Abstract
Purpose:
Colorectal cancer (CRC) is one of the most commonly diagnosed cancers in the world. Early detection would be greatly enhanced if accurate and cost-effective diagnostic biomarkers for CRC were accessible. The development of blood tests would evidently lower the screening cost of CRC detection. The aim of the present study was to examine the prospective of plasma miRNAs as non-invasive biomarkers for CRC screening.
Methods:
The expressions of miR-21 and miR-145 in the plasma of colorectal adenocarcinomas and normal healthy controls were quantified by using TaqMan miRNA assays. MiRNA expression levels were also correlated with commonly used clinicopathological features of CRC.
Results:
Out of 30 CRC patients, 19 were male and 11 were female. The Mean age of patients was 51.3 ±14.6 years. A statistically significant increase in expression of miR-21 was observed in CRC patients’ as compared to healthy controls (p<0.001). A significant association between miR-21 expression and age group (p=0.002) was noticed. Also, a statistically significant difference (p=0.015) between miR-21 expression and tumor location in the proximal and distal sites of the colon was observed in CRC patients. Further, a statistically significant downregulation of miR-145 expression was observed in the plasma of CRC patients as compared to healthy controls (p<0.05). This is the first study to report a significant association between miR-21 expression, age group, and tumor location in CRC patients.
Conclusion:
The present study thus emphasises that the appraisal of miR-21 and miR-145 plasma levels may serve as a promising non-invasive screening tool for the early detection of colorectal cancer.
Key Words: micro-RNAs, real-time polymerase chain reaction, diagnostic, biomarkers, colorectal cancer
Introduction
Colorectal cancer (CRC) is the third most frequently diagnosed malignancy and the second leading cause of cancer-related death worldwide [1, 2]. CRC incidence rates also vary by population, with North America, Australia, and New Zealand having the highest rates and Western Africa having the lowest [3]. In Asia, the prevalence rates of CRC also differ dramatically and are invariably low in almost every South Asian country and high in every developed Asian country. The burden of CRC has risen rapidly, especially in some of the economically developed Asian countries [4]. Although, there has been a little rise in CRC cases in India, it is still the fifth most common cancer death among Indian men and women [5, 6].
CRC incidence rates are rapidly increasing due to the effects of many risk factors, including smoking, physical inactivity, excess weight and obesity, excessive red and processed meat consumption, and excessive alcohol consumption [7]. It has been found that if the disease was diagnosed at an early or premalignant stage, more than 95% of CRC cases would benefit from curative surgery [8, 9]. There have been several early detection procedures developed for the early detection of colorectal cancer that are being used more frequently, including stool-based tests and endoscopic examinations.
Currently, traditional methods like colonoscopy are precise and enable the excision of adenomas, which helps lower the incidence of cancer. However, its use has been impeded by its invasive nature, high cost, and the discomfort it causes to patients [10-12].
The majority of non-invasive molecular diagnosis tests for CRC rely primarily on the analysis of faeces and/or blood samples. The most commonly used test for finding blood in stool is guaiac-based faecal occult blood testing (FOBT). Its screening has been linked to a 15% -33% decline in CRC mortality [13]. However, this test has a number of limitations, including low specificity and sensitivity in the detection of colon adenomas (11%) and CRCs (33–50%) [14]. The faecal immunochemical occult blood test (FIT), which particularly detects human-specific globin, has helped to partially overcome this issue [14]. The sensitivity is quite poor for advanced adenomas, however both of these tests have demonstrated potential for CRC identification in asymptomatic patients if they are performed annually. Further, the proteome of circulating blood has also been used as biomarkers for CRC, such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9), but their sensitivity and specificity, especially for the primary stage of CRC, seem to be insufficient [15].
Therefore, efforts are being made in a focused manner to develop new diagnostic screening biomarkers for the early diagnosis of CRC. Since miRNAs can act as both oncogenes and tumor suppressors, research on them has received a lot of attention in recent years [16]. MiRNAs are a class of endogenous, short (17–22 nucleotides), noncoding, single-stranded, evolutionarily conserved RNA molecules known to be involved in the post-transcriptional regulation of gene expression in protozoa, plants, and vertebrates [17]. Dysregulation of miRNAs may be the ideal tool for the early identification of cancer, according to several studies that have shown that levels of miRNA expression are altered throughout the development of cancer [18–20], thereby suggesting that dysregulation of miRNAs might be the ideal tool for the early detection of cancer.
Although emerging evidence suggests the possibility of miRNA-based biomarkers in tumor tissues, faeces, serum, and urine as a promising method for the early diagnosis of tumors [21, 22], little information is currently known on the utility of plasma miRNAs as diagnostic markers. Hence, the current study has been planned to investigate whether miRNAs have the potential to serve as cutting-edge biomarkers for colorectal cancer screening.
Materials and Methods
Patients
To carry out this study, 30 patients prior to surgery were enrolled after obtaining informed written consent from the study subjects and approval from the Ethics Committee of Postgraduate Institute of Medical Education and Research (PGIMER), Chandigarh. Based on their pathologic diagnosis, CRC patients who underwent treatment at the hospital were recruited for this study. A 5 ml blood sample was obtained from 30 CRC patients, and plasma was immediately isolated from whole blood collected in EDTA vacutainers by centrifugation and then frozen at -80°C for miRNA expression studies. MiRNA isolation was performed within a week. Patients’ information about smokers or non-smokers, alcohol consumption, family history, and clinical history was recorded. 30 apparently healthy subjects, who were not relatives of CRC patients and gave consent for the study, were also recruited.
Plasma RNA Isolation
Plasma miRNAs were extracted from total RNA using the mirVana™ miRNA Isolation Kit (Ambion, Life Technologies, Texas, USA), according to the manufacturer’s instructions. A quantity of 0.25 ml of plasma was used to extract total RNA from each patient and control sample. The Ambion TRIzol LS reagent protocol was optimised to get good quality and quantity of total RNA. Total RNA was eluted into 100 µl of pre-heated (95°C) RNase-free water. A Nanodrop 2000 spectrophotometer (Thermo Scientific, Middlesex, MA) was used to measure total RNA concentrations and purity from plasma samples for further miRNA quantification.
Micro RNA Quantification
Real-time quantification was performed at the central facility of PGIMER, Chandigarh, using StepOnePlus™ Real-Time PCR Systems (Applied Biosystems) for miR-21 and miR-145 expression. Quantification of miR-21 and miR-145 was accomplished using TaqMan miRNA Assays (Applied Biosystems). Approximately 20 ng of RNA was reverse transcribed into miRNA-specific cDNAs, miR-21 and miR-145, using the TaqMan™ MicroRNA Reverse Transcription Kit (Applied Biosystems). The MiR-16 was used as an internal control. MiR-16 was recommended as an endogenous control due to its relatively constant expression levels across many different sample types [23].
The differences between the groups were shown as ΔCt, which represented the difference between the Ct value of the target miRNA and the normalizer miRNA. The objectives for normalisation were chosen based on prior research and the coherence of endogenous Ct signals [24].
Statistical Analysis
The statistical analysis was done with the help of SPSS 20.0 software. A one-sample t-test and a Mann-Whitney test were used to determine whether there was any statistical difference in the expression of miR-21and miR-145 between the two groups. While the Kruskal-Wallis test was used to assess significance among more than two groups. Statistical significance was defined as a p value less than 0.05. Standard error (S.E.) and mean are used to represent all results.
Table 1.
Clinical Characteristic of Patients with Colorectal Cancer
| Clinical Characteristics | N (%) | N (%) | |
|---|---|---|---|
| Patients (n) | 30 | 30 | Controls (n) |
| Age | Age | ||
| ≤50 years | 14 (47%) | 16 (53%) | ≤50 years |
| >50 years | 16 (53%) | 14 (47%) | >50 years |
| Sex | Sex | ||
| Male | 19 (63%) | 18 (60%) | Male |
| Female | 11 (37%) | 12 (40%) | Female |
| Tumor Location | |||
| Proximal | 16 (53%) | ||
| Distal | 14 (47%) | ||
| Clinical Stage (TNM) | |||
| Early (I-II) | 21 (70%) | ||
| Advanced (III-IV) | 9 (30%) | ||
| Histopathological Type | |||
| Adenocarcinoma | 22 (73%) | ||
| Other Types | 8 (27%) | ||
| Histopathological grade | |||
| Well differentiated | 2 (7%) | ||
| Moderately differentiated | 26 (86%) | ||
| Poorly differentiated | 2 (7%) | ||
Colorectal cancer, RQ relative quantification *p<0.05, statistically significant difference
Results
The current study included human subjects from North India, which includes the neighboring states of Punjab, Haryana, and Himachal Pradesh, Jammu and Kashmir, as well as Uttarakhand and Uttar Pradesh. For the study, 30 healthy subjects and 30 patients with colorectal cancer were enrolled. Of the 30 patients with colorectal cancer, 19 were male and 11 were female. while out of 30 healthy controls, 18 (19---75 years.) were males and 12 (40---66 years.) were females. The male patients were in the age group of 19–75 years, with a mean age of 50.21±3.73 years. In the case of females, the age range of patients was 40–68 years, with a mean age of 52.45±3.12 years. The number of male patients was found to be high relative to females (19/30) (63%). The 30 cases included in this study were divided into two groups: patients below 50 years of age and those above 50 years of age. Sixteen subjects (53%) had cancer of the colon and fourteen (47%) had cancer of the rectum. The detailed clinical characteristics of the patients with colorectal cancer are given in Table 1.
MicroRNA-21 and MicroRNA-145 Expression
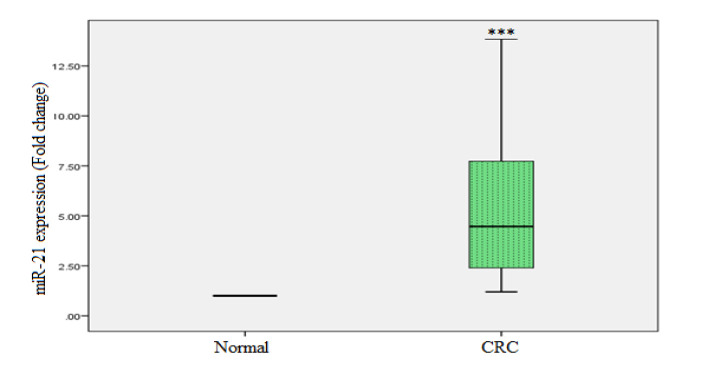
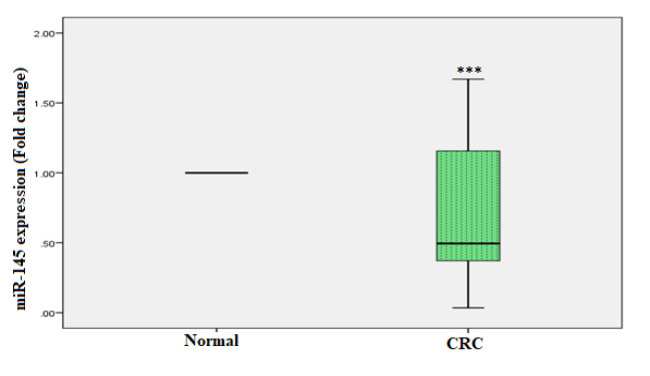
The relative expressions of miR-21 and miR-145 were analysed in the plasma of CRC patients and healthy controls. A statistically significant increased expression of miR-21 was observed in CRC patients, i.e., 4.1 fold as compared to healthy controls (p < 0.001). Where the expression of miR-145 was significantly downregulated in the plasma of CRC patients, i.e., -2.1 fold as compared to healthy controls (p < 0.001). Figures 1 and 2 show real-time PCR analyses of miR-21 and miR-145 in plasma samples, respectively.
Figure 1.
miR-21 in CRC as well as Healthy Controls was Determined by Real-Time PCR. miR-16 was used to normalise miR-21 expression. The one-sample t-test was used for the statistical analysis. ***p < 0.001
Figure 2.
miR-145 in CRC as well as Healthy Controls was Determined by Real-time PCR. miR-16 was used to normalise miR-145 expression. The Mann-Whitney test was used for the statistical analysis. ***p < 0.05
Relationship between miR-21 Expression and Clinicopathological Features of Colorectal Cancer Patients
The associations between miR-21 expression and clinicopathological features are presented in Table 2. The quantitative relative expression of miR-21 was observed to be significantly higher (p< 0.001) in the plasma of CRC patients as compared to healthy controls (Figure 1). There was a significant association between miR-21 expression and age group (p=0.002; Table 2). Further, there was a statistically significant difference (p=0.015) between miR-21 expression and tumor location in CRC patients in the proximal and distal sites of the tumor (Table 2).
Table 2.
Relationship between miR-21 Expression and Clinicopathological Features of Colorectal Cancer Patients
| Clinicopathological parameters | Number of Cases (%) | RQ (Mean±SE) | p-value |
|---|---|---|---|
| Patients (n) | 30 (100 %) | ||
| Age group | |||
| ≤50 years | 14 (47%) | 6.69± 1.04 | 0.002* |
| >50 years | 16 (53%) | 2.90± 0.46 | |
| Sex | |||
| Male | 19 (63%) | 5.76± 0.94 | 0.255 |
| Female | 11 (37%) | 4.14± 0.82 | |
| Tumor Location | |||
| Proximal | 16 (53%) | 6.83± 0.99 | 0.015* |
| Distal | 14 (47%) | 3.61± 0.77 | |
| TNM stage | 0.286 | ||
| I + II | 21 (70%) | 5.77± 0.94 | |
| III | 9 (30%) | 4.12± 0.68 | |
| Lymph node metastasis | 0.286 | ||
| Yes | 9 (30%) | 4.12± 0.68 | |
| No | 21 (70%) | 5.77 ± 0.94 | |
| Histopathological Type | 0.549 | ||
| Adenocarcinoma | 22 (73%) | 5.61± 0.95 | |
| Other Types | 8 (27%) | 4.60 ± 0.83 | 0.86 |
| Histopathological grade | 28 (93%) | 5.16± 0.74 | |
| Good + Moderate | 2 (7%) | 5.67± 0.61 | |
| Poor |
(a): Relationship between miR-21 expression and age group
The level of miR-21 was measured to be 6.69±1.04 (Mean±SE) in colorectal cancer patients younger than 50 years old and 2.90±0.46 (Mean±SE) in colorectal cancer patients older than 50 years old. There was a significant association between miR-21 expression and age group (p = 0.002; Table 2).
(b): Relationship between miR-21 expression and sex
Female CRC patients showed expression of miR-21 (4.14± 0.82, Mean±SE) than male CRC patients (5.76± 0.94, Mean±SE), which was not statistically significant (p = 0.255; Table 2).
(c): Relationship between miR-21 expression and tumor location
In CRC patients, we found a significant difference (p = 0.015) between the amount of miR-21 present and the location of the tumor. The difference was most noticeable at the proximal site (6.83±0.99) than the distal site (3.61±0.77) of the tumor (Table 2).
(d): Relationship between miR-21 expression and TNM stage
Early stages of CRC showed higher expression (5.77±0.94) (Mean±SE) of miR-21 as compared to advanced stages of CRC (4.12± 0.68 (Mean±SE). However, there was no significant (p = 0.286) change in miR-21 expression between the early and advanced stages of CRC (Table 2).
(e): Relationship between miR-21 expression and Histopathological Type
No significant difference (p = 0.549) was found between the amount of miR-21 and the type of cancer in patients with adenocarcinoma (5.61± 0.95 (Mean±SE)) or other types of tumors (4.60± 0.83 (Mean±SE)) (Table 2).
(f): Relationship between miR-21 expression and Lymph node metastasis
There was no significant difference (p = 0.286) between miR-21 expression and lymph node metastasis present 4.12± 0.68 (Mean±SE) and absent 5.77 ± 0.94 (Mean±SE) in patients with CRC (Table 3).
Table 3.
The miRNA-Specific Primer Sequences Used in the Study
| miRNA/Gene | Primer Sequence (5′ → 3′) |
|---|---|
| miR-16 | 5’-UAGCAGCACGUAAAUAUUGGCG - 3’ |
| miR-21 | 5’- UAGCUUAUCAGACUGAUGUUGA -3’ |
| miR-145 | 5’GUCCAGUUUUCCCAGGAAUCCCU-3’ |
Discussion
Studies have demonstrated that molecular methods, such as gene expression profiling, are commonly employed to enhance the classification of CRC, clarify patient prognosis, and predict therapeutic response [25, 26]. Notably, our study has several advantages in terms of the analysis techniques. Methodologically, we employed the sensitive and validated TaqMan qPCR assay, which is more reliable than SYBR Green assays or most of the microarray methods [27]. Early detection of tumors increases the overall survival rate of CRC patients. Thus, there is an imperative need to find sensitive, specific, and non-aggressive molecular biomarkers for the early diagnosis of CRC [28, 29]. MiRNAs have recently received attention from scientists because of their interest in identifying blood biomarkers for cancer screening. This is due to the potential impact miRNAs may have on clinical and translational science with regard to prognosis, treatment, and diagnosis. The regulation of cellular metabolism, development, proliferation, differentiation, and apoptosis is greatly influenced by miRNAs [30]. The natural process of miRNA dysregulation is still mysterious. However, functional studies have specified that the dysregulation of miRNAs may be involved in the process of human carcinogenesis [31]. MiRNAs also play significant roles in tumorigenesis, and the expression level of miRNAs (as anti-oncogenes) is often decreased in cancers because of chromosomal aberrations [32], transcriptional regulation, or methylation [33, 34].
Numerous clinical studies in CRC patients revealed that a large number of miRNAs are actively involved in carcinogenesis, cancer development, and progression by acting both as tumor suppressors (i.e., miR-143 and miR-145) and oncogenes (i.e., miR-20a, miR-21, miR-31, miR-92, and miR-181b) [35, 36]. These epigenetic signatures can now be efficiently assessed in circulating DNA (cfDNA) extracted from plasma, thereby accelerating the diagnosis and optimising disease monitoring. Specific diagnostic advantages of miRNAs also include their rapid release from target tissues into the circulation and their high stability, as these small non-coding RNA sequences are scarcely vulnerable to degradation [37, 38]. MiR-21 is believed to have a crucial role in the control of apoptosis, proliferation, and migration since it primarily exerts its oncogenic activity by targeting the PDCD4 tumor suppressor 3’-UTR, PTEN, Spry-1, and NF-kB [39, 40].There is currently limited knowledge available regarding the diagnostic presentation of plasma miR-21 in CRC patients, despite a large number of studies exploring the potential role of tissue expression of miR-21. In a prior study by Chang et al. [41], they examined the tissue expression of miR-21 in 48 colorectal tumors and discovered that it steadily increased from healthy mucosae to cancer tissues [41]. Schmitz et al. (2009) also measured the expression levels of miR-21 in 18 hyperplastic polyps, 19 sessile serrated adenomas without cytological dysplasia, and 20 normal colonic mucosal specimens, but concluded that assessment of tissue miR-21 did not differentiate the two forms of colorectal disease [42].
The findings of the current investigation made it abundantly evident that plasma samples obtained from CRC patients strongly up-regulate miR-21. The results of present study may also suggest a relationship between the levels of miR-21 expression in the plasma and tumor tissues of CRC patients and those in cancer cells. These findings differ from those of earlier research, which similarly found considerably higher expression levels of miR-21 in CRC cancer tissues [43, 29]. Our results, as well as other studies, have also reported elevated plasma miRNA levels in CRC patients, raising concern about the source of plasma miRNAs. Although in the present study, we did not evaluate the levels of these miRNAs in primary tissue, recently published study has reported elevated levels of miR-21 in CRC tissue as compared to normal mucosa [44]. Further, these results show that increased plasma miRNAs may come from primary tumors, but additional studies are needed to evaluate the association between circulating plasma miRNA levels and the levels in matched CRC tissues.
Interestingly, a substantial association between miR-21 expression levels and patient clinical and demographic traits, like age and cancer site, was discovered. Additionally, earlier research has demonstrated that miR-21 expression is elevated across the TNM stages of CRC, from early to late [44, 29].
However, there were no significant variations in miR-21 expression levels between early stages I, II, and advanced stage III in the current investigation. Instead, miR-21 expression levels were found to be higher in the early stages than in the advanced stages. The fact that there were fewer cases at each stage of the subgrouping could account for this difference.
At present, few studies have examined the pattern of miRNA expression at different stages of cancer. We are among the few early studies that have classified the stages and grades of colon cancer based on the expression of the miR-21 gene. The pattern of miR-21 gene expression in different stages of colon cancer has shown different results. However, most studies have reported a higher expression of miR-21 in stage II compared to other stages, with no significant differences observed in other stages. Some authors showed that if the miR-21 gene expression is high in patients with colon cancer stage II, they are prone to recurrence [45].
According to Kanaan et al. [46], who examined a broader sample size, plasma miR-21 appeared to be particularly promising as a CRC biomarker. Luo et al. [47] reported similar findings [46, 47]. Chen et al. [48] and Sazanov et al. [49] both found that miR-21 in plasma could be a good biomarker for CRC screening [48, 49]. According to Wang et al. [50], there is a connection between the expression levels of circulating miRNAs and the development of CRC, and miRNAs can serve as a non-invasive biomarker for CRC detection with a relatively high sensitivity and specificity. Contrary to our observations, Montagnana et al. [51] observed in their study that plasma miR-21 valuation does not seem to be a useful biomarker for diagnosing and staging CRC.
In the present study, miR-145, which is a tumor suppressor, was down-regulated. An earlier study by Wang et al. [52] observed that miR-145 might have an affirmative function in colorectal cancers development but not in their advancement . Another study by Feng et al. [53] found that miR-145 inhibits the invasive and metastatic abilities of CRC, most likely by targeting Fascin-1 directly; as a result, it appears to be implicated in the initiation and progression of CRC.
Al-Sheikh et al.’s [54] research yielded similar results, amply demonstrating a similar pattern of decreasing miR- 145 miRNA expression levels in the patients’ tissue and plasma. Furthermore, the trends observed by these authors regarding miR-145 expression levels were similar in the patients’ tissue and plasma. A new study by Qin et al. [55] showed for the first time that miR-145 directly targets KLF5 in colon cancer cell lines and stops the cell cycle in the G1 phase. Thus, it is important to emphasise that miR-145 may be a promising therapeutic target for colorectal cancer.
The present study estimated the levels of two microRNAs (miR-21 and miR-145) in the plasma samples of patients with colorectal cancer and healthy controls. Previous studies have demonstrated that blood-based miRNAs are stable and reproducible in expression levels over time or throughout freezing and thawing cycles, as well as resistant to RNase degradation [56]. Also, due to their easy access and non-invasive nature, circulating miRNAs (plasma miRNAs) may have a vital role in the diagnosis and monitoring of colorectal cancer.
In conclusion, these findings will shed light on the molecular development of colorectal cancer and highlight the potential applications of miR-21 and miR-145 as non-invasive, early diagnostic biomarkers. The results of this study demonstrate that measuring the levels of miR-21 and miR-145 expression in plasma could be useful for diagnosing CRC.
As a result, the current work emphasizes that measuring miR-21 plasma levels may be a useful non-invasive screening method for early CRC diagnosis.
Acknowledgements
The authors wish to express their gratitude to the patients and institutions at the Postgraduate Institute of Medical Education and Research (PGIMER) and Panjab University (PU) Chandigarh for their contributions to this study.
Funding
The Indian Council of Medical Research (ICMR), New Delhi, India, provided the funding for this work (Grant No. 3/2/2/112/2012/NCD-III).
Research involving Human Participants
The institutional research committee’s ethical guidelines, the 1964 Helsinki Declaration and its later amendments, or equivalent ethical standards were followed in all procedures carried out during investigations involving human participants.
Informed Consent
All individuals who took part in the study gave their informed consent.
Data availability
The data is available upon request from the author.
Conflict of Interest
The authors declare that they have no conflicts of interest.
Author Contribution Statement
Conceived and designed the experiments: RRN SVR. Performed the experiments: RRN. Analyzed the data: RRN SVR DKD. Contributed reagents/materials/analysis tools: SVR VG RG. Wrote the paper: RRN DKD. Study supervision: RRN, SVR, DKD VG. GUARANTOR: Corresponding author.
References
- 1.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 2.Siegel RL, Miller KD, Fedewa SA, Ahnen DJ, Meester RGS, Barzi A et al. Colorectal cancer statistics, 2017. CA Cancer J Clin. 2017;67(3):177–93. doi: 10.3322/caac.21395. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65(2):87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 4.Cunningham D, Atkin W, Lenz HJ, Lynch HT, Minsky B, Nordlinger B, et al. Colorectal cancer. Lancet. 2010;375(9719):1030–47. doi: 10.1016/S0140-6736(10)60353-4. [DOI] [PubMed] [Google Scholar]
- 5.Ferlay J, Soerjomataram I, Dikshit R, Eser S, Mathers C, Rebelo M, et al. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int J Cancer. 2015;136(5):E359–86. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 6.Mallath MK, Taylor DG, Badwe RA, Rath GK, Shanta V, Pramesh CS, et al. The growing burden of cancer in India: epidemiology and social context. Lancet Oncol. 2014;15(6):e205–12. doi: 10.1016/S1470-2045(14)70115-9. [DOI] [PubMed] [Google Scholar]
- 7.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 8.Gupta AK, Brenner DE, Turgeon DK. Early detection of colon cancer: new tests on the horizon. Mol Diagn Ther. 2008;12(2):77–85. doi: 10.1007/BF03256273. [DOI] [PubMed] [Google Scholar]
- 9.Pawa N, Arulampalam T, Norton JD. Screening for colorectal cancer: established and emerging modalities. Nat Rev Gastroenterol Hepatol. 2011;8(12):711–22. doi: 10.1038/nrgastro.2011.205. [DOI] [PubMed] [Google Scholar]
- 10.Ladabaum U, Song K. Projected national impact of colorectal cancer screening on clinical and economic outcomes and health services demand. Gastroenterology. 2005;129(4):1151–62. doi: 10.1053/j.gastro.2005.07.059. [DOI] [PubMed] [Google Scholar]
- 11.Van Rhijn BW, Van der Poel HG, Van der Kwast TH. Urine markers for bladder cancer surveillance: a systematic review. Eur Urol. 2005;47(6):736–48. doi: 10.1016/j.eururo.2005.03.014. [DOI] [PubMed] [Google Scholar]
- 12.Toloza EM, Harpole L, McCrory DC. Noninvasive staging of non-small cell lung cancer: a review of the current evidence. Chest. 2003;123(1Suppl):137S–146S. doi: 10.1378/chest.123.1_suppl.137s. [DOI] [PubMed] [Google Scholar]
- 13.Mandel JS, Bond JH, Church TR, Snover DC, Bradley GM, Schuman LM, et al. Reducing mortality from colorectal cancer by screening for fecal occult blood Minnesota Colon Cancer Control Study. N Engl J Med. 1993;328(19):1365–71. doi: 10.1056/NEJM199305133281901. [DOI] [PubMed] [Google Scholar]
- 14.Lieberman DA. Clinical practice Screening for colorectal cancer. N Engl J Med. 2009;361(12):1179–87. doi: 10.1056/NEJMcp0902176. [DOI] [PubMed] [Google Scholar]
- 15.Duffy MJ, Van Dalen A, Haglund C, Hansson L, Holinski- Feder E, Klapdor R, et al. Tumor markers in colorectal cancer: European Group on Tumor Markers (EGTM) guidelines for clinical use. Eur J Cancer. 2007;43(9):1348–60. doi: 10.1016/j.ejca.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 16.Esquela-Kerscher A, Slack FJ. Oncomirs - microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–69. doi: 10.1038/nrc1840. [DOI] [PubMed] [Google Scholar]
- 17.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–33. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shenouda SK, Alahari SK. MicroRNA function in cancer: oncogene or a tumor suppressor? Cancer Metastasis Rev. 2009;28(3-4):369–78. doi: 10.1007/s10555-009-9188-5. [DOI] [PubMed] [Google Scholar]
- 19.Patnaik SK, Kannisto E, Mallick R, Yendamuri S. Overexpression of the lung cancer-prognostic miR-146b microRNAs has a minimal and negative effect on the malignant phenotype of A549 lung cancer cells. PLoS One. 2011;6(7):e22379. doi: 10.1371/journal.pone.0022379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kong YW, Ferland-McCollough D, Jackson TJ, Bushell M. microRNAs in cancer management. Lancet Oncol. 2012;13(6):e249–58. doi: 10.1016/S1470-2045(12)70073-6. [DOI] [PubMed] [Google Scholar]
- 21.Ahmed FE, Jeffries CD, Vos PW, Flake G, Nuovo GJ, Sinar DR, et al. Diagnostic microRNA markers for screening sporadic human colon cancer and active ulcerative colitis in stool and tissue. Cancer Genomics Proteomics. 2009;6(5):281–95. [PubMed] [Google Scholar]
- 22.Cortez MA, Calin GA. MicroRNA identification in plasma and serum: a new tool to diagnose and monitor diseases. Expert Opin Biol Ther. 2009;9(6):703–711. doi: 10.1517/14712590902932889. [DOI] [PubMed] [Google Scholar]
- 23.Danese E, Minicozzi AM, Benati M, Paviati E, Lima-Oliveira G, Gusella M, et al. Reference miRNAs for colorectal cancer: analysis and verification of current data. Sci Rep. 2017;7(1):8413 . doi: 10.1038/s41598-017-08784-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mestdagh P, Van Vlierberghe P, De Weer A, Muth D, Westermann F, Speleman F, et al. A novel and universal method for microRNA RT-qPCR data normalization. Genome Biol. 2009;10(6):R64 . doi: 10.1186/gb-2009-10-6-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jacobsen N, Andreasen D, Mouritzen P. Profiling microRNAs by real-time PCR. Methods Mol Biol. 2011;732:39–54. doi: 10.1007/978-1-61779-083-6_4. [DOI] [PubMed] [Google Scholar]
- 26.Kroh EM, Parkin RK, Mitchell PS, Tewari M. Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR) Methods. 2010;50(4):298–301. doi: 10.1016/j.ymeth.2010.01.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pritchard CC, Cheng HH, Tewari M. MicroRNA profiling: approaches and considerations. Nat Rev Genet. 2012;13(5):358–69. doi: 10.1038/nrg3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Huang Z, Huang D, Ni S, Peng Z, Sheng W, Du X. Plasma microRNAs are promising novel biomarkers for early detection of colorectal cancer. Int J Cancer. 2010;127(1):118–26. doi: 10.1002/ijc.25007. [DOI] [PubMed] [Google Scholar]
- 29.Toiyama Y, Takahashi M, Hur K, Nagasaka T, Tanaka K, Inoue Y et al. Serum miR-21 as a diagnostic and prognostic biomarker in colorectal cancer. J Natl Cancer Inst. 2013;105(12):849–59. doi: 10.1093/jnci/djt101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 31.Cho WC. OncomiRs: the discovery and progress of microRNAs in cancers. Mol Cancer. 2007;6:60 . doi: 10.1186/1476-4598-6-60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6(11):857–66. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 33.Bhatavdekar JM, Patel DD, Ghosh N, Chikhlikar PR, Trivedi TI, Suthar TP, et al. Coexpression of Bcl-2, c-Myc, and p53 oncoproteins as prognostic discriminants in patients with colorectal carcinoma. Dis Colon Rectum. 1997;40(7):785–90. doi: 10.1007/BF02055433. [DOI] [PubMed] [Google Scholar]
- 34.Toyota M, Suzuki H, Sasaki Y, Maruyama R, Imai K, Shinomura Y, et al. Epigenetic silencing of microRNA- 34b/c and B-cell translocation gene 4 is associated with CpG island methylation in colorectal cancer. Cancer Res. 2008;68(11):4123–32. doi: 10.1158/0008-5472.CAN-08-0325. [DOI] [PubMed] [Google Scholar]
- 35.Slaby O, Svoboda M, Fabian P, Smerdova T, Knoflickova D, Bednarikova M, et al. Altered expression of miR-21, miR- 31, miR-143 and miR-145 is related to clinicopathologic features of colorectal cancer. Oncology. 2007;72(5-6):397–402. doi: 10.1159/000113489. [DOI] [PubMed] [Google Scholar]
- 36.Slattery ML, Wolff E, Hoffman MD, Pellatt DF, Milash B, Wolff RK. MicroRNAs and colon and rectal cancer: differential expression by tumor location and subtype. Genes Chromosomes Cancer. 2011;50(3):196–206. doi: 10.1002/gcc.20844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kosaka N, Iguchi H, Ochiya T. Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis. Cancer Sci. 2010;101(10):2087–92. doi: 10.1111/j.1349-7006.2010.01650.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A. 2008;105(30):10513–8. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Asangani IA, Rasheed SA, Nikolova DA, Leupold JH, Colburn NH, Post S, et al. MicroRNA-21 (miR-21) post- transcriptionally downregulates tumor suppressor Pdcd4 and stimulates invasion, intravasation and metastasis in colorectal cancer. Oncogene. 2008;27(15):2128–36. doi: 10.1038/sj.onc.1210856. [DOI] [PubMed] [Google Scholar]
- 40.Meng F, Henson R, Wehbe-Janek H, Ghoshal K, Jacob ST, Patel T. MicroRNA-21 regulates expression of the PTEN tumor suppressor gene in human hepatocellular cancer. Gastroenterology. 2007;133(2):647–58. doi: 10.1053/j.gastro.2007.05.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chang KH, Miller N, Kheirelseid EA, Ingoldsby H, Hennessy E, Curran CE, et al. MicroRNA-21 and PDCD4 expression in colorectal cancer. Eur J Surg Oncol. 2011;37(7):597–603. doi: 10.1016/j.ejso.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 42.Schmitz KJ, Hey S, Schinwald A, Wohlschlaeger J, Baba HA, Worm K, et al. Differential expression of microRNA 181b and microRNA 21 in hyperplastic polyps and sessile serrated adenomas of the colon. Virchows Arch. 2009;455(1):49–54. doi: 10.1007/s00428-009-0804-0. [DOI] [PubMed] [Google Scholar]
- 43.Goel A, Boland CR. Epigenetics of colorectal cancer. Gastroenterology. 2012;143(6):1442–60. doi: 10.1053/j.gastro.2012.09.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Liu GH, Zhou ZG, Chen R, Wang MJ, Zhou B, Li Y, et al. Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer. Tumor Biol. 2013;34(4):2175–81. doi: 10.1007/s13277-013-0753-8. [DOI] [PubMed] [Google Scholar]
- 45.Dehghan F, Boozarpour S, Torabizadeh Z, Alijanpour S. miR-21: a promising biomarker for the early detection of colon cancer. Onco Targets Ther. 2019;12:5601–7. doi: 10.2147/OTT.S199508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanaan Z, Rai SN, Eichenberger MR, Roberts H, Keskey B, Pan J, et al. Plasma miR-21: a potential diagnostic marker of colorectal cancer. Ann Surg. 2012;256(3):544–51. doi: 10.1097/SLA.0b013e318265bd6f. [DOI] [PubMed] [Google Scholar]
- 47.Luo X, Stock C, Burwinkel B, Brenner H. Identification and evaluation of plasma microRNAs for early detection of colorectal cancer. PLoS One. 2013;8(5):e62880. doi: 10.1371/journal.pone.0062880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H, Liu H, Zou H, Chen R, Dou Y, Sheng S, et al. Evaluation of Plasma miR-21 and miR-152 as Diagnostic Biomarkers for Common Types of Human Cancers. J Cancer. 2016;7(5):490–9. doi: 10.7150/jca.12351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sazanov AA, Kiselyova EV, Zakharenko AA, Romanov MN, Zaraysky MI. Plasma and saliva miR-21 expression in colorectal cancer patients. J Appl Genet. 2017;58(2):231–237. doi: 10.1007/s13353-016-0379-9. [DOI] [PubMed] [Google Scholar]
- 50.Wang J, Huang SK, Zhao M, Yang M, Zhong JL, Gu YY, et al. Identification of a circulating microRNA signature for colorectal cancer detection. PLoS One. 2014;9(4):e87451. doi: 10.1371/journal.pone.0087451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Montagnana M, Benati M, Danese E, Minicozzi AM, Paviati E, Gusella M, et al. Plasma Expression Levels of Circulating miR-21 are not Useful for Diagnosing and Monitoring Colorectal Cancer. Clin Lab. 2016;62(5):967–70. doi: 10.7754/clin.lab.2015.151015. [DOI] [PubMed] [Google Scholar]
- 52.Wang CJ, Zhou ZG, Wang L, Yang L, Zhou B, Gu J, et al. Clinicopathological significance of microRNA-31, -143 and -145 expression in colorectal cancer. Dis Markers. 2009;26(1):27–34. doi: 10.3233/DMA-2009-0601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Feng Y, Zhu J, Ou C, Deng Z, Chen M, Huang W, Li L. MicroRNA-145 inhibits tumor growth and metastasis in colorectal cancer by targeting fascin-1. Br J Cancer. 2014;110(9):2300–9. doi: 10.1038/bjc.2014.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Al-Sheikh YA, Ghneim HK, Softa KI, Al-Jobran AA, Al-Obeed O, Mohamed MA, et al. Expression profiling of selected microRNA signatures in plasma and tissues of Saudi colorectal cancer patients by qPCR. Oncol Lett. 2016;11(2):1406–12. doi: 10.3892/ol.2015.4068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Qin WW, Zhang R, Chen RA, Li GH, Ji YR, Liu L, et al. MicroRNA-145 induces cell cycle arrest in G1 phase by directly targeting KLF5 in colon cancer. Int J Clin Exp Pathol. . 2016:5197–5209. [Google Scholar]
- 56.Glinge C, Clauss S, Boddum K, Jabbari R, Jabbari J, Risgaard B, et al. Stability of Circulating Blood-Based MicroRNAs- Pre-Analytic Methodological Considerations. PLoS One. 2017;12(2):e0167969. doi: 10.1371/journal.pone.0167969. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data is available upon request from the author.