Abstract
Seven mutations in the C2, V3, and C3 regions of gp120 are implicated in the tropism of the first CD4-independent human immunodeficiency virus type 1 isolate, m7NDK. Site-directed mutagenesis revealed that three amino acids are essential to maintain this tropism, one in the C2 region and two in the V3 loop. Two mutations implied N glycosylation modifications.
The human immunodeficiency virus (HIV) enters its target cells after interaction of its gp120 with cellular receptors. The first step implies CD4 binding, which induces conformational changes in gp120 (23). This allows it to interact with a coreceptor belonging to the family of hepta-spanning, transmembrane G-coupled chemokine receptors, the two well-characterized ones in vivo being CXCR4 and CCR5 (1, 7). Conformational changes in the V1/V2 and V3 regions of gp120 seem to be implicated in the exposition of coreceptor binding sites (2, 22, 25, 27, 28).
However, CD4-independent isolates have been characterized: primary or laboratory-adapted X4- or R5-dependent HIV-2 (6, 11, 20, 21), laboratory-adapted, forced HIV-1 X4 (13) or R5 (15) viruses, and a naturally derived, HIV-1 X4 laboratory-adapted isolate, m7NDK. m7NDK is the first CD4-independent HIV-1 isolate, and it was characterized in our laboratory (10). Since these viruses are CD4 independent, direct interactions between gp120 and the coreceptor might allow virus entry (10, 13, 19). Our aim was to analyze the role of each of the mutations of the m7NDK isolate in this interaction.
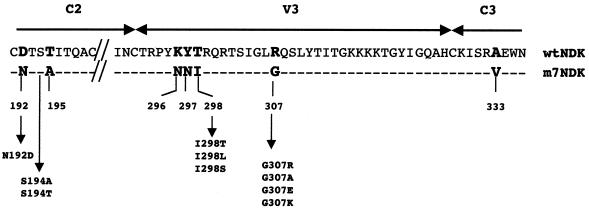
Since seven mutations in the C2, V3, and C3 regions of the env gene are responsible for the phenotype change in the m7NDK isolate (10), we performed site-directed mutagenesis to revert, one by one, each of the mutated amino acids (Fig. 1). For each reverted mutation, we used overlap extension PCR to introduce the mutated base (18). Mutated amplified fragments were then digested using ScaI and HindIII restriction enzymes and cloned in the context of the wild-type NDK (wtNDK) env gene digested by the same enzymes. Those env genes were then cloned in an expression vector (10). As a positive control for CD4 independence, the C2, V3, and C3 regions of the m7NDK env gene were inserted into the ScaI-HindIII sites of the wtNDK virus env gene. Using this strategy, all env genes differed only by the mutation introduced.
FIG. 1.
Comparison of amino acids of the C2, V3, and C3 regions of the wtNDK and m7NDK HIV-1 isolates. Amino acid differences are in boldface, and dashes indicate identical amino acids. Positions of the reverted mutations and the different substitutions and their positions are indicated and numbered according to the HIV-1 wtNDK Env sequence.
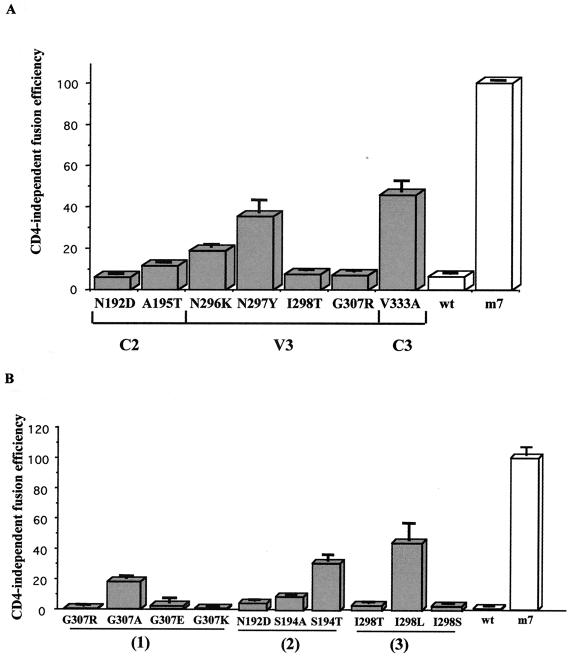
HEK293 cells were then transfected by each Env expression vector by using the calcium phosphate coprecipitation technique (5). Forty-eight hours later, transfected cells were trypsinized and mixed (ratio, 1:1) with HeLaLTRLacZ CD4-positive (P42) or CD4-negative (Z24) indicator cells (9) to analyze fusion efficiencies. CD4-independent fusion was measured 24 h later using a CPRG (chlorophenol red–β-d-galactopyranoside) test (Roche) as previously described (10). For each mutated Env, CD4-independent fusion efficiencies were compared to either those obtained with the CD4-independent control (100% CD4 independent), i.e., the C2, V3, and C3 regions of m7NDK virus cloned in the wtNDK Env protein, or those obtained with the wtNDK Env protein (0% CD4 independent). CD4-independent fusion efficiencies were estimated as the ratio of A570 measured with HeLaLTRLacZ CD4-negative cells (P42)/A570 measured with HeLaLTRLacZ CD4-positive cells (Z24). The result obtained with the CD4-independent control was defined as 100% and was identical to that obtained with the complete m7NDK Env protein (data not shown). We first observed that all Env proteins induced syncytium formation with CD4-positive indicator cells at an equivalent efficiency (data not shown). As expected, compared to the CD4-independent control, the wtNDK Env protein presented a CD4-independent fusion efficiency lower than 5%. Each of the reverted mutations in the C2, V3, and C3 regions of the m7NDK env gene led to a decrease of at least twofold in CD4-independent fusion efficiency (Fig. 2A). This signifies that all amino acids in the m7NDK C2, V3, and C3 regions of gp120 are necessary for providing an optimized CD4-independent fusion. However, they could be classified into three groups: (i) mutants N297Y (Asn at position 297 was mutated into Tyr) and V333A, which presented two- or threefold-lower (33 and 46% efficiency, respectively) CD4-independent fusion efficiencies than the CD4-independent control, (ii) A195T and N296K, which presented five- to ninefold-lower (11 and 18%, respectively) CD4-independent fusion efficiencies than the CD4-independent control, and (iii) N192D, I298T, and G307R, which presented 15- to 20-fold-lower (less than 7%) fusion efficiencies than the CD4-independent control. Mutants N192D, I298T, and G307R thus presented a CD4-dependent fusion; therefore, we focused on these three mutants in the work that followed.
FIG. 2.
Fusion phenotype analysis of mutated gp120s. Mutated env expressors were transiently transfected in HEK293 cells. Fusion analysis was performed after a coculture of the transfected cells with HeLaLTRLacZ cells expressing (P42) or not expressing (Z24) the CD4 protein. Analysis was performed using a quantitative CPRG test (10). The CD4 independence index was stated as the ratio of A570 of Z24 cells/A570 of P42 cells relative to the 100% value corresponding to the m7NDK gp120 CD4-independent fusion efficiency. Experiments were performed in quadruplicate in 96-well plates. The data are representative of three experiments. (A) Each of the seven mutations was reverted one by one in the C2, V3, and C3 regions. These regions were reintroduced in the expression vector. (B) Substitutions of the three essential mutations. Groups 1 to 3 represent revertants G307R, N192D, and I298T and their related mutants, respectively.
The G307R mutation in m7NDK gp120 changes a small and uncharged amino acid (Gly) to a large and charged one (Arg). We therefore changed the Gly into another positively charged (G307K), negatively charged (G307E), or nonpolar (G307A) amino acid (Fig. 1). We observed that the introduction of a charged, either positive or negative, amino acid led to a strictly CD4-dependent fusion phenotype (0 and 2% of CD4-independent fusion efficiency, respectively; Fig. 2B, group 1). Furthermore, introduction of an alanine led to a drastic reduction in CD4-independent fusion efficiency (16%; Fig. 2B, group 1). As all changes had a major influence on the CD4-independent fusion efficiency, we supposed that the CD4-independent tropism required exclusively a noncharged small amino acid at position 307 of the Env protein.
The two other mutations (N192D and I298T) potentially implied modifications of N glycosylation sites (14). Indeed, the N192D reversion eliminated a potential N glycosylation site in the m7NDK C2 region (Fig. 1). We thus mutated this consensus site without modifying the N192 amino acid. We created S194A (no N glycosylation site) and S194T (N glycosylation site) (Fig. 1). We observed that the S194A mutant presented a CD4-independent fusion efficiency more than 10-fold less than that of m7NDK gp120 (9%), whereas a 31% CD4-independent fusion efficiency was obtained with the S194T mutant (Fig. 2B, group 2). These results strongly suggest that the presence of an N glycosylation site is required in the C2 region at position 192 to allow CD4 independence.
The I298T mutation introduces a potential N glycosylation site (NNT) (14) at position 296. This N glycosylation is not present in either of the wtNDK (KYT) or m7NDK (NNI) gp120 proteins (Fig. 1). As K296N is one of the mutations implicated in CD4-independent tropism, we could not mutate this residue directly. Instead, we created I298L (no N glycosylation site) and I298S (N glycosylation site) mutants (Fig. 1). After transfection and fusion analysis, we observed that I298S presented a strict CD4-dependent tropism (2.5% of the CD4-independent fusion efficiency), whereas the I298L mutant presented a 41% CD4-independent fusion efficiency (Fig. 2B, group 3). We thus concluded that the absence of a potential N glycosylation at this position is a prerequisite for CD4-independent tropism.
To confirm the role of N glycosylation site modification, Western blot analysis was performed on HEK293 cells expressing the Env proteins by using a gp120 monoclonal antibody (D7324; Aalto). After washing, bound antibody was detected using a sheep horseradish peroxidase-conjugated secondary antibody followed by enhanced chemiluminescence (ECL; Amersham). Briefly, cells were lysed (1% NP-40, 150 mM NaCl, 10 mM Tris, and 1 mM DTT with 1/100 protease cocktail inhibitors; Sigma) and mixed with an equal volume of 2× Laemmli buffer (8). Extracts were then loaded on a 12% polyacrylamide denaturing gel. Clear differences in migration pattern could be observed between the CD4-independent control (one N glycosylation site) and its revertants N192D (no N glycosylation site) and I298T (two N glycosylation sites) (Fig. 3). Considering that the only difference in these Env proteins is one mutated amino acid, this clearly demonstrated that differences in migration pattern are associated with differences in N glycosylation: in the m7NDK Env protein, residue 192 is N glycosylated whereas residue 298 is not. On the other hand, the mutation N192D suppresses an N glycosylation site, whereas the mutation I298T introduces one.
FIG. 3.
Alteration of migration patterns of mutated gp120 after modification of potential N glycosylation sites. Expression plasmids were transfected in HEK293 cells. Cell extracts were prepared for a Western blot experiment and were analyzed using a monoclonal anti-gp120 antibody (1). C2, V3, and C3 regions from m7NDK virus were inserted in the wtNDK isolate env gene (one N glycosylation site) (2), N192D gp120 mutant (no N glycosylation site) (3), and I298T gp120 mutant (two N glycosylation sites). M, molecular mass marker (in kilodaltons) (BenchMark Prestained Protein Ladder; Life Technologies). Arrow, gp120.
We showed that three mutations are essential for maintaining the CD4-independent phenotype. The first corresponded to a small noncharged amino acid, namely G307. It is worth noting that this amino acid is in the GPG consensus motif (GLG in the case of m7NDK gp120) and that the change occurring in m7DK gp120 is R to G. This motif is probably situated at the tip of the V3 loop, and it is suspected that it forms a type II β-turn (3, 4, 12, 26). As the tip sequence of the V3 domain seems to be implicated in cell tropism (24), the insertion of an electrostatic charge, as well as the presence of a side chain with high steric hindrance at position 307, might constrain the V3 loop conformation and alter CXCR4 binding affinity. On the contrary, the presence of G307 might lead to a better flexibility of the V3 loop, thus allowing a direct and efficient interaction with CXCR4. The two other mutations implied modifications of potential N glycosylation sites. According to the same logic, the existence of an N glycosylation at position 296 might interfere with CXCR4 binding, whereas the N glycosylation in the C2 (amino acid N192) region might stabilize the open conformation of coreceptor binding site. These hypotheses are supported by the recent finding that the presence or absence of a conserved carbohydrate moiety on the V3 region of the gp120 V3 on clade A and B viruses could modulate the CD4 or chemokine binding sites (17).
Although a common mechanism may explain the direct interaction between Env and CXCR4, no consensus region can be evidenced between the different CD4-independent isolates, even though most (16, 21) or all (15) of the modifications concern N glycosylation sites. As alternative conformations of the coreceptor binding site have been suggested (2, 22, 25, 27, 28), a consensus explanation would consider that all CD4-independent viruses present a set of mutations which together provide better accessibility to the coreceptor binding site because of a better flexibility of V3 and/or V1/V2 loops. Taken together, our results strongly suggest that mutations in m7NDK gp120 lead to a high-affinity conformation of the coreceptor binding site since the essential mutations we characterized might have consequences on V3 loop conformation.
Acknowledgments
We are grateful for the technical support of A. Miccinilli. Nicolas Boord edited the manuscript. J.D. holds a fellowship from Ministère de l'Education Nationale, de l'Enseignement Supérieur et de la Recherche.
This work was supported by grants from Agence Nationale de la Recherche contre le SIDA (ANRS), “Ensemble contre le SIDA” AO11 and AO2 “Lutte anti-Sida” from the University of Paris 7-Denis Diderot.
REFERENCES
- 1.Berger E A, Murphy P M, Farber J M. Chemokine receptors as HIV-1 coreceptors: roles in viral entry, tropism, and disease. Annu Rev Immunol. 1999;17:657–700. doi: 10.1146/annurev.immunol.17.1.657. [DOI] [PubMed] [Google Scholar]
- 2.Cao J, Sullivan N, Desjardin E, Parolin C, Robinson J, Wyatt R, Sodroski J. Replication and neutralization of human immunodeficiency virus type 1 lacking the V1 and V2 variable loops of the gp120 envelope glycoprotein. J Virol. 1997;71:9808–9812. doi: 10.1128/jvi.71.12.9808-9812.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Catasti P, Bradbury E M, Gupta G. Structure and polymorphism of HIV-1 third variable loops. J Biol Chem. 1996;271:8236–8242. doi: 10.1074/jbc.271.14.8236. [DOI] [PubMed] [Google Scholar]
- 4.Catasti P, Fontenot J D, Bradbury E M, Gupta G. Local and global structural properties of the HIV-MN V3 loop. J Biol Chem. 1995;270:2224–2232. doi: 10.1074/jbc.270.5.2224. [DOI] [PubMed] [Google Scholar]
- 5.Chen C, Okayama H. High-efficiency transformation of mammalian cells by plasmid DNA. Mol Cell Biol. 1987;7:2745–2752. doi: 10.1128/mcb.7.8.2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Clapham P R, McKnight A, Weiss R A. Human immunodeficiency virus type 2 infection and fusion of CD4-negative human cell lines: induction and enhancement by soluble CD4. J Virol. 1992;66:3531–3537. doi: 10.1128/jvi.66.6.3531-3537.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clapham P R, Reeves J D, Simmons G, Dejucq N, Hibbitts S, McKnight A. HIV coreceptors, cell tropism and inhibition by chemokine receptor ligands. Mol Membr Biol. 1999;16:49–55. doi: 10.1080/096876899294751. [DOI] [PubMed] [Google Scholar]
- 8.Cleveland D W, Fischer S G, Kirschner M W, Laemmli U K. Peptide mapping by limited proteolysis in sodium dodecyl sulfate and analysis by gel electrophoresis. J Biol Chem. 1977;252:1102–1106. [PubMed] [Google Scholar]
- 9.Dragic T, Charneau P, Clavel F, Alizon M. Complementation of murine cells for human immunodeficiency virus envelope/CD4-mediated fusion in human/murine heterokaryons. J Virol. 1992;66:4794–4802. doi: 10.1128/jvi.66.8.4794-4802.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dumonceaux J, Nisole S, Chanel C, Quivet L, Amara A, Baleux F, Briand P, Hazan U. Spontaneous mutations in the env gene of the human immunodeficiency virus type 1 NDK isolate are associated with a CD4-independent entry phenotype. J Virol. 1998;72:512–519. doi: 10.1128/jvi.72.1.512-519.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Endres J M, Clapham P R, Marsh M, Ahuja M, Davis-Turner J, McKnight A, Thomas J F, Stoebenau-Haggarty B, Choe S, Vance P J, Wells T N C, Power C A, Sutterwala S S, Doms R W, Landau N R, Hoxie J A. CD4-independent infection by HIV-2 is mediated by fusin/CXCR-4. Cell. 1996;87:745–756. doi: 10.1016/s0092-8674(00)81393-8. [DOI] [PubMed] [Google Scholar]
- 12.Fontenot J D, Gatewood J M, Mariappan S V, Pau C P, Parekh B S, George J R, Gupta G. Human immunodeficiency virus (HIV) antigens: structure and serology of multivalent human mucin MUC1-HIV V3 chimeric proteins. Proc Natl Acad Sci USA. 1995;92:315–319. doi: 10.1073/pnas.92.1.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoxie J A, LaBranche C C, Endres M J, Turner J D, Berson J F, Doms R W, Matthews T J. CD4-independent utilization of the CXCR4 chemokine receptor by HIV-1 and HIV-2. J Reprod Immunol. 1998;41:197–211. doi: 10.1016/s0165-0378(98)00059-x. [DOI] [PubMed] [Google Scholar]
- 14.Imperiali B, O'Connor S E. Effect of N-linked glycosylation on glycopeptide and glycoprotein structure. Curr Opin Chem Biol. 1999;3:643–649. doi: 10.1016/s1367-5931(99)00021-6. [DOI] [PubMed] [Google Scholar]
- 15.Kolchinsky P, Mirzabekov T, Farzan M, Kiprilov E, Cayabyab M, Mooney L J, Choe H, Sodroski J. Adaptation of a CCR5-using, primary human immunodeficiency virus type 1 isolate for CD4-independent replication. J Virol. 1999;73:8120–8126. doi: 10.1128/jvi.73.10.8120-8126.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.LaBranche C C, Hoffman T L, Romano J, Haggarty B S, Edwards T G, Matthews T J, Doms R W, Hoxie J A. Determinants of CD4 independence for a human immunodeficiency virus type 1 variant map outside regions required for coreceptor specificity. J Virol. 1999;73:10310–10319. doi: 10.1128/jvi.73.12.10310-10319.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malenbaum S E, Yang D, Cavacini L, Posner M, Robinson J, Cheng-Mayer C. The N-terminal V3 loop glycan modulates the interaction of clade A and B human immunodeficiency virus type 1 envelopes with CD4 and chemokine receptors. J Virol. 2000;74:11008–11016. doi: 10.1128/jvi.74.23.11008-11016.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pogulis R J, Vallejo A N, Pease L R. In vitro recombination and mutagenesis by overlap extension PCR. Methods Mol Biol. 1996;57:167–176. doi: 10.1385/0-89603-332-5:167. [DOI] [PubMed] [Google Scholar]
- 19.Potempa S, Picard L, Reeves J D, Wilkinson D, Weiss R A, Talbot S J. CD4-independent infection by human immunodeficiency virus type 2 strain ROD/B: the role of the N-terminal domain of CXCR-4 in fusion and entry. J Virol. 1997;71:4419–4424. doi: 10.1128/jvi.71.6.4419-4424.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reeves J D, Hibbitts S, Simmons G, McKnight A, Azevedo-Pereira J M, Moniz-Pereira J, Clapham P R. Primary human immunodeficiency virus type 2 (HIV-2) isolates infect CD4-negative cells via CCR5 and CXCR4: comparison with HIV-1 and simian immunodeficiency virus and relevance to cell tropism in vivo. J Virol. 1999;73:7795–7804. doi: 10.1128/jvi.73.9.7795-7804.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Reeves J D, McKnight A, Potempa S, Simmons G, Gray P W, Power C A, Wells T, Weiss R A, Talbot S J. CD4-independent infection by HIV-2 (ROD/B): use of the 7-transmembrane receptors CXCR-4, CCR-3, and V28 for entry. Virology. 1997;231:130–134. doi: 10.1006/viro.1997.8508. [DOI] [PubMed] [Google Scholar]
- 22.Rizzuto C D, Wyatt R, Hernandez R N, Sun Y, Kwong P D, Hendrickson W A, Sodroski J. A conserved HIV gp120 glycoprotein structure involved in chemokine receptor binding. Science. 1998;280:1949–1953. doi: 10.1126/science.280.5371.1949. [DOI] [PubMed] [Google Scholar]
- 23.Sattentau Q J, Moore J P, Vignaux F, Traincard F, Poignard P. Conformational changes induced in the envelope glycoproteins of the human and simian immunodeficiency viruses by soluble receptor binding. J Virol. 1993;67:7383–7393. doi: 10.1128/jvi.67.12.7383-7393.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shimizu N, Haraguchi Y, Takeuchi Y, Soda Y, Kanbe K, Hoshino H. Changes in and discrepancies between cell tropisms and coreceptor uses of human immunodeficiency virus type 1 induced by single point mutations at the V3 tip of the Env protein. Virology. 1999;259:324–333. doi: 10.1006/viro.1999.9764. [DOI] [PubMed] [Google Scholar]
- 25.Sullivan N, Sun Y, Sattentau Q, Thali M, Wu D, Denisova G, Gershoni J, Robinson J, Moore J, Sodroski J. CD4-induced conformational changes in the human immunodeficiency virus type 1 gp120 glycoprotein: consequences for virus entry and neutralization. J Virol. 1998;72:4694–4703. doi: 10.1128/jvi.72.6.4694-4703.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu G, MacKenzie R, Durda P J, Tsang P. The binding of a gp120 V3 loop peptide to HIV-1 neutralizing antibodies: structural implications. J Biol Chem. 2000;275:36645–36652. doi: 10.1074/jbc.M005369200. [DOI] [PubMed] [Google Scholar]
- 27.Wyatt R, Kwong P D, Desjardins E, Sweet R W, Robinson J, Hendrickson W A, Sodroski J G. The antigenic structure of the HIV gp120 envelope glycoprotein. Nature. 1998;393:705–711. doi: 10.1038/31514. [DOI] [PubMed] [Google Scholar]
- 28.Wyatt R, Moore J, Accola M, Desjardin E, Robinson J, Sodroski J. Involvement of the V1/V2 variable loop structure in the exposure of human immunodeficiency virus type 1 gp120 epitopes induced by receptor binding. J Virol. 1995;69:5723–5733. doi: 10.1128/jvi.69.9.5723-5733.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]