Abstract
Panicum mosaic virus (PMV) is a recently molecularly characterized RNA virus with the unique feature of supporting the replication of two subviral RNAs in a few species of the family Gramineae. The subviral agents include a satellite RNA (satRNA) that is devoid of a coding region and the unrelated satellite panicum mosaic virus (SPMV) that encodes its own capsid protein. Here we report the association of this complex with a new entity in the RNA world, a defective-interfering RNA (DI) of a satellite virus. The specificity of interactions governing this four-component viral system is illustrated by the ability of the SPMV DIs to strongly interfere with the accumulation of the parental SPMV. The SPMV DIs do not interfere with PMV satRNA, but they do slightly enhance the rate of spread and titer of PMV. The SPMV-derived DIs provide an additional avenue by which to investigate fundamental biological questions, including the evolution and interactions of infectious RNAs.
To date, three types of subviral agents have been described that require a helper virus to complete their life cycles: satellite viruses, satellite RNAs (satRNAs), and defective-interfering RNAs (DIs). Satellite viruses encode a capsid protein (CP) which is used for encapsidation of the RNA genome to form icosahedral particles (1, 15). Four plant satellite viruses (satellite tobacco necrosis virus, satellite maize white line mosaic virus, satellite tobacco mosaic virus, and satellite panicum mosaic virus [SPMV]) and one animal satellite virus (hepatitis delta virus) have been described and characterized (6, 7, 15, 17, 21). satRNAs represent another species of small RNA molecules that are prevalent in plant virus infections (13). DIs differ from satellite viruses and satRNAs in that they represent deletion mutants of the parental viral genomes (4, 20). In contrast to DIs, satellite viruses and satRNAs share very little sequence homology with helper viral genomes. These subviral agents are associated with various biological effects, including amelioration or enhancement of symptoms in mixed infections with the respective helper viruses (13, 15, 20). For example, DIs derived from Tomato bushy stunt virus greatly reduce the virus titer and the concentration of movement-associated proteins and modulate the severe-symptom phenotype on host plants (8, 16, 20).
While many plant RNA viruses have the potential to generate DIs and the capacity to support satRNAs or satellite viruses, panicum mosaic virus (PMV; genus Panicovirus, family Tombusviridae), a plus-sense, single-stranded RNA virus, is unique in its ability to simultaneously support both a satellite virus (SPMV) and satRNAs (2, 3, 14). Although PMV, its satRNAs, and SPMV do not share high sequence similarity, the satRNAs have the unique feature of being preferentially encapsidated by the SPMV CP (5).
The focus of this study was SPMV, an 824-nucleotide (nt) positive-sense single-stranded RNA (9, 12) that encodes a 17.5-kDa CP (Fig. 1). In mixed infections with PMV, SPMV induces a synergism by enhancing the rate of PMV systemic spread and accumulation. It also exacerbates the normally mild PMV-induced disease phenotype, resulting in severe chlorosis and stunting of plants with mixed infections (14). Previous studies determined that the SPMV CP is the major determinant for these severe-symptom phenotypes (11). An additional feature associated with SPMV is that multiple passages of PMV plus SPMV on millet plants result in the accumulation of truncated SPMV RNAs of ca. 400 nt (14). These small RNAs were tentatively designated SPMV defective RNAs (D-RNAs) (12).
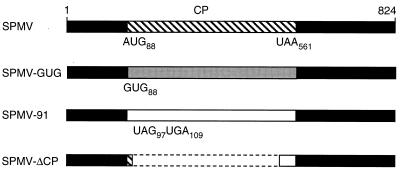
FIG. 1.
Genomic organization of SPMV and mutant derivatives. The black rectangles represent the 5′ and 3′ untranslated regions. The 17.5-kDa SPMV CP gene, extending from an AUG start codon at position 88 to a stop codon at nt 561 (UAA), is indicated on the 824 nt SPMV genome by a hatched rectangle. For the SPMV-GUG mutant, a gray rectangle indicates reduced accumulation of the SPMV CP due to the introduction of a noncanonical GUG start codon (12). For SPMV-91, two premature stop codons were introduced (at positions 97 and 109) to abolish the translation of wild-type SPMV CP, as indicated by the white rectangle. The dashed lines on SPMV-ΔCP represent a deletion from nt 101 to nt 485 on the RNA. A small polypeptide could theoretically be expressed by SPMV-ΔCP RNA as a fusion of the 5′-proximal region of the SPMV CP (hatched region) and an out-of-frame 3′ region (white box).
To further investigate the biology of the D-RNAs, two mutants, SPMV-GUG and SPMV-91, were derived from SPMV type strain cDNAs by site-directed oligonucleotide mutagenesis (Fig. 1). A noncanonical (GUG) start codon at nt 88 was substituted for the authentic AUG start codon to create the mutant SPMV-GUG. In SPMV-91, two stop codons were introduced immediately downstream of the CP start codon to specifically abolish the expression of the 17.5-kDa CP (12). In SPMV-ΔCP, the main portion of the CP open reading frame was deleted to serve as a 434-nt replication-positive control (12). Throughout this paper, we refer to sequences and signature motifs at the RNA level, which were derived from cDNA sequence analyses.
To generate transcription templates, the infectious cDNA constructs of the SPMV type strain and the derived mutants SPMV-GUG, SPMV-91, and SPMV-ΔCP were digested with SmaI. The PMV cDNA was cleaved with EcoICR1 (18). Approximately 0.5 μg of linearized cDNA was used as a template to generate uncapped RNA transcripts in an in vitro transcription reaction based on the conditions specified by the supplier of T7 RNA polymerase (Gibco BRL, Gaithersburg, Md.). PMV and each of four different SPMV-derived transcripts (ca. 6 μg of each) were mixed in RNA inoculation buffer (0.05 M K2HPO4, 0.05 M glycine, 1% bentonite, 1% Celite, pH 9.0) and rubbed onto the leaves of five proso millet (Panicum miliaceum cv. Sunup) or foxtail millet (Setaria italica cv. German R) plants. The inoculated plants were kept at room temperature overnight before being transferred to the greenhouse. The initial inoculation is designated zero passage (Fig. 2). At 12 days postinoculation, the leaves from five systemically infected millet plants were pulverized in virus inoculation buffer (0.05 M K2HPO4, 1% Celite, pH 7.0). This bulked material was subsequently passaged to the leaves of five healthy millet plants. For each construct, five passages were performed at 10- to 12-day intervals. Total RNA was extracted from 50 mg of infected plant leaf tissue 10 to 12 days postinoculation. Approximately 5 μg of total plant RNA was resolved on 1.2% agarose gels and transferred to nylon membranes. The membrane was first hybridized with a [32P]dCTP-labeled SPMV- or satRNA-specific cDNA probe. For plants with three-way infections, duplicate agarose gels and membranes were prepared to assay for SPMV or satRNA. Each membrane was subsequently hybridized with [32P]dCTP-labeled PMV cDNA to confirm the presence of the helper virus, as described previously (12, 14, 18).
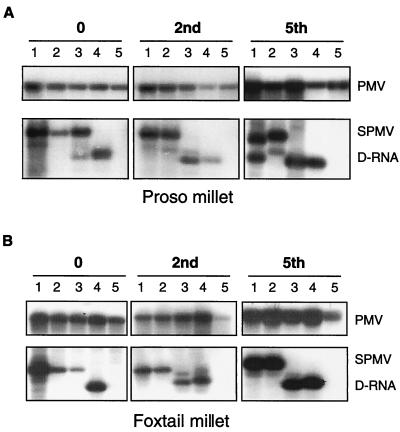
FIG. 2.
Accumulation of SPMV D-RNA during multiple passages on millet plants. Plants were coinoculated with PMV alone or as mixed infections with SPMV or the mutants described in the legend to Fig. 1. Every 10 to 12 days, the infections were transferred to healthy (A) proso millet (P. miliaceum cv. Sunup) or (B) foxtail millet (S. italica cv. German R) plants for a total of five passages. The lane numbers correspond to the following infections: 1, PMV + SPMV; 2, PMV + SPMV-GUG; 3, PMV + SPMV-91; 4, PMV + SPMV-ΔCP; 5, PMV alone. The serial passage numbers (0, 2nd, and 5th) are indicated above each panel. The tops of panels A and B show that PMV was present in all passages, and the bottoms indicate the locations of the ≈400-nt D-RNAs and the 824-nt SPMV RNA. The 434-nt SPMV-ΔCP RNA (lane 4) migrates at a position similar to that of the D-RNA.
RNA blot analyses of the consecutively transferred infections revealed the accumulation of SPMV type strain D-RNAs by the fifth passage on proso millet plants (Fig. 2A) but not on foxtail millet plants (Fig. 2B). D-RNAs were not detected on foxtail millet plants infected with the SPMV-GUG mutant (Fig. 2B), and only low accumulations of D-RNAs were observed on proso millet plants (Fig. 2A). SPMV-91 infections accumulated D-RNAs on the first passage to proso millet plants and the second passage on foxtail millet plants. In subsequent passages, the D-RNA remained prominent while the parental-genome-sized SPMV-91 RNA was no longer detected on either plant species (Fig. 2).
Following the fifth passage of SPMV-91 to proso millet and foxtail millet plants, the D-RNAs were isolated by a gel purification procedure (12). The purified RNAs were used as templates to perform a reverse transcription-PCR with SPMV-specific primers as described previously (11). A reverse SPMV primer (5′-CTGCAGATCTGGGTCCTAGGAGGGGGAGACGC; the PstI site is underlined, and the BglII site is in bold) complementary to 3′-terminal nt 824 to 803 was used for first-strand cDNA synthesis. A forward SPMV primer (5′-GGATCCTAATACGACTCACTATAGGGTATTCCACGCTAGCA AC; the BamHI site is underlined) contained the 5′-terminal 20 nt of SPMV, which was linked to an upstream T7 RNA polymerase promoter sequence (in boldface letters). Following reverse transcription-PCR of the D-RNAs, the products were cloned into the SmaI site of pUC119 and sequenced. A 408-nt D-RNA isolated from foxtail millet (D-RNA-F) was composed of a 5′-terminal domain (I), a 5′-proximal domain (II), and a 3′-terminal domain (III) from the parental SPMV-91 RNA (Fig. 3A) (11). A 399-nt D-RNA (D-RNA-P) isolated from infected proso millet also contained domains I, II, and III from the SPMV-91 RNA (Fig. 3A). In vitro-generated RNA transcripts from D-RNA-P (Fig. 3B) and D-RNA-F (data not shown) replicated and systemically spread through millet plants coinoculated with PMV. Based on the results previously obtained with SPMV deletion mutants, replication and spread of the D-RNAs likely require the maintenance of domains I, II, and III (12).
FIG. 3.
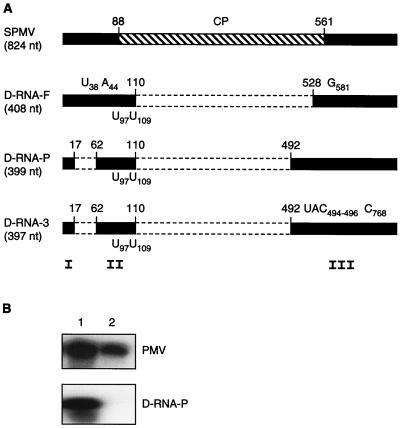
Genomic organization of the SPMV D-RNAs and a test of the biological activity of D-RNA-P. (A) SPMV D-RNA-F and D-RNA-P were isolated from passage experiments done with foxtail millet and proso millet plants, respectively, following inoculation with PMV + SPMV-91. Nucleotide differences between SPMV D-RNAs and wild-type SPMV are indicated on the respective maps. D-RNA-F had a deletion between positions 110 and 528, substitutions of C to U at nt 38 and G to A at nt 44, and the addition of a G at position 581. D-RNA-P had deletions from position 17 to position 62 and from position 110 to position 492 (12). D-RNA-3 was isolated from the third passage of PMV + SPMV + D-RNA-P on proso millet (see Fig. 4). On the 397-nt D-RNA-3, three nucleotides (UAC) were deleted at positions 494 to 496 and a C was inserted at position 768 compared to the parental D-RNA-P. Each of these D-RNAs maintained the two U nucleotide changes introduced into the parental SPMV-91 RNA at positions 97 and 109 (Fig. 1). Beneath the genome map, domains I, II, and III indicate the regions on the SPMV RNA that are necessary for replication and spread in millet plants (12). (B) RNA blots of total RNAs isolated from proso millet plants infected with in vitro transcripts of PMV + D-RNA-P (lane 1) or PMV alone (lane 2). The top and bottom blots were hybridized with 32P-labeled PMV- and SPMV-specific cDNAs, respectively.
The multiple-passage experiments with SPMV-91 suggested that abolishing the expression of the 17.5-kDa SPMV CP triggered the generation and accumulation of SPMV D-RNAs (Fig. 2). Conversely, the presence of some SPMV CP can modulate this effect, as demonstrated by SPMV-GUG, which has ≈80% less CP expression than wild-type SPMV (12). Both SPMV-GUG and SPMV wild-type RNAs were fairly recalcitrant to DI formation when passaged through foxtail or proso millet plants (Fig. 2). This bolsters our hypothesis that the 17.5-kDa CP contributes to the maintenance of the intact genomic SPMV RNA (11).
To determine if the SPMV CP confers a biological advantage on SPMV RNA, millet plants were inoculated with PMV plus SPMV plus D-RNA-P. In these three-way infections, D-RNA-P rapidly replaced SPMV RNA and dominated the viral population as early as the second passage (Fig. 4A). However, D-RNA-P was not noticeably detrimental to PMV accumulation (Fig. 4A). To confirm that the D-RNA was an authentic D-RNA-P and not a newly generated D-RNA from the parental SPMV RNA, we constructed cDNA clones of D-RNAs isolated from third-passage plants from the three-way infection. D-RNA-3 (Fig. 3A) had the two signature U's from SPMV-91 (originally introduced at nt 97 and 109 to generate premature stop codons; Fig. 1). This proved that the parental D-RNA-P displaced the wild-type SPMV RNA during the course of a systemic infection of millet (Fig. 4A).
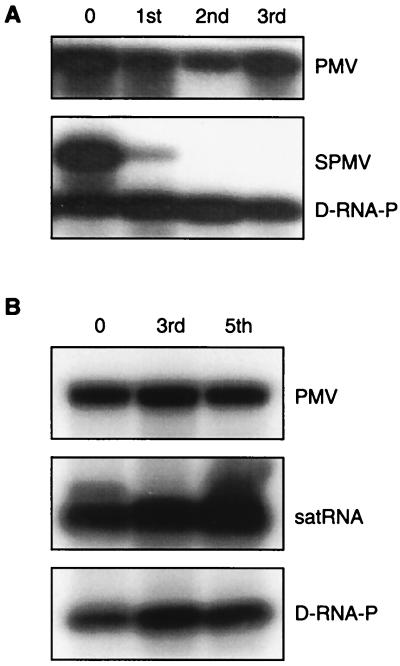
FIG. 4.
Effect of SPMV D-RNA-P on the replication and spread of PMV and its satellites. Proso millet plants were inoculated with transcripts of PMV + SPMV + D-RNA-P (A) or PMV + satRNA + D-RNA-P (B). The initial inoculation is designated zero passage. Each infection was passaged three or five times and sampled as indicated for panels A and B, respectively. In panels A and B, the topmost blots show that PMV is maintained in each passage, as detected by a 32P-labeled PMV cDNA probe. The lower blots indicate the presence of satRNA, SPMV and/or D-RNA-P following hybridization with the appropriate 32P-labeled SPMV- or satRNA-specific cDNA probe.
PMV satRNAs are commonly found in St. Augustine grass (Stenotaphrum secundatum) in mixed infections with PMV + SPMV + satRNA but less commonly with PMV alone (3). This stimulated our interest in determining the effects of the SPMV DIs on the replication of PMV satRNAs in millet plants. Multiple-passage experiments with PMV plus satRNA plus D-RNA-P in millet were readily maintained without perturbing the replication or spread of any of these RNAs (Fig. 4B). In contrast, as indicated above, SPMV was not maintained in mixed infections of PMV + SPMV + D-RNA-P following the first passage (Fig. 4A). Therefore, the D-RNAs possessed an interfering property specific to the replication and movement of SPMV (Fig. 4). These data also support our previous observation that PMV satRNAs can replicate and spread without the presence of, or encapsidation by, SPMV even though there is a strong affinity between the SPMV CP and satRNA (3, 5). Moreover, PMV satRNAs do not interfere with SPMV accumulation (3). Based on these results, the SPMV-derived defective RNAs can be properly classified as authentic and robust DIs with specificity for the parental satellite virus RNA. The data suggest that specific competition may occur between SPMV and its DIs for host factors or sites in the host cell that support replication and spread.
Further investigations of the effects of the host and the environment on the accumulation of the SPMV DIs could provide some new insight into the origins of these RNAs and the cellular requirements for their accumulation in planta. For example, cDNA sequence analysis of the SPMV DIs shows that open reading frames are not maintained. This is in contrast to some helper virus-based DIs which perhaps need host proteins that are associated with the translational machinery to assist in replication (10, 11, 19). In millet protoplasts, we have observed that SPMV-91 and SPMV-ΔCP transcripts appear unstable, based on RNA degradation patterns (12). These SPMV mutants may lack certain structural requirements and/or the ability to associate with PMV replicase or host proteins. This may, in turn, trigger the rapid accumulation of DIs. Perhaps the trans-acting replication or recombination events used to form SPMV DIs from satellites differ from the cis-acting events associated with the generation of DIs from helper viruses.
In summary, the SPMV DIs that accumulate following passage of PMV + SPMV infections of millet plants have the unique feature of foiling the accumulation of SPMV while coexisting with PMV satRNA and slightly stimulating the accumulation of PMV. The satellite virus-derived DIs represent an intriguing new entity in the RNA world and may provide important clues that further our understanding of the origin of plant virus satellites.
Nucleotide sequence accession numbers.
The cDNA sequences of the SPMV-DIs were submitted to the GenBank database and assigned accession numbers AF339446 (D-RNA-F), AF339447 (D-RNA-3), and AF159425 (D-RNA-P).
Acknowledgments
We thank Herman Scholthof, Nancy Keller, and Erik Mirkov for critical reviews and helpful suggestions.
The Texas Agricultural Experiment Station (H-8388) and a grant from the USDA-NRI (99-35303-3714) supported this research.
REFERENCES
- 1.Buzen F G, Niblett C L, Hooper G R, Hubbard J, Newman M A. Further characterization of panicum mosaic virus and its associated satellite virus. Phytopathology. 1984;74:313–318. [Google Scholar]
- 2.Cabrera O, Roossinck M J, Scholthof K-B G. Genetic diversity of panicum mosaic virus satellite RNAs in St. Augustinegrass. Phytopathology. 2000;90:977–980. doi: 10.1094/PHYTO.2000.90.9.977. [DOI] [PubMed] [Google Scholar]
- 3.Cabrera O, Scholthof K-B G. The complex viral etiology of St. Augustine decline. Plant Dis. 1999;83:902–904. doi: 10.1094/PDIS.1999.83.10.902. [DOI] [PubMed] [Google Scholar]
- 4.Damayanti T A, Nagano H, Mise K, Furusawa I, Okuno T. Brome mosaic virus defective RNAs generated during infection of barley plants. J Gen Virol. 1999;80:2511–2518. doi: 10.1099/0022-1317-80-9-2511. [DOI] [PubMed] [Google Scholar]
- 5.Desvoyes B, Scholthof K-B G. RNA: protein interactions associated with satellites of panicum mosaic virus. FEBS Lett. 2000;485:25–28. doi: 10.1016/s0014-5793(00)02177-3. [DOI] [PubMed] [Google Scholar]
- 6.Dodds J A. Satellite tobacco mosaic virus. Curr Top Microbiol Immunol. 1999;239:145–157. doi: 10.1007/978-3-662-09796-0_8. [DOI] [PubMed] [Google Scholar]
- 7.Francki R I B. Plant virus satellites. Annu Rev Microbiol. 1985;39:151–174. doi: 10.1146/annurev.mi.39.100185.001055. [DOI] [PubMed] [Google Scholar]
- 8.Hillman B I, Carrington J C, Morris T J. A defective interfering RNA that contains a mosaic of a plant virus genome. Cell. 1987;51:427–433. doi: 10.1016/0092-8674(87)90638-6. [DOI] [PubMed] [Google Scholar]
- 9.Masuta C, Zuidema D, Hunter B G, Heaton L A, Sopher D S, Jackson A O. Analysis of the genome of satellite panicum mosaic virus. Virology. 1987;159:329–338. doi: 10.1016/0042-6822(87)90471-5. [DOI] [PubMed] [Google Scholar]
- 10.Molenkamp R, Greve S, Spaan W J M, Snijder E J. Efficient homologous RNA recombination and requirement for an open reading frame during replication of equine arteritis virus defective interfering RNAs. J Virol. 2000;74:9062–9070. doi: 10.1128/jvi.74.19.9062-9070.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Qiu W, Scholthof K-B G. Genetic identification of multiple biological roles associated with the capsid protein of satellite panicum mosaic virus. Mol Plant-Microbe Interact. 2001;14:21–30. doi: 10.1094/MPMI.2001.14.1.21. [DOI] [PubMed] [Google Scholar]
- 12.Qiu W, Scholthof K-B G. In vitro- and in vivo-generated defective RNAs of satellite panicum mosaic virus define cis-acting RNA elements required for replication and movement. J Virol. 2000;74:2247–2254. doi: 10.1128/jvi.74.5.2247-2254.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roossinck J M, Sleat D, Palukaitis P. Satellite RNAs of plant viruses: structures and biological effects. Microbiol Rev. 1992;56:265–279. doi: 10.1128/mr.56.2.265-279.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scholthof K-B G. A synergism induced by satellite panicum mosaic virus. Mol Plant-Microbe Interact. 1999;12:163–166. [Google Scholar]
- 15.Scholthof K-B G, Jones R W, Jackson A O. Biology and structure of plant satellite viruses activated by icosahedral helper viruses. Curr Top Microbiol Immunol. 1999;239:123–143. doi: 10.1007/978-3-662-09796-0_7. [DOI] [PubMed] [Google Scholar]
- 16.Scholthof K-B G, Scholthof H B, Jackson A O. The effect of defective interfering RNAs on the accumulation of tomato bushy stunt virus proteins and implications for disease attenuation. Virology. 1995;211:324–328. doi: 10.1006/viro.1995.1410. [DOI] [PubMed] [Google Scholar]
- 17.Taylor J M. Human hepatitis delta virus: an agent with similarities to certain satellite RNAs of plants. Curr Top Microbiol Immunol. 1999;239:107–122. doi: 10.1007/978-3-662-09796-0_6. [DOI] [PubMed] [Google Scholar]
- 18.Turina M, Maruoka M, Monis J, Jackson A O, Scholthof K-B G. Nucleotide sequence and infectivity of a full-length cDNA clone of panicum mosaic virus. Virology. 1998;241:141–155. doi: 10.1006/viro.1997.8939. [DOI] [PubMed] [Google Scholar]
- 19.White K A, Bancroft J B, Mackie G A. Coding capacity determines in vivo accumulation of a defective RNA of clover yellow mosaic virus. J Virol. 1992;66:3069–3076. doi: 10.1128/jvi.66.5.3069-3076.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.White K A, Morris T J. Defective and defective interfering RNAs of monopartite plus-strand RNA plant viruses. Curr Top Microbiol Immunol. 1999;239:1–17. doi: 10.1007/978-3-662-09796-0_1. [DOI] [PubMed] [Google Scholar]
- 21.Zhang L, Zitter T A, Palukaitis P. Helper virus-dependent replication, nucleotide sequence and genome organization of the satellite virus of maize white line mosaic virus. Virology. 1991;180:467–473. doi: 10.1016/0042-6822(91)90060-o. [DOI] [PubMed] [Google Scholar]