [DOI] [PubMed] [Google Scholar]
- 21.McElroy, H. E., Sisson, G. W., Schoettlin, W. E., Aust, R. M. & Villafranca, J. E. (1992) J. Crystal Growth 122, 265–272. [Google Scholar]
- 22.Campbell, J. W., Duee, E., Hodgson, G., Mercer, W. D., Stammers, D. K., Wendell, P. L., Muirhead, H. & Watson, H. C. (1972) Cold Spring Harbor Symp. Quant. Biol. 36, 165–170. [DOI] [PubMed] [Google Scholar]
- 23.Castle, L. A., Siehl, D. L., Gorton, R., Patten, P. A., Chen, Y. H., Bertain, S., Cho, H. J., Duck, N., Wong, J., Liu, D. & Lassner, M. W. (2004) Science 304, 1151–1154. [DOI] [PubMed] [Google Scholar]
- 24.Stemmer, W. P. (1994) Proc. Natl. Acad. Sci. USA 91, 10747–10751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stemmer, W. P. (1994) Nature 370, 389–391. [DOI] [PubMed] [Google Scholar]
- 26.Van Duyne, G. D., Standaert, R. F., Karplus, P. A., Schreiber, S. L. & Clardy, J. (1993) J. Mol. Biol. 229, 105–124. [DOI] [PubMed] [Google Scholar]
- 27.Leslie, A. G. W. (1992) Joint CCP4 ESF-EAMCB Newsl. Protein Crystallogr., no. 26.
- 28.Collaborative Computational Project No. 4 (1994) Acta Crystallogr. D 50, 760–763.15299374 [Google Scholar]
- 29.Abrahams, J. P. & Leslie, A. G. W. (1996) Acta Crystallogr. D 52, 30–42. [DOI] [PubMed] [Google Scholar]
- 30.Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997) Acta Crystallogr. D 53, 240–255. [DOI] [PubMed] [Google Scholar]
- 31.Crameri, A., Raillard, S. A., Bermudez, E. & Stemmer, W. P. (1998) Nature 391, 288–291. [DOI] [PubMed] [Google Scholar]
- 32.Ness, J. E., Kim, S., Gottman, A., Pak, R., Krebber, A., Borchert, T. V., Govindarajan, S., Mundorff, E. C. & Minshull, J. (2002) Nat. Biotechnol. 20, 1251–1255. [DOI] [PubMed] [Google Scholar]
- 33.DeLano, W. L. (2002) The pymol Molecular Graphics System (DeLano Scientific, San Carlos, CA).