Abstract
Maroons in Suriname and French Guiana descend from enslaved Africans who escaped the plantations during colonial times. Maroon farmers still cultivate a large diversity of rice, their oldest staple crop. The oral history and written records of Maroons by colonial authorities provide contrasting perspectives on the origins of Maroon rice. Here, we analyzed the genomic ancestry of 136 newly sequenced Maroon rice varieties and found seven genomic groups that differ in their geographical associations. We interpreted these findings in light of ethnobotanical and archival investigations to reconstruct the historical contexts associated with the introduction of rice varieties to the Guianas. We found that two rice groups trace to West Africa, which we propose are linked to the transatlantic slave trade (c. 1526 to 1825). We posit that the Maroon rice stock additionally contains varieties that derive from rice introduced by indentured laborers from Java (1890 onwards), USA rice breeders (1932 onwards), and Hmong refugees who fled the Vietnam War (1991). Furthermore, on the Maroon fields, we found rice types never documented before that were derived from crosses. Overall, our results demonstrate that the Maroon farmers prioritize maintenance of a high stock diversity, which we posit reflects the expertise they inherited from their (African) ancestors. Ignored by agricultural modernization initiatives, Maroon farmers today are custodians of a unique cultural heritage. Notably, the genomic findings underline many Maroon stories about their past. We anticipate that a similar study approach can be applied to other heirloom crops of (Indigenous) communities that may have preserved their history on their farms to reconstruct, acknowledge, and honor the past.
Keywords: rice, Maroon history, domestication, traditional landraces, agriculture, transatlantic slave trade, Columbian exchange, genomics, ethnobotany, West Africa, Suriname, French Guiana
Introduction
Marronage, the temporary or permanent escape from slavery, was a widespread act of resistance against the plantation economies in the Americas during colonial times (∼1526 to 1867). Today, Maroon communities exist in Suriname and French Guiana, accounting for c. 239,000 people, divided into six distinct groups (Price 2018). Maroon farmers, almost all of them women, grow various staple crops using shifting cultivation. This method consists of multi-cropping on a cleared piece of forest field, rarely using agrochemicals (Fleury 1994; Van Andel et al. 2016a). The farmers grow various crops that are originally from Africa. These African crops have remained central to both the nutrition and the preservation of the spiritual and cultural practices of the Maroon community (Price 1991; Van Andel et al. 2019). The Maroon farmers obtain planting material from their own fields, and through interhousehold exchange within and between communities (Van Andel et al. 2019; Pinas et al. 2023).
A crop of great significance to the Maroons is rice. They grow many rice varieties, sometimes up to 21 per farmer. These rice varieties are landraces (Pinas et al. 2024): cultivated, genetically heterogeneous varieties that have evolved in a certain ecogeographical area and therefore adapted to the edaphic and climatic conditions, and traditional management and uses (Casañas et al. 2017). Landraces retain ecogeographical relevant information in their genomes. This has allowed for reconstructing crop dispersal and domestication in the past, such as for wheat (Scott et al. 2019), rice (Gutaker et al. 2020), and tropical fruits (Zerega et al. 2004; Larranaga et al. 2021). Many Maroon landraces retain adaptive traits of wild rice that make them more resilient to ecosystem changes and predation compared to commercial cultivars, such as shattering seeds, red pericarp, and long awns (Pinas et al. 2023). Maroons consider some of their rice landraces as African, and their oral history has preserved numerous stories that provide context to the naming of their rice varieties and origins (Carney 2005; Ramdayal et al. 2021; Pinas et al. 2023; Van Andel et al. 2023).
Here, we leverage the global rice landrace diversity presented by the 3,000-rice genome (3K-RG) project (The 3000 Rice Genomes Project 2014; Wang et al. 2018) to clarify the origins of the various Maroon rice varieties. The 3K-RG project distinguished nine major genomic groups among the global varieties of Asian rice (Oryza sativa L.), including across its two subspecies indica and japonica, most of which are broadly associated with specific (eco-)geographical regions (Han and Xue 2003; Agrama et al. 2010; Wang et al. 2018). Ancestry characterization of Maroon rice varieties in this global context of landrace diversity may clarify where in the world their closest genomic relatives are located. By integrating genomic ancestry with ethnobotanical field studies and archival research we aim to reconstruct the historical contexts associated with the introduction of rice varieties to the Guianas and reconstruct events of agronomic innovation in the Maroon past.
Results
We obtained whole-genome resequencing data for 136 rice varieties collected from Saamaka and Okanisi Maroon communities across Suriname and French Guiana (Fig. 1a and supplementary fig. S1, Supplementary Material online). In addition, we built a comparative dataset that includes ∼1,400 globally distributed O. sativa landraces selected from the 3K-RG project (Wang et al. 2018), early commercial cultivars from the USA (registered: 1905 to 1963) (Vaughn et al. 2021), varieties of African rice (Oryza glaberrima Steud.) (Meyer et al. 2016; Van Andel et al. 2019), and relevant diploid wild Oryza species (AA genomes) from Africa (Oryza barthii A. Chev., Oryza longistaminata A. Chev & Roehr), and the Americas (Oryza glumaepatula Steud.) (Fig. 1a). Subsequently, we called genotypes for the newly sequenced varieties based on single-nucleotide polymorphism (SNP) overlapping with the 3K-RG 404k coreSNP Dataset (available via https://snp-seek.irri.org/_download.zul) and intersected the Maroon rice data with the comparative dataset (supplementary fig. S2, Supplementary Material online). After SNP- and individual-level filtering, our final genotype dataset for population genomic analyses consists of 109,203 SNPs (‘∼110k panel’) that are highly ancestry informative for a total of 1,390 rice varieties (supplementary fig. S2, extended data S1a and b, Supplementary Material online). Our sequenced Maroon rice varieties cover 95.0 ± 5.0% (mean ± SD) of the SNPs with an average genotype depth of 10.3 ± 6.1×, which overlaps with that for the 3K-RG landraces (14.4 ± 6.9×) (supplementary fig. S2, Supplementary Material online).
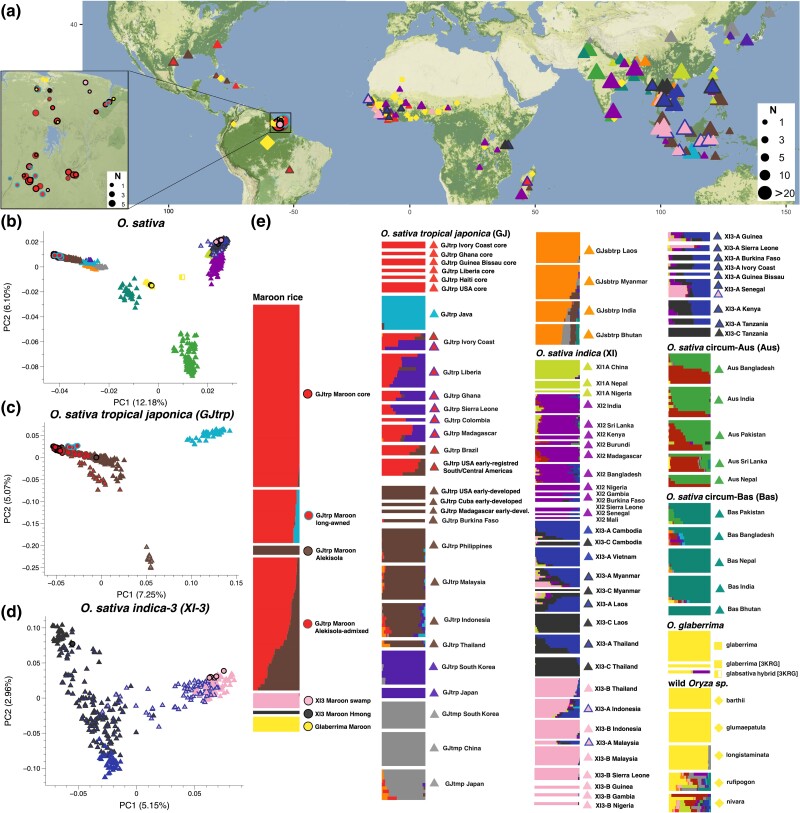
Fig. 1.
Global ancestry characterization of Maroon rice varieties in relation to worldwide rice diversity. Circles indicate Maroon varieties. Other symbols represent different rice species: O. sativa (triangles), O. glaberrima (squares) and wild rice (diamonds). Colors reflect the ancestry clusters and profiles assigned by ADMIXTURE at K = 15. a) Sampling locations of the rice varieties analyzed here, detailing the Maroon communities in the Guianas (box). Symbol size corresponds to sampling size (number of accessions). Basemap: © Stadia Maps. b) PC space constructed from global O. sativa landrace diversity. The O. glaberrima and wild rice varieties are projected onto the O. sativa PC space. PC1 separates O. sativa japonica varieties from indica varieties, and PC2 the circum-Aus varieties. Most of the Maroon rice varieties (n = 126) cluster among O. sativa tropical japonica varieties, a few (n = 6) among O. sativa indica-3 varieties, and a few (n = 4) we identified as O. glaberrima varieties in additional genomic analyses. c) PC space constructed from global tropical japonica (GJtrp) varieties. The GJtrp Maroon core varieties cluster outside of the Southeast-Asian landrace diversity. The GJtrp Maroon long-awned varieties are shifted on a cline toward GJtrp Java, and the GJtrp Maroon Alekisola-admixed varieties plot between the GJtrp Maroon core and GJtrp Maroon Alekisola varieties. d) PC space constructed from global O. sativa indica-3 (XI3) varieties. The first two PCs discern three distinct XI3 groups, and the XI3 Maroon varieties cluster among two of them. e) ADMIXTURE plot for K = 15. A maximum of ten randomly selected accessions is shown per rice group. For the full dataset with detailed individual labels, see extended data S2, Supplementary Material online. The ancestry profiles of the Maroon rice varieties cluster in seven genomic groups; four can be discerned among the O. sativa tropical japonica varieties, two among the O. sativa indica-3 varieties, and one non-sativa group that after additional ancestry characterization we identified as O. glaberrima, African rice.
Seven Rice Genomic Groups
Then, we investigated how the Maroon varieties relate to the diversity of global rice landraces, and what genomic groups as defined in the 3K-RG project are represented (Wang et al. 2018). For this we used Principal Component Analysis (PCA, Patterson et al. 2006) (Fig. 1b–d), Partitioning Around Medoids (PAM, see Materials & Methods, extended data S1c, Supplementary Material online), and ADMIXTURE (Alexander et al. 2009) for the unsupervised model-based clustering of global rice diversity (Fig. 1e, extended data S1d, Supplementary Material online). Overall, we identified seven Maroon rice genomic groups. By far the majority (∼93%) of the Maroon varieties are O. sativa of the tropical japonica group (“GJtrp Maroon”, n = 126), whereas O. sativa indica-3 varieties account for a small subset (∼4%) (“XI3 Maroon”, n = 6) of the diversity (Fig. 1b–d). A few Maroon landraces (∼3%) have ancestry that overlaps poorly with O. sativa diversity (Fig. 1b). Maroons call these varieties baaka alisi (“black rice”) and matu alisi (“forest rice”), and they are morphologically similar to O. glaberrima (African rice). We hypothesized that these are Maroon varieties of O. glaberrima and labeled this group “Glaberrima Maroon” (n = 4).
Within the Maroon O. sativa diversity, the ADMIXTURE clustering model further distinguishes two tropical japonica and two indica-3 Maroon rice groups with non-admixed ancestry profiles (Fig. 1e, extended data S2, Supplementary Material online). The largest group consists of upland rainfed tropical japonica varieties with diverse landrace traits (supplementary table S1, Supplementary Material online), which we labeled “GJtrp Maroon core” (n = 62, including one previously published 3K-RG variety). The second group consists of upland rainfed tropical japonica varieties with elongated white grains referred to by the Maroons as Alekisola (supplementary table S1, Supplementary Material online), which we labeled as “GJtrp Maroon Alekisola” (n = 3). The largest indica-3 genomic group consists of wetland landraces with tall stems > 2 m that are nearly all grown by Okanisi Maroons in deep swamps in the Cottica region (supplementary table S1, Supplementary Material online). We labeled these as “XI3 Maroon swamp” (n = 5). One indica-3 variety, an upland rainfed landrace with elongated grains, white husk and short white hairs, has an unadmixed ancestry profile showing a different ancestry component (supplementary table S1, Supplementary Material online). This variety is referred to by the Maroons as Hmong, and we labeled “XI3 Maroon Hmong” (n = 1). Importantly, these Maroon rice genomic groups show distinct affinities to landraces in different world regions (Fig. 1e, supplementary extended data S2, Supplementary Material online). Therefore, we hypothesized that these rice groups reflect various historical events during which rice was introduced to the Americas. Over time, several of these introduced rice varieties were incorporated into Maroon agriculture and grown and maintained as heirloom varieties by Maroon farmers with the seeds passed down from generation to generation.
In addition, the ADMIXTURE clustering model distinguishes two tropical japonica Maroon rice groups with ancestry profiles that have two admixed ancestry components (Fig. 1e). The largest group contains varieties for which one ancestry component is found in a non-admixed form in the GJtrp Maroon core group and the other in the GJtrp Maroon Alekisola group. The varieties in this group are heterogenous in admixture proportions, and their diverse traits overlap with those found in the GJtrp Maroon core and GJtrp Maroon Alekisola groups (supplementary table S1, extended data S1e, Supplementary Material online). We labeled this group “GJtrp Maroon Alekisola-admixed” (n = 46). The other admixed group consists of varieties with distinctively long, stiff awns or brittle black awns (supplementary table S1, Supplementary Material online), which we labeled “GJtrp Maroon long-awned” (n = 18, including two previously published 3K-RG varieties). These varieties carry one ancestry component that only appears in non-admixed form in tropical japonica landraces from Java. Since both admixed GJtrp Maroon groups have at least one ancestry component that is also found in non-admixed form among the Maroon varieties, we hypothesize that these rice groups resulted from local admixture. Maroon farmers may have discovered off-types that resulted from (spontaneous) crosses, and selected varieties with desirable traits.
Maroon African Rice (O. glaberrima)
First, we investigated our hypothesis that the four landraces that we labeled “Glaberrima Maroon” are indeed Maroon varieties of African rice. In genomic distance analyses based on shared identity-by-state (IBS), the Glaberrima Maroon varieties form a cluster with a previously published O. glaberrima variety obtained from Maroons in Suriname (Van Andel et al. 2016b), and fall within a larger cluster with West African O. glaberrima varieties (supplementary fig. S3, Supplementary Material online). Outgroup f3-statistics cannot conclusively discern whether the Maroon baaka alisi and matu alisi varieties share more ancestry with O. glaberrima varieties or O. barthii (supplementary fig. S4, Supplementary Material online). However, the results of various f4-statistics decisively show that the Maroon varieties form a clade with O. glaberrima varieties from West Africa to the exclusion of O. barthii (supplementary table S2 and S3, Supplementary Material online).
Maroon Core Rice (tropical japonica)
Subsequently, we investigated the geographical association for the morphologically diverse landraces in the GJtrp Maroon core group. In ADMIXTURE, the ancestry component for this group is also found maximized in a few landraces from West Africa (Guinea Bissau, Ghana, Ivory Coast, and Liberia), one from Haiti and four early-registered cultivars from the USA (Fig. 1e, supplementary extended data S2, Supplementary Material online). To investigate the genomic relation among these rice varieties in more detail, we combined individual-level genomic distance analyses based on shared-IBS with various population-level symmetry f4-statistics. We found that the landraces in the GJtrp Maroon core group show the highest levels of shared genetic drift with “core” landraces in West Africa, Haiti and some of the early-registered USA cultivars, and lower levels with other varieties from the Americas (e.g. Brazil, Colombia), (West) Africa and Asia (Fig. 2a, supplementary fig. S5, Supplementary Material online). Congruently, in the admixture graph the GJtrp Maroon core and West Africa core groups are fitted on a sister branch to the GJtrp Brazil group (supplementary fig. S6, Supplementary Material online). Taken together, these findings indicate a direct genomic link between the GJtrp Maroon core and other “core” varieties from the Americas to the “core” landraces in West Africa.
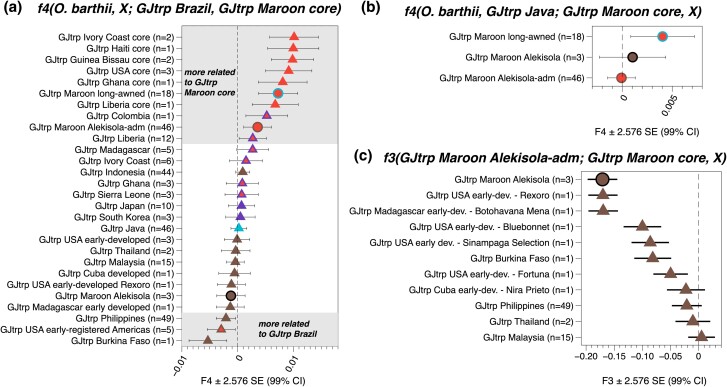
Fig. 2.
F-statistics for the ancestry characterization of the various tropical japonica (GJtrp) Maroon rice groups to trace their geographical origins. Error bars reflect 2.576 SEs, corresponding to 99% confidence intervals (CIs). a) F4-statistics that compare the ancestry of the GJtrp Maroon core and GJtrp Brazil varieties to measure their differences in shared genetic drift to other GJtrp groups (X). The GJtrp Maroon core group significantly shares more alleles with other “core” subsets in the Americas and West Africa, indicating a close genomic link. b) Admixture f4-statistics that test for directional admixture of GJtrp Java ancestry in the various GJtrp Maroon groups. GJtrp Maroon core represents the ancestry of the progenitor population, i.e. the tropical japonica diversity cultivated by the Maroons prior to admixture. Only the Maroon GJtrp long-awned group shows a significantly positive f4-statistic (Z = 3.272), indicating that the Java ancestry is specific to this group. c) Admixture f3-statistics for the GJtrp Maroon Alekisola-admixed group. The statistics test what combination of ancestry from the GJtrp Maroon core group and another GJtrp group X approximates the gene pool of the GJtrp Maroon Alekisola-admixed group best. The most extreme negative f3-values indicate the strongest admixture signals and are found for the local GJtrp Maroon Alekisola varieties (99% CI: −0.197 < f3 < −0.144) and the closely related early-developed cultivars Rexoro and Botohavana Mena. The admixture f3-values are significantly less negative for more distantly related landraces from the Philippines (99% CI −0.048 < f3 < −0.005) and other regions in Southeast Asia.
Maroon Long-Awned Rice (tropical japonica)
The major ancestry component in the varieties of the GJtrp Maroon long-awned group is shared with the GJtrp Maroon core and other “core“ varieties. However, ADMIXTURE also distinguishes an ancestry component (5.9% to 13.3%) that is characteristic of tropical japonica landraces from Java (Fig. 1e, supplementary extended data S2, Supplementary Material online). Since many Javanese tropical japonica have similar long awns (Iskandar and Ellen 1999) this raises the possibility that this trait admixed into some of the Maroon rice varieties. Indeed, using various admixture f4-statistics we found a significant admixture signal for ancestry from Javanese landraces only in the GJtrp Maroon long-awned group (Z = 3.272) and not in other (Maroon) landrace groups (Fig. 2b, supplementary fig. S7a, Supplementary Material online). Moreover, a significant admixture signal is obtained only for landraces from Java, and not for sources from other geographical regions (supplementary fig. S7b, Supplementary Material online). Congruently, in the admixture graph the GJtrp Maroon long-awned group is fitted on an admixed branch with a 90% ancestry contribution from the GJtrp Maroon core group and a 10% ancestry contribution from a source for which GJtrp Java is the closest proxy (supplementary fig. S6, Supplementary Material online). However, we note that the GJtrp Maroon core group can be replaced as a source for admixture by another group with core ancestry, including GJtrp West Africa core or early-registered varieties from the USA with “core” ancestry (supplementary fig. S7c, Supplementary Material online). As such, we cannot determine if the admixture occurred locally (with the Maroons) in Suriname.
Maroon Alekisola Rice (tropical japonica)
A similar ancestry profile is found for the varieties in the GJtrp Maroon Alekisola group, and landraces in Southeast Asia, predominantly the Philippines, and some early-developed USA cultivars (Fig. 1e, supplementary extended data S2, Supplementary Material online). The latter includes the glabrous-hulled Rexoro cultivar that was developed in 1926 in Louisiana (Khush and Brar 2008). We investigated whether the Maroon rice varieties named Alekisola are local variants of Rexoro and/or other early-registered USA cultivars, or more related to landraces in the Philippines. The outgroup f3-statistics indicate the GJtrp Maroon Alekisola varieties share significantly more genetic drift with the USA early-developed cultivar Rexoro and the Botohavana Mena variety from Madagascar than with other early-developed landraces from the Philippines (supplementary fig. S8, Supplementary Material online). Similar results are obtained with individual-level genomic distance analyses based on shared-IBS (supplementary fig. S9, extended data S3, Supplementary Material online), i.e. the shared-IBS among these varieties is considerably higher than among the Maroon Alekisola (or Rexoro) and any of the other early (USA) developed cultivars. These results indicate that the closest genetic relatives for the Maroon Alekisola varieties are the Rexoro variety from the USA and the Botohavana Mena variety from Madagascar.
Maroon Alekisola-Admixed Rice (tropical japonica)
In the ADMIXTURE analysis the varieties in the GJtrp Maroon Alekisola group appear as admixed between varieties that fall in the GJtrp Maroon core and GJtrp Maroon Alekisola groups with the Alekisola component seemingly accounting for up to ∼90% (Fig. 1e, supplementary extended data S2, Supplementary Material online). However, tropical japonica landraces from Brazil, as well as five early-registered USA cultivars that were collected as landraces in Central and South America, and a few landraces from West Africa, similarly show an admixed ancestry profile. Therefore, we investigated whether the varieties in the GJtrp Maroon Alekisola-admixed group more likely have resulted from admixture between varieties in the local Maroon core and Alekisola groups, or were introduced from places outside the Guianas, such as Brazil.
Admixture f4-statistics, taking either GJtrp Brazil or GJtrp Maroon core as a progenitor source, show that the GJtrp Maroon Alekisola-admixed group has significant additional ancestry that best matches GJtrp Maroon Alekisola or the closely related GJtrp USA early-developed cultivars (e.g. Rexoro) (supplementary figs. S10a and b, Supplementary Material online). The local GJtrp Maroon core landraces are a closer progenitor source than those from Brazil, since for the statistic f4(O. barthii, GJtrp Maroon Alekisola-admixed; GJtrp Brazil, GJtrp Maroon core) the GJtrp Maroon Alekisola-admixed and GJtrp Maroon core group show a significant excess of shared genetic drift (Z = 3.27) (Fig. 2a). The differential ancestry between GJtrp Brazil and GJtrp Maroon core group that is driving this statistic is associated with ancestry from GJtrp Philippines (Z = −3.08), and not GJtrp Maroon Alekisola (Z = −1.05) or the USA early-registered cultivars (Z = −0.29) (Fig. 2a). Subsequently, we tested the hypothesis that the GJtrp Maroon Alekisola-admixed gene pool resulted from local admixture more directly, using conservative admixture f3-statistics (Fig. 2c, supplementary fig. S11, Supplementary Material online). Indeed, the strongest signals for admixture are found for the local GJtrp Maroon Alekisola varieties (or the closely related early-developed Rexoro cultivar from the USA) (Fig. 2c), and GJtrp (Maroon) core varieties (supplementary figs. S10 and S11, Supplementary Material online). This indicates that the GJtrp Maroon Alekisola-admixed group on average can be considered a result of local admixture between varieties in the GJtrp Maroon core and GJtrp Maroon Alekisola groups. Congruently, in the admixture graph the GJtrp Maroon Alekisola-admixed group is fitted on an admixed branch with an ancestry contribution of 61% from the GJtrp Maroon core group and 39% from a source for which Alekisola/Rexoro is the closest proxy (supplementary fig. S6, Supplementary Material online).
Maroon Swamp Rice (indica-3)
We found similar unadmixed ancestry profiles for the XI3 Maroon swamp varieties and XI3-B landraces from Southeast Asia (Indonesia, Malaysia, Thailand) and West Africa (Sierra Leone, Guinea, Nigeria, Gambia) (Fig. 1e, supplementary extended data S2, Supplementary Material online). To narrow down the geographical association for the XI3 Maroon swamp group we investigated whether they are more related to varieties in Southeast Asia or West Africa. With individual-level genomic distance analyses based on shared-IBS we find that the XI3 Maroon swamp varieties form a cluster among XI3-B landraces from Indonesia, in particular Java, Sumatra, and Sulawesi, and not among West African varieties (supplementary extended data S4, Supplementary Material online). Congruently, symmetry f4-statistics indicate that the Maroon XI3 swamp varieties are more closely related to XI3-B landraces in Indonesia, whereas those from West Africa are more closely related to XI3-B varieties from Malaysia and Thailand (supplementary figs. S12a and b, Supplementary Material online). In addition, in the admixture graph the XI3 Maroon swamp group is not fitted on a branch that derives from the XI3-B West Africa group (supplementary fig. S13, Supplementary Material online). Instead, these two groups are fitted on sister branches that both split from XI3-B varieties in Asia. This makes it unlikely that the Maroon varieties derived directly from varieties in West Africa.
Maroon Hmong Rice (indica-3)
Lastly, we characterized the ancestry further of the upland indica-3 rice variety referred to by the Maroons as Hmong rice (“XI3 Maroon Hmong”). In ADMIXTURE, this variety is maximized for an ancestry component that is predominantly found in XI3-C landraces in Laos, Cambodia and Thailand (Fig. 1e, supplementary extended data S2, Supplementary Material online). We investigated the geographical association of this variety further with individual-level genomic distance analyses based on shared-IBS. We found that the XI3 Maroon Hmong variety falls in a cluster that is characterized by a high level of shared-IBS among one variety from Thailand (Khoa Nahng Prung/Dawk Lao), one from Myanmar (Pakistanhmwe) and three from Tanzania (ES-21; Moshi, Kilombero) (supplementary fig. S14 and extended data S4, Supplementary Material online). This cluster falls among a much larger diversity of XI3-C landraces from Thailand, Laos and Cambodia. In the admixture graph the Maroon Hmong and varieties from Tanzania are placed on the same sister branch to XI3-C Asia (predominantly varieties from Laos, Thailand and Cambodia) (supplementary fig. S13, Supplementary Material online). These findings suggests that the Maroon Hmong variety, as well as the very closely varieties from Tanzania and Myanmar, most likely all ultimately have derived from XI3-C varieties in Thailand, Cambodia or Laos.
Reconstructing the Historical Contexts of Rice Introductions to the Guianas
To interpret the geographical associations inferred from the Maroon rice group ancestry profiles, we placed them in a historical perspective, informed by ethnobotanical and archival investigations. This allowed us to reconstruct the historical contexts associated with the introduction of rice varieties to the Guianas. Taken together, we can link the origin of the Maroon rice varieties to at least four different historical events (Fig. 3a).
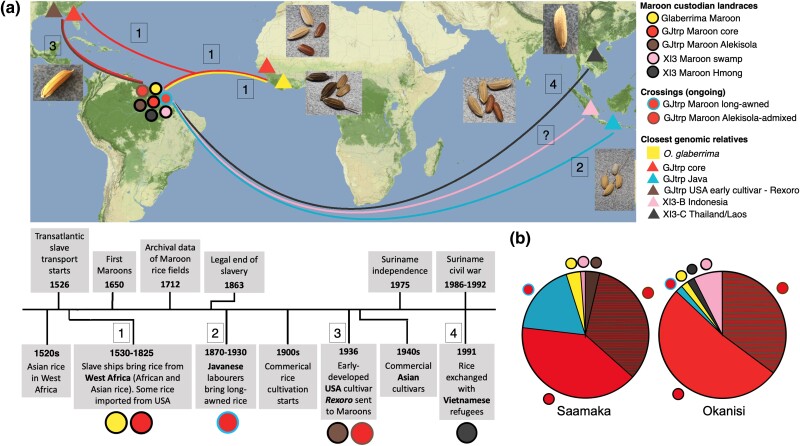
Fig. 3.
Maroon farmers have preserved rice varieties from at least four introduction events to the Guianas during (post-)colonial history. a) Summary map and timeline showing the geographical connections and chronology of the historical contexts associated with the Maroon rice genomic groups. [1]: Tropical japonica and African “black” rice (O. glaberrima) varieties from West Africa during the transatlantic slave trade (1526 to 1825). [2]: Long-awned tropical japonica rice by indentured laborers from Java (1890 to 1980). The Javanese ancestry is only detected in admixed form in the Maroon varieties. [3]: Tropical japonica USA cultivar Rexoro (1936 onwards). [4]: An upland indica-3 variety from Hmong refugees (1991). [?]: Cottica Maroons receive deep water indica-3 landraces that ultimately come from Indonesia. Basemap: © Stadia Maps. b) Relative proportions of the various Maroon rice genomic groups observed among Saamaka and Okanisi farmers. Despite their preference difference for certain rice traits, farmers of both Maroon groups cultivate varieties from nearly all genomic groups, demonstrating a shared motivation for maintaining crop diversity.
Transatlantic Slave Trade (1650 to 1825)
Firstly, we posit that the similar ancestry found in the landraces of the GJtrp Maroon core group and varieties in other regions of the Americas and West Africa reflects a relict genomic signature that traces back to the transatlantic slave trade. Intensive trade networks connected these areas during colonial times, starting in the early 16th century (Eltis and Richardson 1995). (Tropical) japonica varieties from the Philippines and Malaysia were introduced by the Portuguese to West Africa around 1520 (Nawani 2013; Gilbert 2015), and shipped from West Africa together with enslaved Africans to Brazil from the 1530 s onwards (Carney 1998) (a review on the history of rice dispersal is provided in supplementary Text S1, Supplementary Material online). In addition, both Maroon oral accounts and colonial archival records provide evidence of rice imports from West Africa to the Guianas (Fleury 1994; Carney 2005; Van Andel 2010). Dutch archival records about rice in Suriname, dating from 1639 to 1719, discuss the planting of the cereal as bulk food for soldiers and the enslaved Africans on the plantations (supplementary Text S2, Supplementary Material online: Evidence #1 to 4). The archival record also indicates that different types of rice were present in Suriname in the late 1600s and explicitly mention white rice, suggesting O. sativa (Asian rice), and black-husked rice with a red pericarp, suggesting O. glaberrima (African rice) (supplementary Text S2, Supplementary Material online: Evidence #1). According to the Maroons, among their oldest rice landraces are those that are named after specific female ancestors or early runaway groups that brought rice to the Maroon villages (Pinas et al. 2023). The genomic analyses show that these varieties, e.g. named Ma Paanza, Alisi Seei, Baakapautjaka and Agbosó, fall either in the GJtrp Maroon core or GJtrp Maroon Alekisola-admixed group (supplementary extended data S2 and S3, Supplementary Material online). The landraces in the GJtrp Maroon core group have a large trait diversity (supplementary extended data S1e, Supplementary Material online). Several retain natural defense traits associated with wild rice, which allows the Maroons to farm them with little maintenance. In addition, these landraces typically have a short maturation period of circa three to four months (Van Andel 2010; Meyer et al. 2016; Van Andel et al. 2019; Pinas et al. 2024). According to the Maroons, this made these rice varieties particularly suitable during marronage when frequent harvesting was necessary for unpredictable resettlements to escape recapturing efforts by the colonial authorities (Van Andel 2010).
Another rice group for which we can link the origin to this time in history are the four landraces named baaka alisi and matu alisi in the Glaberrima Maroon group. The genomic analyses confirmed that these are indeed varieties of African rice (O. glaberrima). O. glaberrima was domesticated from wild O. barthii ∼3,000 yr ago along the Niger River (Meyer et al. 2016), and except for Maroon communities (Van Andel et al. 2016a) is nowadays cultivated only in Africa. Around 1700, O. glaberrima likely was also cultivated in the Carolinas (USA) and elsewhere in the Americas, grown from seed stock that had been taken aboard of slave ships (Carney 2001). After the introduction of mechanical milling devices in the 18th century, O. sativa replaced O. glaberrima as an export crop (Carney 2001, 2005). Hence, the O. glaberrima varieties most likely were a part of the Maroon rice stock before this. Similar to the varieties in the GJtrp Maroon core group, the Maroon O. glaberrima varieties have a short maturation time of three to four months and retain many natural defense traits (Van Andel 2010; Pinas et al. 2024). Taken together, the genomic ancestry analyses, rice traits, and Maroon nomenclature all underline that the GJtrp Maroon core and Maroon O. glaberrima landraces have their origin in the time of transatlantic slave trade and marronage. Congruent with Maroon oral history and historical written documentation, we pose that these landraces represent a part of the oldest Maroon rice stock.
Javanese Indentured Laborers (1890 to 1930)
Secondly, we posit that the characteristic Java ancestry in the GJtrp Maroon long-awned varieties resulted from admixture with landraces that were brought to Suriname by Javanese indentured laborers. After the abolishment of slavery in 1863, the Dutch tried to keep their plantation economy in Suriname viable by contracting ∼70,000 laborers from India and Java (Hoefte 1998).These contract laborers brought along crops from their home areas, including rice, and played an important role in the emergence of a commercial rice sector in the early 1900s. After their contract ended, the majority of the Asian workers settled in Suriname as smallholders along the coast and continued cultivating rice for subsistence (Maat and Van Andel 2018). Young Maroon men, searching for wage labor in coastal Suriname, also ended up working on these farms (Price 1993). Given that Maroon farmers bought tubers from Javanese farmers (Van Andel et al. 2016a), a similar exchange of rice varieties between Javanese and Maroons is plausible. Alternatively, the Maroon long-awned rice may descend from early cultivars that resulted from crossing experiments at a breeding station. In the early 1900s, the colonial Department of Agriculture imported new rice types into Suriname to support rice farming, also from Java (Maat and Van Andel 2018). Archival records indicate that at least one Javanese variety, Boeloeh poetih, was multiplied on the plantation Guineesche-Vriendschap (Guinean Friendship) in Suriname because of its popularity (Maat and Van Andel 2018). In 1949, the Dutch government created the Stichting Machinale Landbouw (SML) that in cooperation with the Van Dijk company conducted rice breeding programs in Suriname. Their archival records indicate that experimental crosses included Javanese and Maroon rice (Van der Meulen 1941; Maat and Van Andel 2018). These improved rice cultivars were often distributed to farmers (Van der Meulen 1941; Maat and Van Andel 2018), possibly also to Maroon farmers. It is important to note, however, that awns were among the major traits that commercial rice breeders started selecting against during the Green Revolution (Hua et al. 2015; Svizzero et al. 2019; Amarasinghe et al. 2020; Luong et al. 2022).
Our ancestry analyses cannot discern if the crossing(s) that resulted in the Maroon long-awned rice were done by the Maroons, early breeders in Suriname or elsewhere. However, complementary insights are provided by the Maroon rice naming system. If the origin story of a particular rice landrace is known to the Maroon farmer, they typically name that rice type after the (female) ancestor, Maroon or other ethnic group that they received the rice from (Pinas et al. 2023). The Maroon names for the long-awned varieties all refer to the awn phenotype—i.e. through analogies with animal characteristics, such as the long hair of the bush pig (e.g. pakia), stiff whiskers of the river otter (watadagu) or the soft cat's tail (puspusi) (supplementary extended data S1e, Supplementary Material online) (Pinas et al. 2023). Since none of the names for the long-awned varieties refer to Javanese people, this suggests that the Maroons most likely did not obtain these rice varieties from Javanese farmers directly.
USA Early Rice Breeders (1932 Onwards)
Thirdly, we posit that the Maroon Alekisola varieties are locally adapted varieties of the early cultivar Rexoro that was developed by rice breeders in 1926 in Louisiana (Khush and Brar 2008). Rexoro became one of the most popular and economically lucrative rice varieties of that time and was shipped to various places over the world for trade and breeding experiments (Lu, et al. 2005). The genomic analyses indicate that the Alekisola varieties are more closely related to Rexoro than to another early-developed (USA) cultivar or landrace in our dataset. A notable exception is the Bhotohavana Mena variety from Madagascar, which shows a comparable close genetic similarity to Rexoro. As such, based on the genomic results alone we cannot determine whether the Alekisola varieties descent from Rexoro that was sent directly from the USA, or from other early cultivars closely related to Rexoro elsewhere in the world. However, historical records indicate that Rexoro rice was imported from Louisiana to Suriname's agricultural experimental station in the spring of 1932 (Stahel 1932). In addition, there is also written evidence from the director (Stahel) of the Suriname agricultural experimental station that “a bale of Rexora seed padi” was sent to the “Saramaccan village Ganzee in 1936” (Stahel 1944). Additional support for a direct introduction of Rexoro to the Maroon community is preserved in the Maroon rice naming system. Maroons often switch the “R” into an “L”. Hence, Lexola, Alekisola and Sola are Maroon name versions for rice that was named Rexoro/Rexora by early breeders in Louisiana (Pinas et al. 2023).
Hmong Refugees in French Guiana (1991)
Fourth, we posit that the upland indica-3 variety that the Maroons refer to as Hmong was obtained directly from the Hmong, as reflected in its rice name. The Hmong are an ethnolinguistic group from southern China and the northern mountains of Laos, Vietnam, and Thailand. During the Vietnam War, many Hmong fled their native area and some settled in French Guiana around 1977 where they had their own farms (Clarkin 2005). Our genomic analyses indicate that the Maroon Hmong variety, together with three closely related varieties from Tanzania and one from Myanmar, have an ancestry profile that is characteristic for XI3-C landraces from Thailand, Laos and Cambodia. This geographical association is in line with the possibility that Hmong farmers brought this rice variety with them during their migration to French Guiana. However, since we could not find historical records that mention that the Hmong people migrated to Tanzania or Myanmar, it is also possible that these varieties were distributed through rice breeding stations.
Additional insights for the origin of the Maroon Hmong rice variety are provided by first-handed recollections by several of the elder (60+ years) Okanisi Maroon farmers in the Cottica region (Pinas et al. 2023). They explained that they had spent time as refugees in French Guiana during the civil war in Suriname between 1986 and 1992. Maroons were not allowed to grow their own food in the French refugee camps, but worked illegally on Hmong provision fields that were situated close to the refugee camps (Hoogbergen and Polimé 2002). After the civil war, several Maroons returned to the Cottica. The Maroon elders said they had lost many rice varieties because of the war, but they also received some new ones, like the Hmong rice. They said they also called this rice “anambu” (egret) or “rice of the water Chinese” and explained that they chose this name “because the Hmongh in French Guiana live in houses on long stilts above the swamp, just like egrets stand with their long thin legs in the swamps” (Pinas et al. 2023). Taken together, we posit that the Maroon Hmong rice was indeed obtained directly from the Hmong, and that the closely related varieties in Tanzania and Myanmar were likely exchanged through breeding stations.
The Origin of the Maroon Swamp Varieties Remains a Mystery
Lastly, we found that the historical context for how the Okanisi Maroons in the Cottica obtained the swamp landraces is difficult to establish. Some farmers said they received the swamp rice from Creoles, descendants of enslaved Africans who became smallholder farmers after emancipation and were addressed respectfully as masaa (master) (Pinas et al. 2023). Other farmers said they received this rice from city Creoles with a high social position, who are also addressed as kioo (Creole) or masaa, and who visited Maroon villages as teachers, missionaries, and agricultural extension officers in the period 1930 to 1980. Our genomic analyses show that the ancestry profile of the deep water indica-3 landraces is most similar to that of XI3-B landraces in Indonesia. How the genomic ancestry of the swamp varieties connects to the first-hand testimonies by Maroon farmers for how they obtained them, remains to be resolved by future genomic, ethnobotanical, and archival investigations.
Saamaka and Okanisi Maroon Rice Diversity and Trait Preferences
We found that farmers from both the Saamaka and Okanisi Maroon community cultivate rice varieties from all genomic groups, except for the single indica-3 Hmong variety which is cultivated only by the Okanisi (Fig. 3b). Hence, the farmers from both communities are similarly focused on creating and maintaining rice diversity. A crucial Maroon cultural practice for maintaining this rice diversity is the seed exchange of successful and desired varieties. This mostly takes place between families, neighbors, and friends. Village ceremonies, such as funerals, are typical events where gifts and tokens are exchanged, including seeds (Van Andel et al. 2019; Pinas et al. 2023).
Moreover, we also found that the Saamaka and Okanisi farmers have some distinct preferences for certain rice traits that can be attributed to differences in farming practices and adaptation to local agroecology. Several Okanisi rice varieties are named alulu (“shatterer”) and shatter easily, which facilitates their way of threshing in which they tread barefoot on the panicles (supplementary extended data S1e, Supplementary Material online) (Pinas et al. 2023, 2024). In contrast, the Saamaka thresh with a wooden mortar and pestle, and tend not to grow shattering varieties, except for the Maroon O. glaberrima varieties. In addition, a larger number of Saamaka varieties have awns (Fig. 3b), which may better protect against bird predation (Hua et al. 2015; Svizzero et al. 2019; Pinas et al. 2024). Since the Saamaka do not thresh barefoot, the awns do not bother them. Lastly, most rice varieties found with the Maroons are upland rainfed tropical japonica types, but Okanisi that live near the Cottica, the deepest river in Suriname, also farm several deep water indica-3 swamp varieties (Fig. 3b).
Discussion
Here, we posit that a substantial part of the Maroon rice stock diversity traces back to the transatlantic slave trade from West Africa and marronage, and includes both Asian rice (tropical japonica landraces with core ancestry) and African rice (O. glaberrima landraces). The Maroon farmers acquired and developed other rice types after emancipation, by including rice that was introduced to the Guianas by contract laborers from Java, early rice breeders in the USA and Hmong people that fled the Vietnam War.
Archival records indicate that laborers from India also brought rice with them to the Guianas (Maat and Van Andel 2018). Congruently, some Maroon farmers attested that their grandparents obtained several varieties named kuli alisi (“coolie” or “Hindustani rice”) and watralanti (‘water land’) from Hindustani people (Ramdayal et al. 2021). However, the kuli alisi and watralanti varieties in our dataset fall in the GJtrp Maroon core and Alekisola-admixed groups, and show no indication that they share ancestry with landraces in India. Possibly, these are rice varieties from Maroon farmer fields that ended up in Hindustani-run agricultural shops and were named “Hindustani rice” by the Maroon buyers.
Moreover, additional layers of complexity may be hidden in the diversity of the GJtrp Maroon Alekisola-admixed group. The varieties in this group are highly heterogenous in terms of genomic ancestry, traits and Maroon rice names. Although the ancestry of this group on average is best approximated as admixed between local Maroon tropical japonica core and Alekisola ancestry (Fig. 3b), there may be additional population substructure. A subset of varieties in the GJtrp Alekisola-admixed group seem relatively more closely related to landraces from Brazil and some early-registered cultivars from South America, as well as some landraces from West Africa and Madagascar (supplementary extended data S3, Supplementary Material online). It is possible that these varieties share an additional ancestry that falls outside of the tropical japonica core diversity, and that in our (ADMIXTURE) analyses appears as Southeast-Asian ancestry. Notably, the Portuguese were shipping rice from Malaysia and the Philippines via Madagascar to West Africa, and from there to Brazil as early as 1530 (Nawani 2013) (supplementary Text S1, Supplementary Material online). When the Portuguese expelled the Dutch from Brazil in 1654, some 100 Jewish planters moved to Suriname, bringing along their plantation inventory and workforce (Carney 2005; Vink 2010). The additional Southeast-Asian ancestry that is observed in rice varieties from these regions, including perhaps in a subset of the GJtrp Maroon Alekisola-admixed varieties, may reflect this additional rice dispersal event during early colonial history. Additional genomic data for (historical) rice landraces from these regions will be crucial for investigating this hypothesis further.
The results of our integrated research approach indicate that the Maroons farmers conserve but are not conservative in their farming practices. Notably, among both the Saamaka and Okanisi farmers the comparatively recent Alekisola(-admixed) and indica varieties never fully replaced the older tropical japonica landraces that represent the Maroon communities’ roots to West Africa. The genomic analyses also identified types of rice never documented before, i.e. long-awned tropical japonica varieties with Java ancestry and some of the Alekisola-admixed tropical japonicas. This shows that Maroon farmers are not only avid collectors but also active selectors of new rice varieties, derived from intentional and/or spontaneous crosses, to further increase their stock diversity. The varieties grown by the Maroons are continuously evolving in tandem with their environment. Based on the genomic ancestry characterization of the Maroon rice groups, we propose to classify the Maroon varieties in two landrace categories. Some varieties are so-called allochthonous landraces (Zeven 1998) and descent from landraces that were locally adapted to a foreign region before being introduced in the Maroon farmer fields and continuing their local adaptation there. Among the allochthonous landraces are the tropical japonica varieties with core ancestry and most likely also the Hmong indica-3 variety. Other varieties (partly) descent from non-locally developed early cultivars that were introduced to the Maroon farming system. These varieties have regained landrace traits over time and in the Maroon farmers’ fields agroecologically behave like the allochthonous landraces. These include the Alekisola(-admixed) tropical japonica varieties, and possibly the tropical japonica long-awned and the indica-3 swamp varieties.
Since the Green Revolution in the 1960s a significant proportion of landraces has disappeared from farmer's fields worldwide, resulting in severe genetic erosion (Zeven 1998). Largely ignored by the agricultural modernization initiatives of the colonial government and the independent Surinamese state, the Maroons have followed their own trajectory of landrace conservation and agricultural improvement. We posit that the high landrace diversity currently grown by Maroons reflects the farming expertise they inherited from their (African) ancestors. Enslaved Africans were often allowed to grow their own food on small provision fields (Fleury 1994; Van Andel et al. 2016a) and may also have planted rice as an anti-commodity crop (Maat 2015), hidden between other crops as an act of protest against the slave-based plantation system and in memory of their African heritage (Carney 2005; Maat and Van Andel 2018). After their escape to freedom and once safely settled in the forested interior, Maroons were free to invest time and space into the farming systems of their choice, multi-cropping landraces of various familiar African food crops (e.g. rice, okra, plantains) with crops of indigenous origin like cassava, tobacco, and sweet potato (Price 1991; Van Andel et al. 2016a).
This study demonstrates the long history of the (rice) farming expertise of the enslaved Africans and Maroons, and more recently also the Asian contract laborers and Creoles, in the Guianas and elsewhere in the Americas. Colonial authorities may frequently have overstated their involvement in the discovery and development of productive rice varieties and farming innovations that were economically lucrative (Carney 2001). Early archival documents on the rice crop in Suriname and Brazil just mention “rice that grows here” (supplementary Text S2, Supplementary Material online: Evidence #1 to 2) and remain silent on those who found the seeds and knew how to plant and harvest them. However, the fact that colonial authorities in the Guianas sent out military raids for almost two centuries to destroy Maroon provision fields (Dragtenstein 2002) (supplementary Text S2, Supplementary Material online: Evidence #5) attests to the vast knowledge of Maroon farmers to establish highly productive farming systems in different ecosystems. Therefore, this study provides a case study that demonstrates the power of cross-disciplinary crop genomics research to obtain a more comprehensive perspective on a time of the human past for which the historical record may be biased or incomplete. Similar studies can be envisioned for landraces of other cultural heritage crops of (Indigenous) communities who may have preserved their history on their own farms, to both reconstruct and honor the past.
Materials and Methods
Ethnobotanical Surveys
Fieldwork on Maroon rice farming was carried out in French Guiana and Suriname in 2017, 2021, and 2022. Before the start of each fieldwork period, we obtained permission from the traditional Maroon authorities and prior informed consent from each rice farmer. In addition, we obtained consent and permission of the Suriname National Rice Research Institute (SNRI/ADRON). We interviewed 99 Maroon rice farmers (21 in 2017, 67 in 2021 to 2022 11 in 2023 to 2024, almost all women), in Dutch, French, or one of the Maroon languages. We asked questions about the rice varieties grown, their local names, the meaning of these names, specific varietal traits, farmers’ motivation for rice cultivation, specific rituals involving rice, and oral history associated with rice and its origins (Van Andel et al. 2019; Pinas et al. 2023; Van Andel et al. 2023; Pinas et al. 2024).
Rice Samples
Our primary focus was to collect as much Maroon rice diversity as possible, rather than to obtain an accurate account of the diversity preserved by individual farmers. Hence, when we encountered a rice variety with morphological characteristics and a Maroon name identical to that of a variety we had already collected, we did not sample it again. Herbarium vouchers of rice plants were collected using standard botanical methods. Duplicates were deposited at the Herbier IRD du Guyane (CAY) in Cayenne (French Guiana), the National Herbarium of Suriname (BBS) in Paramaribo, and at Naturalis Biodiversity Center (L) in Leiden, the Netherlands. Living seeds of most varieties have been deposited with the associated data on locality, varietal information, and farmer's name at the Suriname National Rice Institute (SNRI/ADRON) at Nickerie for further propagation, phenotyping, and germplasm storage. Dead seed samples were stored at the Naturalis Economic Botany Collection. For each collected variety, we reported morphological and agronomical characteristics.
Historical Data Collection
The digital archives of the Dutch West India Company and the Society of Suriname, both housed by the National Archives in the Hague, the Netherlands, were queried for the search terms “rice”, “rijst”, “rys(t)”, “reis”, “kost” (provision), “gronden” (grounds). A literature survey was carried out on rice cultivation in the Guianas and Dutch Brazil during the early colonial period, and archival records mentioned in these studies were verified.
Sequencing Data
DNA extracts and libraries for the rice specimens were generated from leaf, stem, or seed material from living rice plants that were either collected in the farmer's field or fresh leaf from germinated seeds in the Netherlands (supplementary extended data S1a, Supplementary Material online). Preferably, we used leaf material when available. The tissue samples were dried with silica gel beads and stored in dark containers for conservation. The tissue samples were ground using the Retsch Mixer Mill MM400 while frozen in liquid nitrogen. For each rice specimen, a DNA extract was generated from typically 10 to 20 mg powder with a standard CTAB-based method (https://openwetware.org/wiki/Maloof_Lab: 96well_CTAB). We added 2% PVP-40 to the CTAB buffer to remove phenols and polysaccharides. DNA yields in the extracts were 12 to 140 ng μl−1. Then 5 uL of (normalized) DNA extract was converted into double-stranded DNA libraries with unique dual indexes (supplementary extended data S1a, Supplementary Material online) using tagmentation with an in-house protocol for the Illumina DNA Prep kit (formerly Nextera DNA Flex). Our in-house protocol is based on a previously published “hack” protocol (Gaio, et al. 2022), but we added normalization steps before and after tagmentation to balance the DNA concentration, hence raw sequence read output, among the rice accessions. The hack protocol allows us to use the Illumina kit for 1,450 instead of 24 library preps. Sequencing was done on the Illumina Novoseq6000 platform with 2 × 150 bp paired-end read configuration to a target genome-wide sequencing depth of ∼14× at the Core Facility of Novogene Europe in Cambridge or GenomeScan in Leiden. The sequenced reads were demultiplexed according to the expected index pair for each library, allowing one mismatch per 8 bp index. Sequencing data for these accessions are available from the SRA under Bioproject accession number PRJNA1019116.
Read Processing, Quality Control and Genotyping
The grenepipe v0.7.0 snakemake pipeline was used for read processing, quality control and calling SNPs (Czech and Exposito-Alonso 2022) (https://github.com/moiexpositoalonsolab/grenepipe). Prior to starting the read processing pipeline, the fastq format files from different sequencing runs for the same library were merged. We used fastp v0.20.0 (Chen, et al. 2018) to clip adapters, Ns stretches and PolyG tails of the reads. We merged paired-end reads into a single sequence for regions with a minimal overlap of 30 bp, and only kept merged sequences ≥55 bp. Sequence reads were mapped against the Nipponbare IRGSP-1.0 reference genome (Kawahara et al. 2013), a temperate japonica rice cultivar, using the global aligner BWA v.0.7.17 in “mem2” mode (Vasimuddin et al. 2019) producing .bam files. The mapped reads were sorted, indexed, and filtered (mapQ 60) using samtools v1.3 (Li et al. 2009). Duplicate reads, identified by comparing sequences in the 5′ positions of both reads and read pairs, were removed with MarkDuplicates in PICARD v2.23.0 (http://broadinstitute.github.io/picard/). The .bam files were used to call diploid haplotypes across all rice accessions with Freebayes v1.3.1 (Garrison and Marth 2012), parallelized for contigs (e.g. chromosomes), while forcing allele calls at 29 million known SNP positions (https://snp-seek.irri.org/_download.zul) using the option –haplotype-basis-alleles combined with BEDTOOLS v2.30 (Quinlan and Hall 2010) for region intersection (bedtools intersect -a). These 29 million SNP positions were previously validated as segregating among >3,000 rice accessions of the 3K-RG project for reads mapped to the Nipponbare IRGSP-1.0 reference genome and include the 404k core set. The VCF files for each contig were concatenated, indexed and compressed with PICARD MergeVcfs, resulting in a single VCF file with raw genotype data for a set of 29 M genome-wide SNPs across all Maroon rice accessions. From this raw set of SNPs we removed all non-biallelic sites. Subsequently, with GATK v4.1.4.1 (McKenna, et al. 2010) we subjected it to a series of hard filtering steps (−filter-name snv-hard-filter) to remove SNP sites with low-quality genotypes normalized by depth (QD < 2.0), mapping quality (MQ < 60.0 and MQRankSum < −12.5), read position bias from Wilcoxon's test (ReadPosRankSum −8.0) and strand bias from Fisher's test (FS > 60.0). Using an in-house python script we extracted the genotypes for all SNP positions in the 3K-RG 404k core SNP panel (https://snp-seek.irri.org/_download.zul), and transformed the VCF file into packedped files with PLINK v1.90b (Purcell et al. 2007; Chang et al. 2015), and subsequently into eigenstrat files using convertf from the EIGENSOFT package v7.2.0 (https://github.com/DReichLab/EIG).
Comparative Dataset
We made a core comparative dataset for global rice landrace diversity that we based on the 3K-RG 404k CoreSNP Dataset in PLINK format provided by the Toronto International Data Release Workshop Authors, available via https://snp-seek.irri.org/_download.zul. The 404,388 biallelic SNPs in the 3K-RG 404k CoreSNP panel were selected to optimally discern between the major rice genomic groups of Asian (O. sativa) rice, as well as discern some (eco-)geographical structure among rice varieties within each of the major rice genomic groups. From the 3,024 rice accessions in this 3K-RG 404k CoreSNP Dataset we selected the rice landraces from Asia (n = 1,064) for which landrace status was previously validated by Gutaker et al. (2020). In addition, using a similar approach to Gutaker et al., we selected 3K-RG rice accessions from Africa (n = 97) and the Americas (n = 11) that in Genesys (https://www.genesys-pgr.org) were annotated with “traditional” status or as an inbred lineage descendent thereof, to validate them as putative landraces in downstream analyses. To expand the comparative dataset further we downloaded sequencing data for USA early-registered cultivars (Vaughn et al. 2021) (n = 12 [SRA project accession PRJNA603026]), O. glaberrima varieties from Africa (Meyer et al. 2016) (n = 20 [PRJNA315063, PRJDB4713]) and from Suriname (Van Andel et al. 2016b) (n = 1 [PRJNA514989]). Moreover, we downloaded sequence data for representative varieties for diploid wild Oryza species (AA genomes) found in the Americas (O. glumaepatula: n = 20 [PRJDB4703], O. longistaminata: n = 9 [PRJDB4705]), Africa (O. barthii: n = 8 [PRJNA30379]), and Asia (O. nivara: n = 6 [PRJNA202926]), O. rufipogon (Huang et al. 2012): n = 6 [PRJEB2829]). The sequencing data was downloaded in fastq format from the Short Read Archive (SRA) using the FASTQ-DUMP tool v.2.8.2 with the option to split reads into forward, reverse and trimmed. Then the reads were processed and genotyped following the same pipeline as the newly sequenced Maroon rice varieties. The resulting 404k SNP genotype datasets were merged with that for the Maroon rice, and subsequently with that of the (candidate) landraces extracted from 3K-RG 404k CoreSNP Dataset.
SNP Filtering
From the 404k core dataset, we filtered out SNP markers with PLINK v.1.90b (Purcell et al. 2007; Chang et al. 2015) that were determined as outliers with the 1.5xIQR rule (more extreme than the 3th Quartile + 1.5 * IQR) regarding genotype missingness (Fmiss) and heterozygosity ratio (HetRateObs). The latter is to remove SNPs that we expect are the result from spuriously mapped reads that are from tandem duplicated genomic regions (Gutaker et al. 2020). These filtering steps were applied to the full dataset, as well as separately for (1) Maroon landraces (2) indica landraces, (3) japonica landraces, (4) circum-Aus landraces, and (5) O. glaberrima/wild Oryza species. Genotype missing rates for markers (Fmiss) were determined with –missing, and SNPs with excess rates (typically Fmiss > 0.25) were removed from the dataset with –geno. To determine the threshold for an excess heterozygosity ratio, we first calculated the genotype counts per SNP with PLINK using –freqx. From the data in the .freqx file we subsequently calculated the heterozygosity ratio per SNP, here defined as: HetRateObs = C(Het)/(C(Hom A1) + C(Het) + C(Hom A2)). This corresponds to the heterozygosity count divided by the total genotype count (not including missing genotype counts). The IDs of SNP markers with outlier heterozygosity rates (typically > 0.20) were collected in a bad_snp_list, and subsequently removed from the dataset with –exclude. Then the SNP set was subjected to a two-step LD pruning procedure. The first step was carried out with the INDEP-PAIRWISE function in windows of 10 kb with variant shift = 1 and r2 = 0.8. The second step was carried out with the same function in windows of 50 variants. These filtering steps resulted in 109,203 highly informative SNPs (“110k panel”). In contrast to the 404k core panel (supplementary fig. S2a, Supplementary Material online), the calling rates for non-missing genotypes in the 110k panel are similar for the Maroon (95.0 ± 4.3%) and 3K-RG rice landraces (97.0 ± 4.3%) (supplementary fig. S2b, Supplementary Material online). The average number of reads supporting a SNP genotype call (genotype depth) for the Maroon varieties is 10.3 ± 6.1 × (mean ± SD), which overlaps with the 14.4 ± 6.9 × mean genotype depth for the 3K-RG project accessions (Wang, et al. 2018) (supplementary fig. S2c, Supplementary Material online). We caution that, like the 404k core panel, also the 110k core panel may not be as effective in resolving genomic affinities with non-sativa rice species, because a relatively large fraction of the SNPs is non-segregating.
Individual Filtering
We removed rice varieties from the dataset with high marker missingness, using –remove. Like the SNP filtering steps, thresholds for outliers were based on the 1.5xIQR rule and determined separately for the different subsets of data. In addition, we filtered out landraces with mixed ancestry based on their joint genetic clustering in PCA and ADMIXTURE. These in general include, but were not limited to, 3K-RG varieties labeled as admix, GJadm or XIadm, as well as some of the 3K-RG rice varieties from Africa and the Americas that we had selected as candidate landraces.
Population Assignment
Clustering
We conducted an unsupervised genomic clustering of global rice diversity using ADMIXTURE v.1.3.0 (Alexander et al. 2009). To reduce the impact of different inbreeding levels among the rice varieties on the clustering results we reduced diploid to haploid calls by randomly sampling one allele. We ran ADMIXTURE with fivefold cross-validation varying the number of ancestral populations between K = 2 and K = 20 in 100 bootstraps with different random seeds (supplementary extended data S1d and S2, Supplementary Material online). The model did not converge to a global optimum (lowest cross-validation error) and continued to marginally improve with increasing k-means because of further breakdowns in geographical structure (supplementary fig. S15, Supplementary Material online). This results from the rationale behind the design of the SNP panel to capture a high proportion of common variance within each of the 3K-RG major genomic groups to discern additional geographical structure. As such, the ADMIXTURE model starts to discern broad ecotypes within indica-3 (from K = 7) and tropical japonica (from K = 8) varieties before it has discerned the nine major rice genomic groups as defined in the 3K-RG project (at K = 12). From K = 16 the breakdown into increasingly smaller ecotype subsets becomes detailed to a level that is no longer meaningful for the global ancestry characterization of rice varieties at a population level.
To obtain an alternative indication for the optimal number of genomic clusters we evaluated our dataset using PAM with the fviz_nbclust() function from the “factoextra” package (Kassambara and Mundt 2020) in R for k-values in the range 2:20. We limited this analysis to O. sativa varieties. The PAM-based clustering tendency was assessed both with average silhouette scores (“silhouette”) and gap statistic (“gap_stat”) (supplementary figs. S16a and b, Supplementary Material online). The former method showed a global optimum at K = 3, and a local optimum at K = 13. We used the pam() and fviz_cluster() functions to plot a multidimensional scaling plot (MDS) based on Euclidian distanced to visualize clusters for K =13, and fviz_silhouette() to generate silhouette plots for cluster validation (supplementary fig. S16c and d, Supplementary Material online). Details on the silhouette scores for individuals and average silhouette scores for assigned PAM-clusters for K2-20 are provided in supplementary extended data S1c, Supplementary Material online. The 13 PAM-clusters show a clear geographical association (supplementary figs. S17 and S18, Supplementary Material online). Taken together, we chose K = 15 as our preferred local optimum for the ADMIXTURE model since it is the lowest k-value that includes the thirteen clusters also discerned with the PAM-based method (supplementary table S4, Supplementary Material online). The two additional ancestry components discerned at K = 15 in ADMIXTURE maximize an ancestry component in the O. glaberrima, O. barthii and O. longistaminata varieties (these were excluded from the PAM-based analyses), and a second ecotype among circum-Aus varieties, respectively.
Grouping and labeling
We grouped rice landraces that were collected in the same country and with a similar ADMIXTURE ancestry profile (and assigned PAM cluster) in a single population. As such, we followed a similar approach for population assignment as in human population genomic studies based on ancient DNA, which frequently rely on small population sizes or even single individuals as proxies for the ancestry of human groups associated with an archaeological context at a given area and moment in time (Sikora et al. 2019; Mao et al. 2021; Posth et al. 2023; Villalba-Mouco et al. 2023). We considered an ADMIXTURE ancestry profile to be unadmixed when less than 5% ancestry is assigned to another ancestry component.
The varieties in the GJtrp Maroon core, GJtrp Maroon Alekisola, and XI3 Maroon swamp and XI3 Maroon Hmong groups fall in different PAM-clusters and show unadmixed ancestry profiles in ADMIXTURE for distinct components (supplementary table S1, Supplementary Material online). The grouping of the varieties in the GJtrp Maroon long-awned and GJtrp Maroon Alekisola-admixed groups is based on the similarity of their admixed ancestry profile in ADMIXTURE, as well as similarities in morphological and agronomic traits, and Maroon rice names (supplementary table S1, extended data S1e, Supplementary Material online).
Discretization
We also made a “discretized 110k” dataset for the f-statistic tests and admixture graph analyses in which we filtered out accessions from the rice groups that cluster poorly, using the DISCRETIZE algorithm in R provided by Gutaker et al. (2020) (for method details, see supplementary Text S3, Supplementary Material online). This approach filtered out around half of the O. sativa landraces, including some smaller populations in the Americas and Africa that are central to our research questions (supplementary extended data S1a, Supplementary Material online). Hence, throughout the manuscript we report the f-statistics for the unfiltered groups. However, a reanalysis using the discretized populations does not affect the conclusions, and all f-statistics are reported in supplementary extended data S5, Supplementary Material online.
Principal Component Analyses
PCA analyses were run with the program smartpca from the EIGENSOFT package v7.2.0 (https://github.com/DReichLab/EIG) on the global diversity of O. sativa landraces, as well as on the subsets of tropical japonica and indica-3 landrace diversity. In the PCA for global O. sativa diversity the O. glaberrima and wild Oryza varieties were projected on the PCs with the option “lsqproject: YES”. The variance explained by each principal component (PC) was calculated as the dimension's eigenvalue divided by the sum of all positive eigenvalues, and the first two PCs were plotted.
Individual-Level Pairwise Genomic Distances
Genomic distances within O. glaberrima, tropical japonica, and indica-3 rice subgroups were calculated between all pairs of rice landraces with formulation 1-DST. DST was determined with the –genome and –genome-full options in PLINK v1.90b (Purcell et al. 2007; Chang et al. 2015), which is calculated from IBS measurements: DST (IBS distance) = IBS2 + 0.5 * IBS1)/(IBS0 + IBS1 + IBS2). Heat plots of 1-DST matrixes were made with the pheatmap() function from the pheatmap v1.0.12 package in R, which relies on the hclust() function from the stats package for hierarchical clustering.
F-statistics
For ancestry analyses on the population level, we used various f3 and f4-statistics methods (Reich et al. 2009; Patterson et al. 2012; Raghavan et al. 2014). Standard errors (SE) were estimated using the 5 cM block jackknife method (Busing et al. 1999) as implemented in the qpDstat and qp3Pop programs in the ADMIXTOOLS v7.0 package (Patterson et al. 2012). We define statistical significance as [−2.576 < Z < 2.576], corresponding to a 99% confidence interval (CI). For a more detailed description of the various f-statistics, see supplementary Text S3.S1, Supplementary Material online.
Admixture Graphs
To investigate the topological positions of the various Maroon rice groups, we made separate admixture graphs for relevant worldwide tropical japonica and indica-3 groups. We generated the admixture graphs with the ADMIXTOOLS 2 package in R (Maier, et al. 2023). For this analysis, we used the 110k discretized dataset and pooled some of the rice groups into meta-groups that have similar population ancestries and are from the same broader geographical region (supplementary table S5, Supplementary Material online). For a detailed description of how the admixture graphs were constructed and evaluated, see supplementary Text S3.S2, Supplementary Material online. A summary of the non-significantly worse fitting alternative admixture graphs for the top graphs we preferred are provided in supplementary fig. S19, Supplementary Material online.
Supplementary Material
Acknowledgments
We would like to express our gratitude to the 99 Maroon farmers in Suriname and French Guiana who shared their knowledge with us and provided rice varieties. In addition, we thank Paul Richards and Paul Struik for sharing their anthropological and historical perspectives on the genomic results, Siva Selvanayagam for providing advice on genotype QC, and Rafal Gutaker and one anonymous reviewer for insightful comments that improved the manuscript further.
Contributor Information
Marieke S van de Loosdrecht, Biosystematics Group, Wageningen University, Wageningen, The Netherlands.
Nicholaas M Pinas, Biosystematics Group, Wageningen University, Wageningen, The Netherlands; Naturalis Biodiversity Center, Leiden, The Netherlands.
Evanne Dongstra, Biosystematics Group, Wageningen University, Wageningen, The Netherlands.
Jerry R Tjoe Awie, Anne van Dijk Rijst Onderzoekscentrum Nickerie (SNRI/ADRON), Nickerie, Suriname.
Frank F M Becker, Biosystematics Group, Wageningen University, Wageningen, The Netherlands; Laboratory of Genetics, Wageningen University, Wageningen, The Netherlands.
Harro Maat, Knowledge, Technology & Innovation group, Wageningen University, Wageningen, The Netherlands.
Robin van Velzen, Biosystematics Group, Wageningen University, Wageningen, The Netherlands.
Tinde van Andel, Biosystematics Group, Wageningen University, Wageningen, The Netherlands; Naturalis Biodiversity Center, Leiden, The Netherlands; Clusius chair in History of Botany and Gardens, Leiden University, Leiden, The Netherlands.
Michael Eric Schranz, Biosystematics Group, Wageningen University, Wageningen, The Netherlands.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Funding
This research was funded by Dutch Research Council (NWO), grant number OCENW.KLEIN.419. Open access funding provided by Wageningen University & Research.
Author Contributions
T.v.A. and M.E.S. conceptualized the study. T.v.A., M.E.S., and R.v.V. acquired funds. N.M.P. and T.v.A. contributed to sample collection and ethnobotanical surveys. J.R.T.A. contributed to sample propagation, phenotyping and storage in germplasm bank. F.F.M.B. and M.S.v.d.L. did genomic lab work. M.S.v.d.L. contributed to genomic analyses. M.S.v.d.L., T.v.A., N.M.P., H.M., M.E.S., R.v.V., and R.J.T.A. contributed to data integration. M.S.v.d.L. contributed to data visualization. T.v.A., M.E.S., H.M. supervised the study. M.S.v.d.L., T.v.A., M.E.S. wrote the original draft. All authors wrote, reviewed, and edited the article.
Data Availability
Genomic data (BAM format) are available through the Sequence Read Archive (accession number PRJNA1019116). The genotype data for the Maroon rice 110k panel can be downloaded via https://osf.io/qxw5y/.
References
- Agrama H, Yan W, Jia M, Fjellstrom R, McClung A. Genetic structure associated with diversity and geographic distribution in the USDA rice world collection. Nat Sci. 2010:2:247–291. 10.4236/ns.2010.24036. [DOI] [Google Scholar]
- Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009:19(9):1655–1664. 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amarasinghe YPJ, Kuwata R, Nishimura A, Phan PDT, Ishikawa R, Ishii T. Evaluation of domestication loci associated with awnlessness in cultivated rice, Oryza sativa. Rice. 2020:13(1):26. 10.1186/s12284-020-00386-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busing FMTA, Meijer E, Leeden RVD. Delete-m Jackknife for unequal M. Stat Comput. 1999:9(1):3–8. 10.1023/A:1008800423698. [DOI] [Google Scholar]
- Carney JA. The role of African rice and slaves in the history of rice cultivation in the Americas. Hum Ecol. 1998:26(4):525–545. 10.1023/A:1018716524160. [DOI] [Google Scholar]
- Carney JA. Black rice: the African origins of rice cultivation in the Americas. Cambridge: Harvard University Press; 2001. [Google Scholar]
- Carney JA. Rice and memory in the age of enslavement: Atlantic passages to Suriname. Slavery Abol. 2005:26(3):325–348. 10.1080/01440390500319562. [DOI] [Google Scholar]
- Casañas F, Simó J, Casals J, Prohens J. Toward an evolved concept of landrace. Front Plant Sci. 2017:8:145. 10.3389/fpls.2017.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CC, Chow CC, Tellier LC, Vattikuti S, Purcell SM, Lee JJ. Second-generation PLINK: rising to the challenge of larger and richer datasets. GigaScience. 2015:4(1):7. 10.1186/s13742-015-0047-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S, Zhou Y, Chen Y, Gu J. Fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018:34(17):i884–i890. 10.1093/bioinformatics/bty560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkin P. Hmong resettlement in French Guiana. Hmong Stud J. 2005:6:1–27. https://api.semanticscholar.org/CorpusID:131051531. [Google Scholar]
- Czech L, Exposito-Alonso M. Grenepipe: a flexible, scalable and reproducible pipeline to automate variant calling from sequence reads. Bioinformatics. 2022:38(20):4809–4811. 10.1093/bioinformatics/btac600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragtenstein F. ‘De ondraaglijke stoutheid der wegloopers’: Marronage en koloniaal beleid in Suriname, 1667–1768. Utrecht: Utrecht University; 2002. [Google Scholar]
- Eltis D, Richardson D. Atlas of the transatlantic slave trade. New Haven: Yale University Press; 1995. [Google Scholar]
- Fleury M. Impact de la traite des esclaves sur la phytogéographie: exemple chez les aluku (Boni) de Guyane française. J d'Agric Trad Bot Appl. 1994:36:113–137. 10.3406/jatba.1994.3537. [DOI] [Google Scholar]
- Gaio D, Anantanawat K, To J, Liu M, Monahan L, Darling AE. Hackflex: low-cost, high-throughput, Illumina Nextera Flex library construction. Microb Genom. 2022:8(1):000744. 10.1099/mgen.0.000744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert E. Asian rice in Africa: plant genetics and crop history. In: Bray F, Coclanis PA, Fields-Black EL, Schäfer D, editors. Rice: global networks and new histories. New York: Cambridge University Press; 2015. p. 212–228. [Google Scholar]
- Gutaker RM, Groen SC, Bellis ES, Choi JY, Pires IS, Bocinsky RK, Slayton ER, Wilkins O, Castillo CC, Negrão S, et al. Genomic history and ecology of the geographic spread of rice. Nat Plants. 2020:6(5):492–502. 10.1038/s41477-020-0659-6. [DOI] [PubMed] [Google Scholar]
- Han B, Xue Y. Genome-wide intraspecific DNA-sequence variations in rice. Curr Opin Plant Biol. 2003:6(2):134–138. 10.1016/S1369-5266(03)00004-9. [DOI] [PubMed] [Google Scholar]
- Hoefte R. In place of slavery: a social history of British Indian and Javanese laborers in Suriname. Gainesville: University Press of Florida; 1998. [Google Scholar]
- Hoogbergen W, Polimé T. Oostelijk Suriname 1986-2002. OSO—Tijdschrift voor Surinaamse taalkunde, letterkunde en geschiedenis. Nijmegen, The Netherlands: Stichting Instituut ter Bevordering van de Surinamistiek; 2002. p. 225–242. [Google Scholar]
- Hua L, Wang DR, Tan L, Fu Y, Liu F, Xiao L, Zhu Z, Fu Q, Sun X, Gu P, et al. LABA1, a domestication gene associated with long, barbed awns in wild rice. Plant Cell. 2015:27(7):1875–1888. 10.1105/tpc.15.00260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Kurata N, Wei X, Wang Z-X, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, et al. A map of rice genome variation reveals the origin of cultivated rice. Nature. 2012:490(7421):497–501. 10.1038/nature11532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iskandar J, Ellen R. In situ conservation of rice landraces among the Baduy of west Java. J Ethnobiol. 1999:19:97–125. https://api.semanticscholar.org/CorpusID:90562568. [Google Scholar]
- Kassambara A, Mundt F. Factoextra: extract and visualize the results of multivariate data analyses. R Package Version 1.0.7; 2020 [accessed 2023 Apr 13]. https://CRAN.R-project.org/package=factoextra.
- Kawahara Y, de la Bastide M, Hamilton JP, Kanamori H, McCombie WR, Ouyang S, Schwartz DC, Tanaka T, Wu J, Zhou S, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013:6(1):4. 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khush GS, Brar DS. Rice genetics from Mendel to functional genomics. In: Rice Genetics IV; 2008. p. 3–25. [Google Scholar]
- Larranaga N, van Zonneveld M, Hormaza JI. Holocene land and sea-trade routes explain complex patterns of pre-Columbian crop dispersion. New Phytol. 2021:229(3):1768–1781. 10.1111/nph.16936. [DOI] [PubMed] [Google Scholar]
- Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R. The sequence alignment/map format and SAMtools. Bioinformatics. 2009:25(16):2078–2079. 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu H, Redus MA, Coburn JR, Rutger JN, McCouch SR, Tai TH. Population structure and breeding patterns of 145 U.S. rice cultivars based on SSR marker analysis. Crop Sci. 2005:45(1):66–76. 10.2135/cropsci2005.0066. [DOI] [Google Scholar]
- Luong NH, Balkunde SG, Shim K-C, Adeva C, Lee H-S, Kim H-J, Ahn S-N. Characterization of domestication loci associated with awn development in rice. Rice. 2022:15(1):61. 10.1186/s12284-022-00607-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maat H. Commodities and anti-commodities: rice on Sumatra, 1915–1925. In: Schäfer D, Fields-Black E, Bray F, Coclanis P, editors. Rice: global networks and new histories. New York: Cambridge University Press; 2015. p. 335–354. [Google Scholar]
- Maat H, van Andel T. The history of the rice gene pool in Suriname: circulations of rice and people from the eighteenth century until late twentieth century. Hist Agrar. 2018:75:69–91. 10.26882/histagrar.075e04m. [DOI] [Google Scholar]
- Maier R, Flegontov P, Flegontova O, Işıldak U, Changmai P, Reich D. On the limits of fitting complex models of population history to f-statistics. Elife. 2023:12:e85492. 10.7554/eLife.85492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao X, Zhang H, Qiao S, Liu Y, Chang F, Xie P, Zhang M, Wang T, Li M, Cao P, et al. The deep population history of northern East Asia from the Late Pleistocene to the Holocene. Cell. 2021:184(12):3256–3266.e13. 10.1016/j.cell.2021.04.040. [DOI] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010:20(9):1297–1303. 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RS, Choi JY, Sanches M, Plessis A, Flowers JM, Amas J, Dorph K, Barretto A, Gross B, Fuller DQ, et al. Domestication history and geographical adaptation inferred from a SNP map of African rice. Nat Genet. 2016:48(9):1083–1088. 10.1038/ng.3633. [DOI] [PubMed] [Google Scholar]
- Nawani S. The Portuguese in archipelago Southeast Asia (1511–1666). Proc Indian Sci Congr. 2013:74:703–708. http://www.jstor.org/stable/44158873. [Google Scholar]
- Patterson N, Moorjani P, Luo Y, Mallick S, Rohland N, Zhan Y, Genschoreck T, Webster T, Reich D. Ancient admixture in human history. Genetics. 2012:192(3):1065–1093. 10.1534/genetics.112.145037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006:2(12):e190. 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pinas NM, Tjoe Awie JR, Dongstra RE, Maat H, Schranz ME, van de Loosdrecht MS, van Andel T. Yield and growth duration of Maroon rice landraces measured in traditional settings. Genet Resour Crop Evol. 2024. 10.1007/s10722-024-02093-1. [DOI] [Google Scholar]
- Pinas NM, van de Loosdrecht MS, Maat H, van Andel T. Vernacular names of traditional rice varieties reveal the unique history of Maroons in Suriname and French Guiana. Econ Bot. 2023:77(2):117–134. 10.1007/s12231-023-09571-0. [DOI] [Google Scholar]
- Posth C, Yu H, Ghalichi A, Rougier H, Crevecoeur I, Huang Y, Ringbauer H, Rohrlach AB, Nägele K, Villalba-Mouco V, et al. Palaeogenomics of upper palaeolithic to neolithic European hunter-gatherers. Nature. 2023:615(7950):117–126. 10.1038/s41586-023-05726-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price R. Subsistence on the plantation periphery: crops, cooking, and labour among eighteenth-century Suriname Maroons. Slavery Abol. 1991:12(1):107–127. 10.1080/01440399108575025. [DOI] [Google Scholar]
- Price R. Maroons in Guyane: getting the numbers right. Nieuwe West Indische Gids. 2018:92(3–4):275–283. 10.1163/22134360-09203001. [DOI] [Google Scholar]
- Price S. Co-wives and calabashes. Michigan: University of Michigan Press; 1993. [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007:81(3):559–575. 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010:26(6):841–842. 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghavan M, Skoglund P, Graf KE, Metspalu M, Albrechtsen A, Moltke I, Rasmussen S, Stafford TW, Orlando L, Metspalu E, et al. Upper Palaeolithic Siberian genome reveals dual ancestry of Native Americans. Nature. 2014:505(7481):87–91. 10.1038/nature12736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramdayal M, Maat H, van Andel T. The legacy of traditional rice cultivation by descendants of Indian contract laborers in Suriname. J Ethnobiol Ethnomed. 2021:17(1):60. 10.1186/s13002-021-00485-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Thangaraj K, Patterson N, Price AL, Singh L. Reconstructing Indian population history. Nature. 2009:461(7263):489–494. 10.1038/nature08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MF, Botigué LR, Brace S, Stevens CJ, Mullin VE, Stevenson A, Thomas MG, Fuller DQ, Mott R. A 3,000-year-old Egyptian emmer wheat genome reveals dispersal and domestication history. Nat Plants. 2019:5(11):1120–1128. 10.1038/s41477-019-0534-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikora M, Pitulko VV, Sousa VC, Allentoft ME, Vinner L, Rasmussen S, Margaryan A, de Barros Damgaard P, de la Fuente C, Renaud G, et al. The population history of northeastern Siberia since the Pleistocene. Nature. 2019:570(7760):182–188. 10.1038/s41586-019-1279-z. [DOI] [PubMed] [Google Scholar]
- Stahel G. Rexora rijst. Suriname: Koloniaal Nieuws- en Advertentieblad; 1932. [Google Scholar]
- Stahel G. De nuttige planten van Suriname. Paramaribo: Landbouwproefstation in Suriname. Bulletin. 1944:59:239. [Google Scholar]
- Svizzero S, Ray A, Chakraborty D. Awn reduction and the domestication of Asian rice: a syndrome or crop improvement trait? Econ Bot. 2019:73(4):477–488. 10.1007/s12231-019-09465-0. [DOI] [Google Scholar]
- The 3,000 rice genomes project . The 3,000 rice genomes project. GigaSci. 2014:3(1):7. 10.1186/2047-217X-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. Unpublished data. 2012. http://arxiv.org/abs/1207.3907. Last accessed May 7, 2024.
- van Andel TR. African rice (Oryza glaberrima Steud.): lost crop of the enslaved Africans discovered in Suriname. Econ Bot. 2010:64(1):1–10. 10.1007/s12231-010-9111-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Andel TR, Maat H, Pinas N. Maroon women in Suriname and French Guiana: rice, slavery, memory. Slavery Abol. 2023:45(2):187–211. 10.1080/0144039X.2023.2228771. [DOI] [Google Scholar]
- van Andel TR, Meyer RS, Aflitos SA, Carney JA, Veltman MA, Copetti D, Flowers JM, Havinga RM, Maat H, Purugganan MD, et al. Tracing ancestor rice of Suriname Maroons back to its African origin. Nat Plants. 2016b:2(10):16149. 10.1038/nplants.2016.149. [DOI] [PubMed] [Google Scholar]
- van Andel TR, van der Velden A, Reijers M. The ‘Botanical Gardens of the Dispossessed’ revisited: richness and significance of Old World crops grown by Suriname Maroons. Genet Resour Crop Evol. 2016a:63(4):695–710. 10.1007/s10722-015-0277-8. [DOI] [Google Scholar]
- van Andel TR, Veltman MA, Bertin A, Maat H, Polime T, Hille Ris Lambers D, Tjoe Awie J, De Boer H, Manzanilla V. Hidden rice diversity in the Guianas. Front Plant Sci. 2019:10:1161. 10.3389/fpls.2019.01161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van der Meulen JGJ. De rijstselectie in Nederlandsch Indië. Landbouwkundig tijdschrift voor Nederlandsch Indië. Buitenzorg: Vereeniging van Landbouwconsulenten in Nederlandsch-Indië; 1941. p. 943–1029. https://resolver.kb.nl/resolve?urn=MMUBWA02:001898001:00483. [Google Scholar]
- Vasimuddin M, Misra S, Li H, Aluru S. Efficient architecture-aware acceleration of BWA-MEM for multicore systems. In: 2019 IEEE international parallel and distributed processing symposium (IPDPS). Rio de Janeiro, Brazil: IEEE; 2019. p. 314–324. 10.1109/IPDPS.2019.00041. [DOI] [Google Scholar]
- Vaughn JN, Korani W, Stein JC, Edwards JD, Peterson DG, Simpson SA, Youngblood RC, Grimwood J, Chougule K, Ware DH, et al. Gene disruption by structural mutations drives selection in US rice breeding over the last century. PLoS Genet. 2021:17(3):e1009389. 10.1371/journal.pgen.1009389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villalba-Mouco V, van de Loosdrecht MS, Rohrlach AB, Fewlass H, Talamo S, Yu H, Aron F, Lalueza-Fox C, Cabello L, Cantalejo Duarte P, et al. A 23,000-year-old southern Iberian individual links human groups that lived in Western Europe before and after the Last Glacial Maximum. Nat Ecol Evol. 2023:7(4):597–609. 10.1038/s41559-023-01987-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vink W. Creole Jews: negotiating community in colonial Suriname. Leiden, The Netherlands: Brill; 2010. [Google Scholar]
- Wang W, Mauleon R, Hu Z, Chebotarov D, Tai S, Wu Z, Li M, Zheng T, Fuentes RR, Zhang F, et al. Genomic variation in 3,010 diverse accessions of Asian cultivated rice. Nature. 2018:557(7703):43–49. 10.1038/s41586-018-0063-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerega NJC, Ragone D, Motley TJ. Complex origins of breadfruit (Artocarpus altilis, Moraceae): implications for human migrations in Oceania. Am J Bot. 2004:91(5):760–766. 10.3732/ajb.91.5.760. [DOI] [PubMed] [Google Scholar]
- Zeven AC. Landraces: a review of definitions and classifications. Euphytica. 1998:104(2):127–139. 10.1023/A:1018683119237. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Garrison E, Marth G. Haplotype-based variant detection from short-read sequencing. Unpublished data. 2012. http://arxiv.org/abs/1207.3907. Last accessed May 7, 2024.
Supplementary Materials
Data Availability Statement
Genomic data (BAM format) are available through the Sequence Read Archive (accession number PRJNA1019116). The genotype data for the Maroon rice 110k panel can be downloaded via https://osf.io/qxw5y/.