Abstract
Fluorescent in situ hybridization (FISH) enables the visualization of the position and abundance of nucleic acid molecules in fixed cell and tissue samples. Many FISH-based methods employ sets of synthetic, computationally designed DNA oligonucleotide (oligo) FISH probes, including massively multiplexed imaging spatial transcriptomics and spatial genomics technologies. Oligo probes can either be designed de novo or accessed from an existing database of pre-discovered probe sequences. This chapter describes the use of PaintSHOP, a user-friendly, web-based platform for the design of sets of oligo-based FISH probes. PaintSHOP hosts large collections of pre-discovered probes from many model organisms and also provides collections of functional sequences such as primers and readout domains and interactive tools to add these functional sequences to selected probes. Detailed examples are provided for three common experimental scenarios.
Keywords: Cytogenetics, Oligonucleotide, FISH, Chromosomes, Chromatin, Spatial Genomics, Spatial Transcriptomics
1. Introduction
Fluorescent in situ hybridization (FISH) techniques enable the detection and visualization of cellular RNA and DNA molecules using fluorescently labeled probes. Since the introduction of in situ hybridization in 1969 (1) and the development of FISH (2) shortly thereafter, many advancements have been made to improve the detection efficiency and sensitivity of FISH methods. Advanced FISH techniques leverage DNA synthesis technologies to use synthetic oligonucleotide (oligo) libraries as a source for probe material (3). These synthetic oligo-FISH probes offer several advantages: they can be custom-designed against any sequenced genome, engineered to have specific thermodynamic properties, programmed to include additional sequences or barcodes, and are easily manufactured. Massively parallel array synthesis techniques have given rise to complex oligo libraries that enable FISH technologies to target single-copy genomic regions. These complex oligo libraries have resulted in massively multiplexed DNA-FISH experiments and the growth of spatial genomic and spatial transcriptomic approaches.
Experimental advances in the use of oligo FISH probes has led to the development of computational tools to support the design of oligo probes and probe sets such as OligoArray (4), PROBER (5), Chorus (6), mathFISH (7), OligoMiner (8), iFISH (9), ProbeDealer (10), Chorus2 (11), and PaintSHOP (12). The Beliveau lab developed the paint server and homology optimization pipeline, known as PaintSHOP, to facilitate all aspects of the oligo probe design process, making it accessible to those with limited bioinformatic knowledge. In this chapter, we describe PaintSHOP, a freely accessible, user-friendly platform for the interactive design of oligo-based FISH experiments at the transcriptome- and genome-scale. We provide an overview of the probe mining process, discuss critical considerations for probe design, and introduce the tools used within PaintSHOP. Furthermore, we provide detailed protocols for generating ready-to-order probe sets with user-specifications against any target in the genome or transcriptome, covering three common use cases: (1) single-target RNA-FISH probe design, (2) multi-target RNA-FISH probe design, and (3) multi-target DNA-FISH probe set design.
2. Materials
The PaintSHOP web application (https://paintshop.io) facilitates all aspects of the probe set design process. The web application can be used to: (1) retrieve probes covering a RNA or DNA target(s) of interest; (2) confirm the correct strand orientation of the probe sequences; (3) consider trimming or unifying the number of probes per target in a probe set; and (4) append primers and barcode sequences for a given experimental design. Here, we describe three simple design schemes that rely on the preconstructed genome-scale probe collections hosted at the PaintSHOP resources page. The probe set collection and genomic or transcriptomic targets are the minimum information required to design an RNA or DNA oligo probe.
3. Methods
3.1. Probe Mining and Important Considerations
There are two phases in the computational design of oligo-FISH probes. First, a probe discovery algorithm processes the sequence of a genome assembly to identify short windows of genomic sequence with suitable thermodynamic and sequence properties to serve as FISH probes. Probe discovery tools commonly used can include those such as OligoArray, OligoPicker—which was used to create the iFISH probe database, the command-line OligoMiner tool, or its web-based version OligoMinerApp (13). These design algorithms typically consider key aspects that influence the effect of oligo-probes: oligonucleotide length, %G+C content, melting temperature, tendency to form secondary structures and dimers, homopolymeic stretches, and the presence of annotated repetitive sequences. Most importantly, oligo probes should be specific to their intended target, so probes are screened for specificity to predict if they will have specific binding. Second, the ‘primary’ probes are processed into probe sets that can be ordered from a vendor. PaintSHOP can support both design phases; however, this volume will focus on the second phase of the probe design process that is the most common use of PaintSHOP.
3.2. PaintSHOP
PaintSHOP is a computational framework for the interactive design of transcriptome- and genome-scale oligo-based FISH experiments. It consists of two main components: (1) a large-scale collection of primary oligo probe sequences targeting the genomes and transcriptomes of 16 experimental organisms, and (2) an interactive web application for the automated creation of ready to order probe sets. The PaintSHOP webpage can be found at paintshop.io shown in Fig. 1. The website includes links to the PaintSHOP resources page, the PaintSHOP pipeline page, and a web application for interactive probe design. The PaintSHOP Resources page contains a collection of downloadable genome-wide DNA and RNA FISH probe sets and appending sequence files. The PaintSHOP Pipeline link leads to a GitHub page with the Snakemake workflow used to generate all PaintSHOP probe collections. The blue button labeled ‘Launch PaintSHOP’ opens the interactive web application used for designing oligo-FISH probe libraries for any experiment. The interactive web application is shown in Fig 2. Here, we will focus on the user-friendly web application and describe the general workflow, probe collections, input files and specifications, expected outputs, and include instructions for three common use cases.
Fig 1.
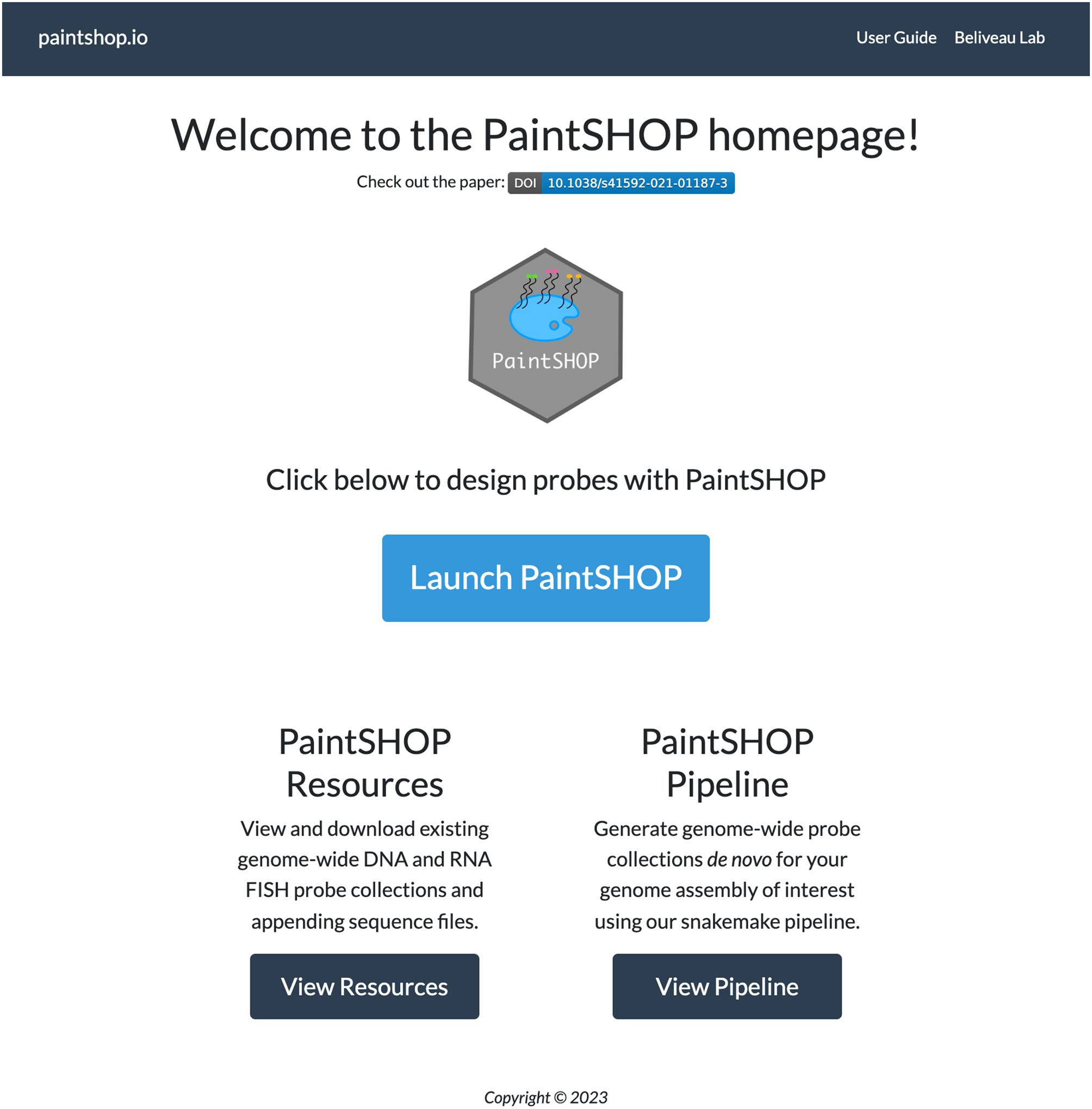
PaintSHOP homepage.
Fig 2.
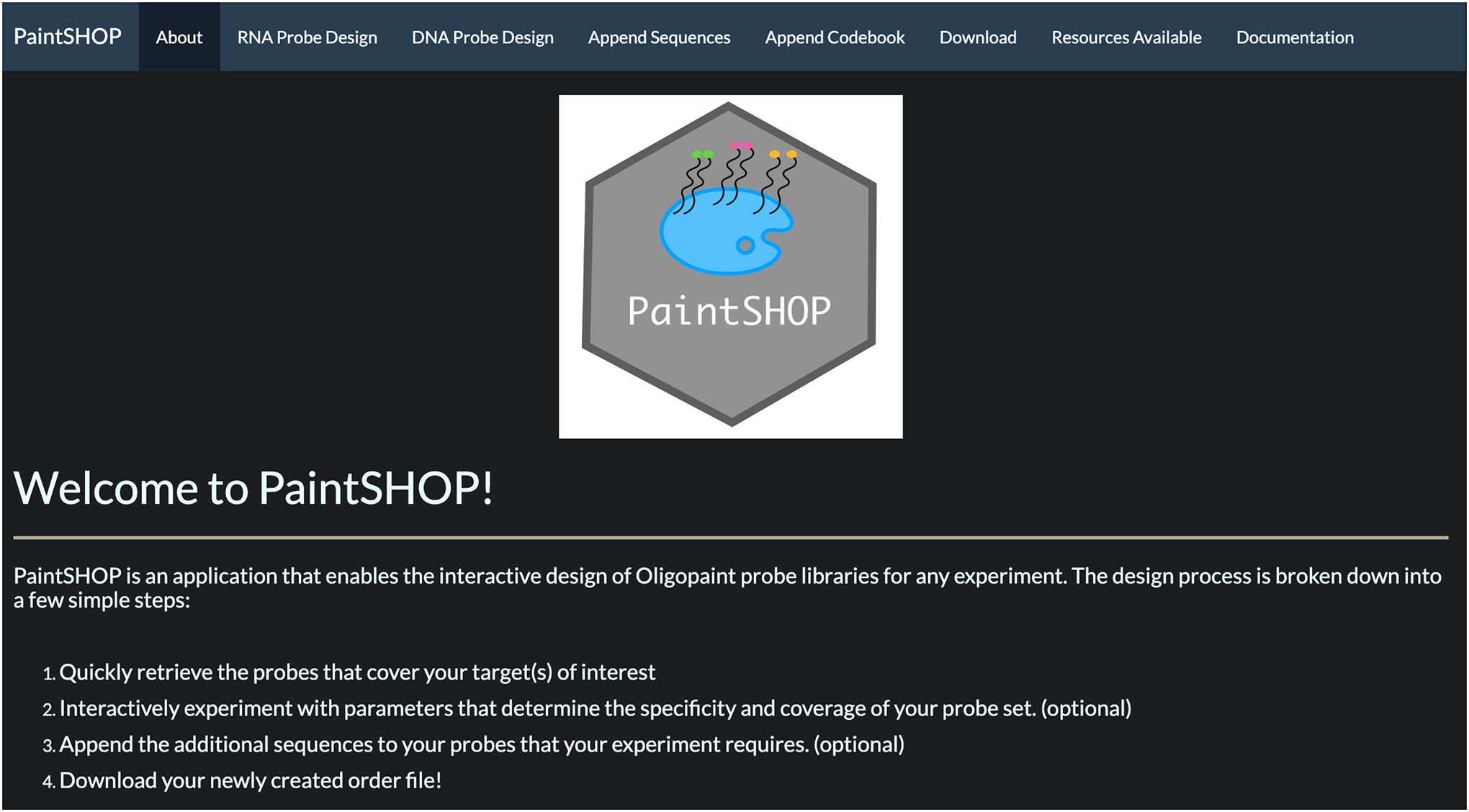
PaintSHOP interactive web application.
The PaintSHOP design process can be broken down into four main steps: (1) retrieve probes that cover your target(s) of interest, (2) interactively experiment with parameters that determine the specificity and coverage of your probe set (optional), (3) append additional sequences to your probes, and (4) download your designed library. The application workflow is shown in Fig 3. The PaintSHOP web application can be used to retrieve probes covering a single DNA or RNA target of interest or to design probe sets for multi-target experiments. The probes are retrieved from preconstructed genome-scale probe collections that can be found at the PaintSHOP resources page.
Fig 3.
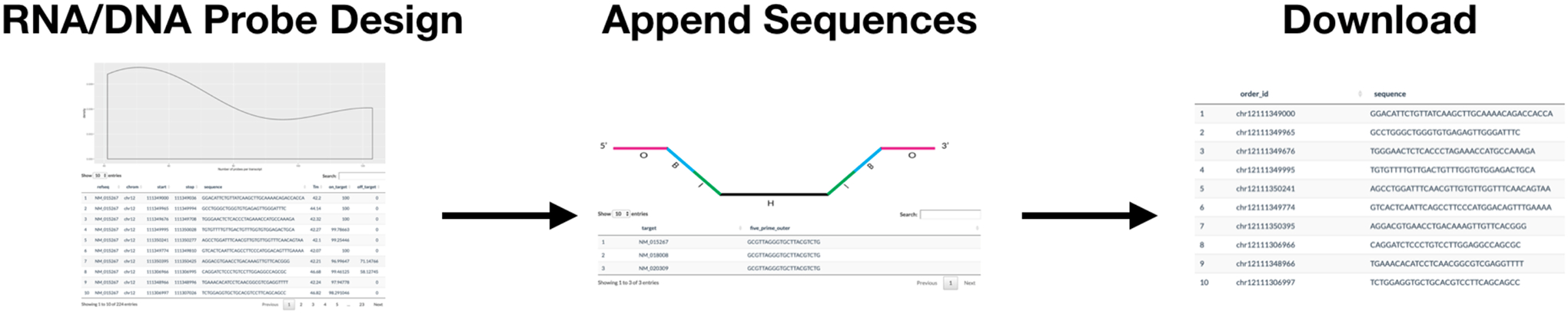
PaintSHOP web application probe design workflow.
3.3. PaintSHOP Resources
PaintSHOP includes large-scale collections of genome-wide probe sets for 16 experimental organisms. These probe sets were created using OligoMiner, a probe discovery utility for the genome-scale design of oligo probes for FISH. OligoMiner considers probe length, thermodynamic behavior, and sequence specificity to mine an input sequence for probes. It accepts a genomic sequence in FASTA format and identifies regions suitable to serve as FISH probes. They are then screened for specificity to exclude probes likely to hybridize to off-target, or unintended, sites. The probes are passed to an alignment program to find regions with high sequence similarity to the candidate probes to assess off-target binding potential. Probes predicted to have high off-target binding potential are filtered out which results in the final primary probe set which can be accessed by the PaintSHOP Resources page.
3.4. PaintSHOP FISH probe sets
There are multiple PaintSHOP RNA probe sets that have different characteristics. All probe sets found on the RNA Probe Design tab have mapped the probe coordinates to reference annotations from RefSeq for quick retrieval of probes for annotations. The RNA FISH probe sets found in the PaintSHOP Resources have both isoform-resolved and isoform-flattened versions. The isoform-flattened set of RefSeq annotations for each genome assembly prioritizes shared transcript intervals between isoforms to maximize the chance of detecting isoforms for a given gene. The isoform-flattened probe sets should be queried with a gene symbol of interest (ex. MYC). The isoform-resolved sets should be used to target a particular transcript or isoform using the RefSeq IDs (ex. RefSeq annotation for the MYC transcript variant 1: NM_002467.6). To retrieve probes target regions outsides these annotations, please use the DNA Probe Design tab. The probe collection types are described below.
-
newBalance (RNA)
These are probe sets for 20 genomes with a probe length window of 30–37 nt and a Tm window of 41–72 degrees. These parameters were selected to optimize probe coverage and hybridization. More information on this probe set can be found at the PaintSHOP publication.
-
OligoMiner (RNA)
The ‘balance’ probe sets generated by OligoMiner for the hg38 and hg19 reference genomes. The probes have a length window of 35–41 nt and a Tm window of 42–47 degrees. More information on this probe set can be found at the OligoMiner publication.
-
2012 Oligopaints (RNA)
The original genome-scale probe set from the Beliveau et al. 2012 PNAS publication (14). The probes have a length of 32 bases and an approximate Tm window of 34–42 degrees. More information on this probe set can be found at the 2012 Oligopaints publication.
-
iFISH4U (RNA)
The full hg19 40-mer probe set from iFISH4U. These ‘spotting probe’ sets tile along individual chromosomes. They can also be used to retrieve probe sets targeting individual regions from a collection of prediscovered probes which are the ones used in this probe set collection.
3.4. RNA FISH single target probe design
This protocol describes the process of designing probes for a single RNA target and creating an order file of the probe set. RefSeq annotations can be used to retrieve probes for a target or list of targets. Annotations can be set manually into the “Enter RefSeq IDs manually” text box or uploaded as a file with one annotation per line. For this demo, we will use the hg38 newBalance (isoform-flattened) probe set to design probes targeting the mRNA transcript of MYC, a known proto-oncogene. We will also append short DNA sequences to the 3’ends of the selected probes that can subsequently be extended in vitro by the primer exchange reaction (PER) (15) to add long tails of concatemeric single-stranded DNA. These long concatemers in turn can recruit many copies of a complementary oligo that is fluorescently labeled and amplify the resulting FISH signal via a method termed ‘signal amplification via exchange reaction’ (SABER) (16). We will then export all information required to make an order of the designed probe set.
Go to the PaintSHOP website found at paintshop.io and open the interactive web application using the “Launch PaintSHOP” button.
Select the RNA Probe Design tab at the top left of the screen seen in Fig 2.
In the probe set dropdown, select the hg38 newBalance (isoform-flattened) option.
Enter the gene symbol MYC either manually in the text box or as a file with the RefSeq annotation on a single line. (see Note 1)
Once the annotation has been entered, indicate which input type was used (Manual or File).
Press the Submit button to generate and retrieve the probe set. The table returned describes the probe set that covers the MYC gene. The plot above will be empty.
To append sequences to the generated probe set, select the Append Sequences tab at the top of the screen seen in Fig 2.
Select the RNA Probe Design for the Design Scheme.
Select SABER for the 3’ Appending Scheme option and use the default settings shown in Fig 4.
Press the Append button at the bottom of the page. The table returned describes the target of interest and information about the sequences that were appended.
Select the Download tab.
Select Appended Sequences under the Used Appending section and RNA Probe Design for the Design Scheme section.
Under the Choose a file to download option, choose Order File. The expected output is provided as an accompanying Supplementary File 1.
Select the Download button to save a text file with the designed probe set locally. The expected output is provided as an accompanying Supplementary File 2.
Additionally, download the Appending File for downstream bridge oligo design. The expected output is provided as an accompanying Supplementary File 3.
Fig 4.
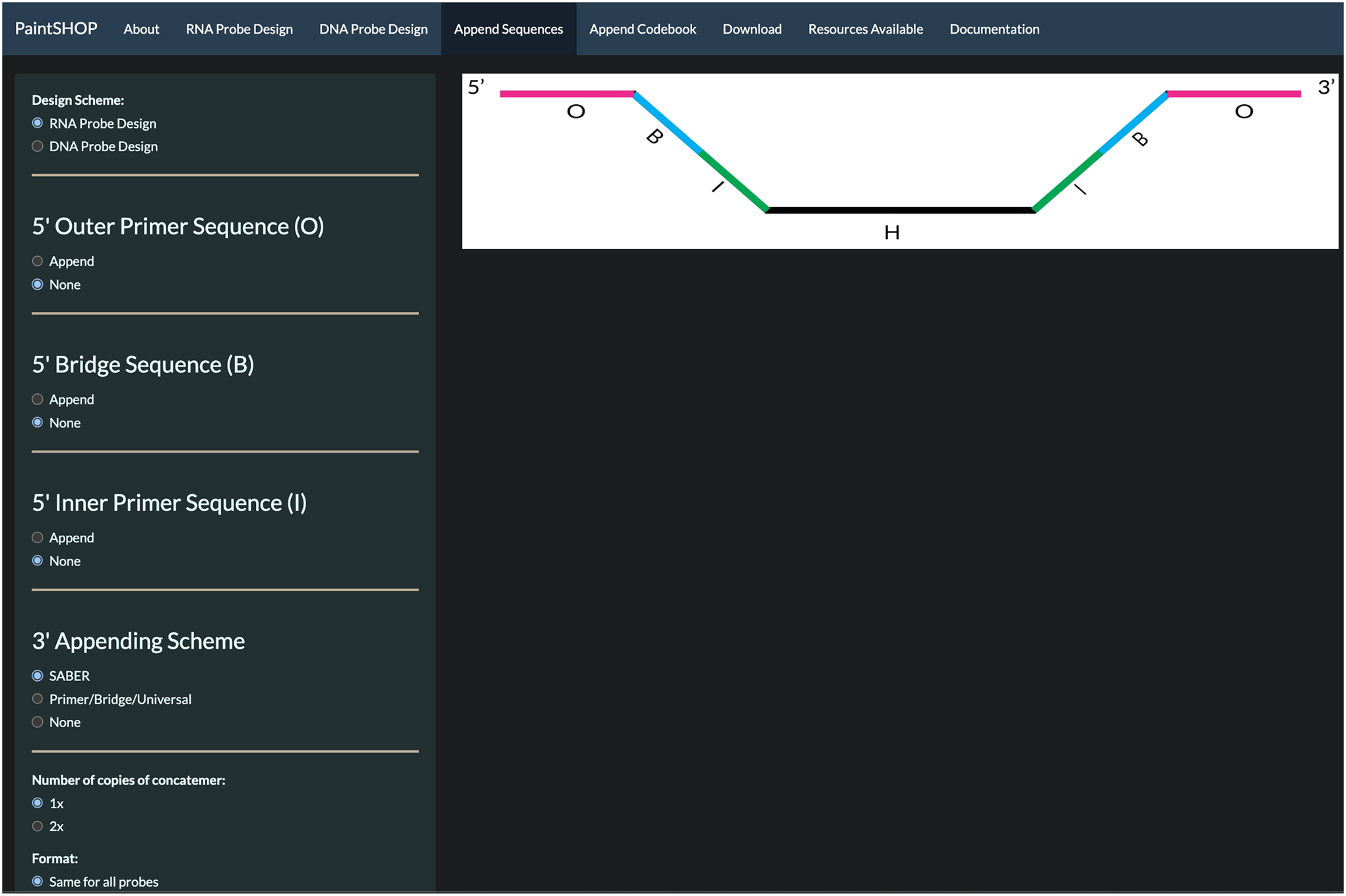
Appending sequences page for RNA single copy probe design using SABER 3’ Appending Scheme.
3.5. RNA FISH multi-target probe design
This protocol describes the process of designing probes for a panel of multiple RNA targets and creating an order file of the probe set. For this demo, we will use the hg38 newBalance (isoform-flattened) RNA probe set to design probes targeting the mRNA transcripts of the Human ADAMTS1, CBX5, UMOD genes, and we will append a unique 42mer bridge sequence to the probes of each target. We will then export all information required to make an order of the designed probe set. Separately, secondary oligos with reverse-complementary bridge sequences and appended SABER concatemer sequences are required to amplify fluorescent signals and visualize targets.
Go to the PaintSHOP website found at paintshop.io and open the interactive web application using the “Launch PaintSHOP” button.
Select the RNA Probe Design tab at the top left of the screen.
In the probe set dropdown, select the hg38 newBalance (isoform-flattened) option.
Enter the RefSeq annotation for the targets of interest (e.g. “ADAMTS1, CBX5, UMOD”) manually in the text box. Alternatively, upload a text file with one gene symbol ID per line. (see Note 2)
Once the annotation has been entered, indicate which input type was used (Manual or File).
Press the Submit button to generate and retrieve the probe set. The table returned describes the probe set that covers the target transcripts. A plot of the distribution of probes per target appears above the table and is shown in Fig. 5.
Optionally trim probe sets to 30 probes per target. On the left menu bar, show the “Optimize Set” options. Select Trim with 30 probes per target selected using the slider.
To append bridge sequences to the generated probe set, select the Append Sequences tab at the top of the screen.
Select the RNA Probe Design for the Design Scheme option.
Select Primer/Bridge/Universal for the 3’ Appending Scheme option.
Select Append for the 3’ Bridge Sequence option.
Select Forward for orientation.
Select Unique for each target for the format option.
Select Kishi et al. 2019 Bridges for the Select Sequence Set option. These configurations are shown in Fig. 6.
Press the Append button at the bottom of the page. The table returned shown in Fig. 7 describes the target of interest and information about the sequences that were appended.
Select the Download tab.
Select Appended Sequences under the Used Appending section and RNA Probe Design for the Design Scheme section.
Under the Choose a file to download option, choose Order File. The expected output is provided as an accompanying Supplementary File 4.
Select the Download button to save a text file with the designed probe set locally. The expected output is provided as an accompanying Supplementary File 5.
Additionally, download the Appending File for downstream bridge oligo design. The expected output is provided as an accompanying Supplementary File 6.
Fig 5.
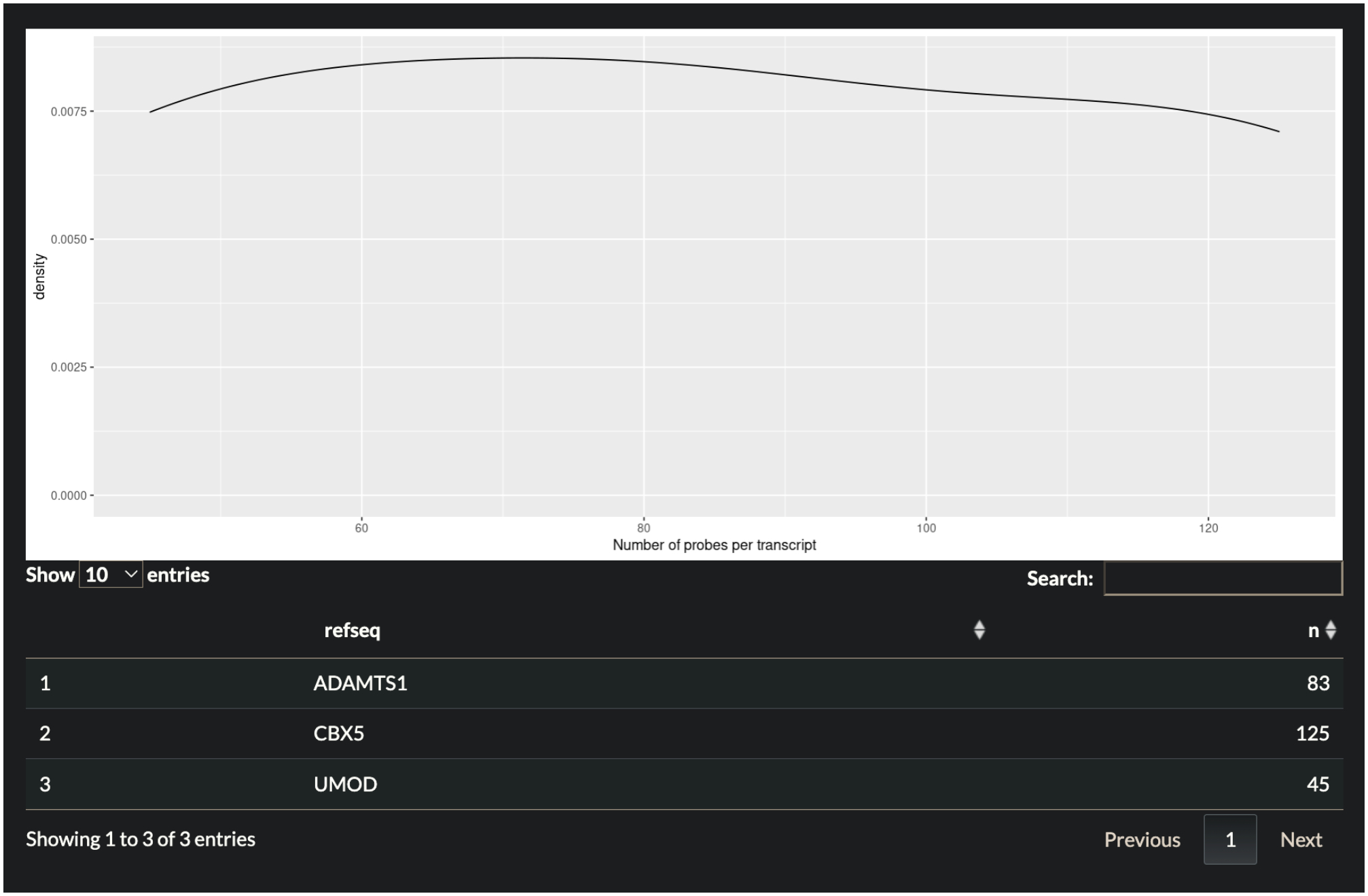
Plot showing distribution of probes per gene target. Table below describes information shown in the plot separated by RefSeq gene.
Fig 6.
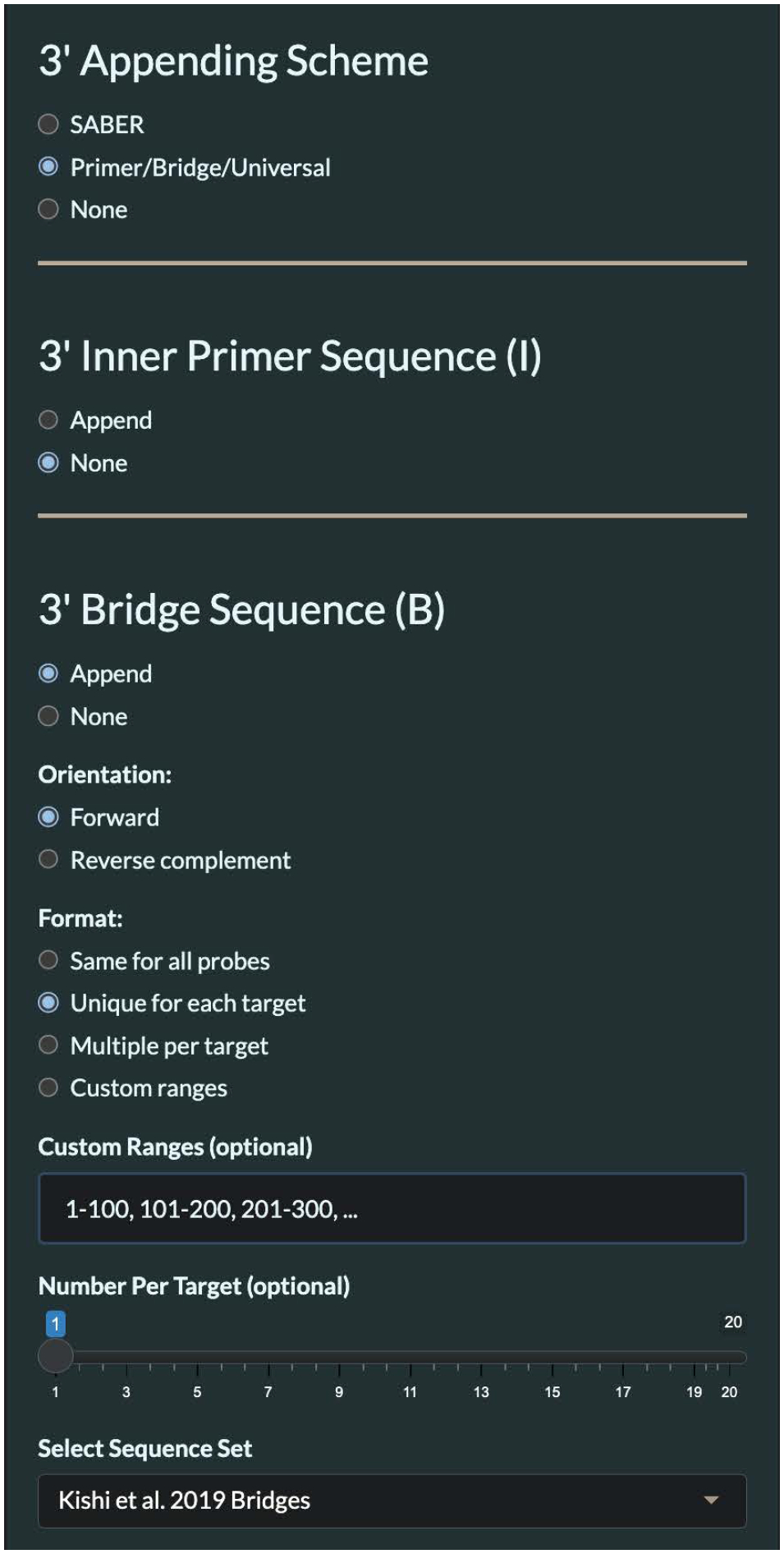
Appending Sequences user specifications for Demo 2.
Fig 7.
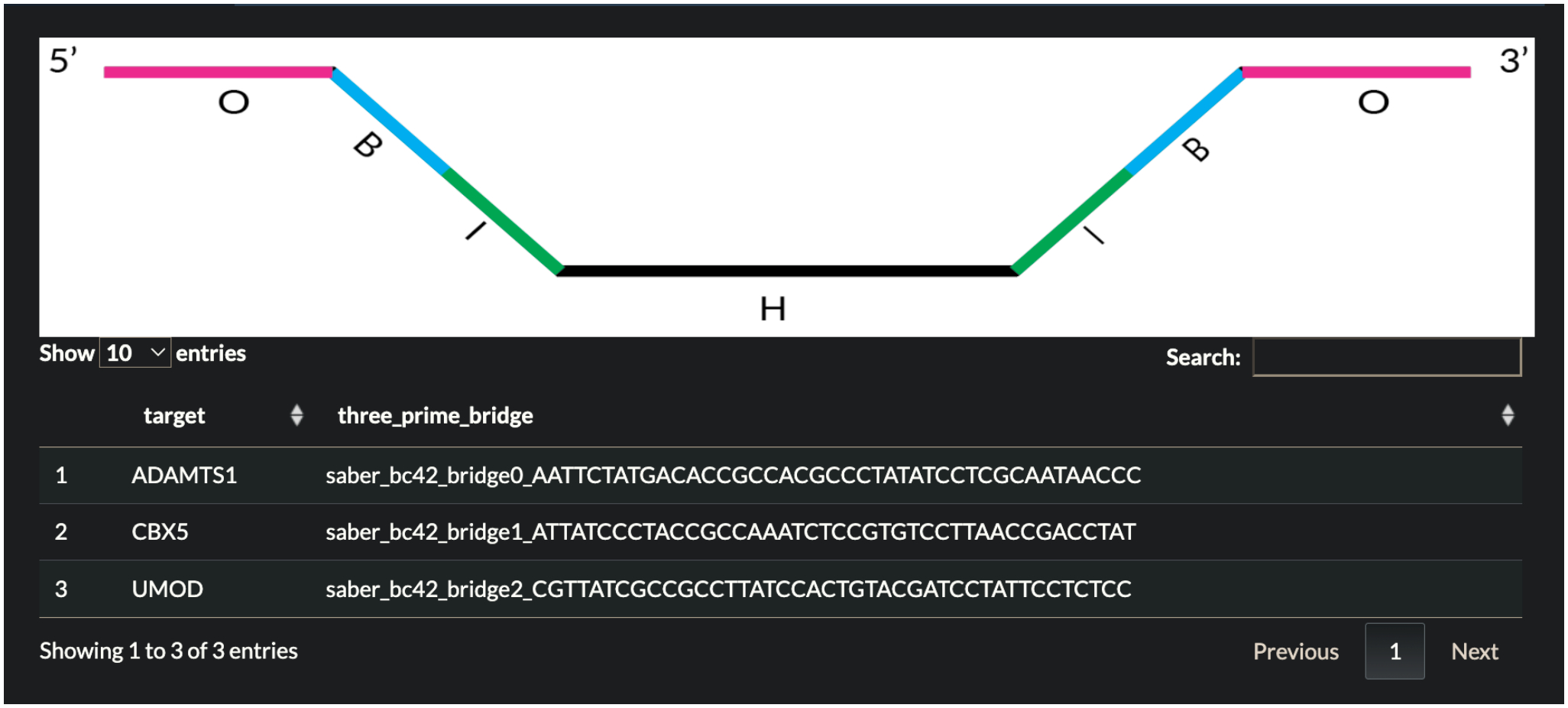
Table returned in Demo 2 describing each target of interest and the sequences that were appended.
3.6. DNA FISH multi-target probe design
This protocol describes the process of designing probes for a panel of multiple DNA targets and creating an order file of the probe set. For this demo, we will use the hg38 newBalance probe set to design probes targeting three loci on Human chromosome X, and we will append a unique 42mer bridge sequence to the probes of each target. Additionally, we will append PCR priming sites for PCR amplification of the probe library. We will then export all information required to make an order of the designed probe set. Separately, secondary oligos with reverse-complementary bridge sequences and appended SABER concatemer sequences are required to amplify fluorescent signals and visualize targets, and PCR primers are required to amplify the oligo library.
Go to the PaintSHOP website found at paintshop.io and open the interactive web application using the “Launch PaintSHOP” button.
Select the DNA Probe Design tab at the top left of the screen.
In the probe set dropdown, select the hg38 newBalance option.
Enter the chromosomal coordinates for the genomic loci of interest (e.g. “chrX:2800000–3000000, chrX:124400000–124600000, chrX:155200000–155400000”) manually in the text box. Alternatively, upload a text file in BED format with one locus per line.
Once the targets have been entered, indicate which input type was used (Manual or File).
Press the Submit button to generate and retrieve the probe set. The table returned describes the probe set that covers the target transcripts.
Optionally trim probe sets to 500 probes per target. On the left menu bar, show the “Optimize Set” options. Select Trim with 500 probes per target selected using the slider. Fine adjustments in the slider value can be made using arrow keys once the slider is selected.
To append bridge sequences to the generated probe set, select the Append Sequences tab at the top of the screen.
Select the DNA Probe Design for the Design Scheme option.
To add a forward PCR primer site, select Append under 5’ Outer Primer Sequence option and use the default settings.
To add target-specific bridge sequences, select Primer/Bridge/Universal for the 3’ Appending Scheme option. Select Append for the 3’ Bridge Sequence option. Select Forward for orientation. Select Unique for each target for the format option. Select Kishi et al. 2019 Bridges for the Select Sequence Set option.
To add a reverse PCR primer site, select Append under 3’ Outer Primer Sequence option and use the default settings.
Press the Append button at the bottom of the page. The table returned describes the target of interest and information about the sequences that were appended.
Select the Download tab.
Select Appended Sequences under the Used Appending section and DNA Probe Design for the Design Scheme section.
Under the Choose a file to download option, choose Order File. The expected output is provided as an accompanying Supplementary File 7.
Select the Download button to save a text file with the designed probe set locally. The expected output is provided as an accompanying Supplementary File 8.
Additionally, download the Appending File for downstream bridge oligo design. The expected output is provided as an accompanying Supplementary File 9.
These probes can be amplified using PCR with the following PCR primers: 5’-GCGTTAGGGTGCTTACGTCTG-3’ is the forward primer sequence and 5’-CACCTCCGTCTCTCACCTCTC-3’ is the reverse primer.
5. Notes
RefSeq IDs are case sensitive and should be matched with the RefSeq database.
When manually entering multiple annotations for RNA probe design, each annotation should be separated with a comma and a space. If a file is provided as input, there should only be one annotation per line.
All probe database files used within PaintSHOP are available from the PaintSHOP resources repository (https://github.com/beliveau-lab/PaintSHOP_resources), including homology probe sequences for DNA- and RNA-FISH as well as synthetic sequence domains used in appending (e.g. bridge and primer sequences). These files are useful for selecting primary keys (i.e. gene symbol) to query PaintSHOP and for downstream oligo design.
Supplementary Material
References
- 1.Pardue ML and Gall JG (1969) Molecular hybridization of radioactive DNA to the DNA of cytological preparations. Proc Natl Acad Sci U S A 64:600–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rudkin GT and Stollar BD (1977), High resolution detection of DNA-RNA hybrids in situ by indirect immunofluorescence [29], 10.1038/265472a0 [DOI] [PubMed] [Google Scholar]
- 3.Lewis ME, Sherman TG, and Watson SJ (1985) In situ hybridization histochemistry with synthetic oligonucleotides: strategies and methods. Peptides 6 Suppl 2:75–87 [DOI] [PubMed] [Google Scholar]
- 4.Rouillard J-M, Zuker M, and Gulari E (2003) OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res 31:3057–3062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Navin N, Grubor V, Hicks J, et al. (2006) PROBER: oligonucleotide FISH probe design software. Bioinformatics 22:2437–2438 [DOI] [PubMed] [Google Scholar]
- 6.Han Y, Zhang T, Thammapichai P, et al. (2015) Chromosome-Specific Painting in Cucumis Species Using Bulked Oligonucleotides. Genetics 200:771–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yilmaz LS, Parnerkar S, and Noguera DR (2011) mathFISH, a web tool that uses thermodynamics-based mathematical models for in silico evaluation of oligonucleotide probes for fluorescence in situ hybridization. Appl Environ Microbiol 77:1118–1122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beliveau BJ, Kishi JY, Nir G, et al. (2018) OligoMiner provides a rapid, flexible environment for the design of genome-scale oligonucleotide in situ hybridization probes. Proc Natl Acad Sci U S A 115:E2183–E2192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelali E, Girelli G, Matsumoto M, et al. (2019) iFISH is a publically available resource enabling versatile DNA FISH to study genome architecture. Nat Commun 10:1636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hu M, Yang B, Cheng Y, et al. (2020) ProbeDealer is a convenient tool for designing probes for highly multiplexed fluorescence in situ hybridization. Sci Rep 10:22031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang T, Liu G, Zhao H, et al. (2021) Chorus2: design of genome-scale oligonucleotide-based probes for fluorescence in situ hybridization. Plant Biotechnol J 19:1967–1978 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hershberg EA, Camplisson CK, Close JL, et al. (2021) PaintSHOP enables the interactive design of transcriptome- and genome-scale oligonucleotide FISH experiments. Nat Methods 18:937–944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Passaro M, Martinovic M, Bevilacqua V, et al. (2020) OligoMinerApp: a web-server application for the design of genome-scale oligonucleotide in situ hybridization probes through the flexible OligoMiner environment. Nucleic Acids Res 48:W332–W339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Beliveau BJ, Joyce EF, Apostolopoulos N, et al. (2012) Versatile design and synthesis platform for visualizing genomes with Oligopaint FISH probes. Proc Natl Acad Sci U S A 109:21301–21306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kishi JY, Schaus TE, Gopalkrishnan N, et al. (2017) Programmable autonomous synthesis of single-stranded DNA. Nat Chem [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kishi JY, Lapan SW, Beliveau BJ, et al. (2019) SABER amplifies FISH: enhanced multiplexed imaging of RNA and DNA in cells and tissues. Nat Methods [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


