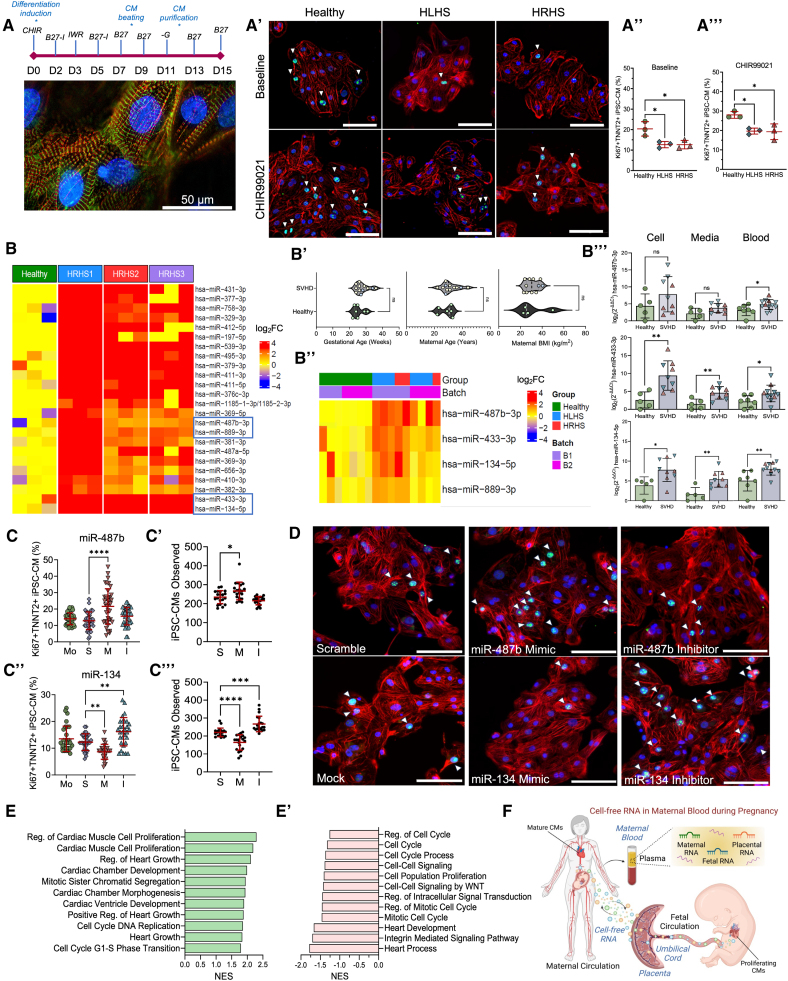
Single ventricle heart defects (SVHDs) are the most severe type of congenital heart disease.1 Through complex surgical procedures, patients with SVHDs can survive but ultimately have only one functional ventricle as either the left (hypoplastic left heart syndrome [HLHS]) or right (hypoplastic right heart syndrome [HRHS]) ventricle is hypoplastic. Prenatal diagnosis of SVHDs relies on ultrasound screening and fetal echocardiography. This complex imaging technology requires experienced physicians and sonographers along with costly equipment, which inevitably elevates the burden of health care expenditures and deepens the risk of health care inequity for socioeconomically disadvantaged groups. Cell-free DNA/RNA in maternal blood can serve as a noninvasive diagnostic test to detect and predict fetal and maternal health conditions; however, their ability to identify a fetus with an SVHD is unknown.2,3 In this study, we leveraged deep sequencing and patient-specific induced pluripotent stem cells (iPSCs) to identify cell-free RNA that could potentially be developed as noninvasive maternal biomarkers for prenatal detection of SVHDs.
Meet the First Author, see p 973
We hypothesized cardiac proliferation is dysregulated in patients with SVHDs. We compared the proliferation of iPSC-derived cardiomyocytes (iPSC-CMs) from individuals with SVHDs and healthy individuals. Under an institutional review board–approved protocol, we recruited healthy individuals (n=5) and patients with SVHD with either HLHS (n=4) or HRHS (n=5) to generate iPSC lines and differentiated into cardiomyocytes via Wnt signaling modulation.4 iPSC-CMs displayed a typical sarcomere structure with intercalated TNNT2 (cardiac troponin T) and α-actinin (Figure [A]). Cardiac proliferation between a subset of iPSC lines (n=3) from each group was assessed by quantifying coexpression of TNNT2 and cell cycle marker Ki67 (Figure [A′]). Healthy iPSC-CMs expressed a greater population of TNNT2+Ki67+ cells compared with SVHD cardiomyocytes (Figure [A″]). We further assessed cardiomyocyte proliferation by adding CHIR99021, which activates Wnt signaling and stimulates cardiomyocyte expansion5 (Figure [A‴]). Though CHIR99021 promoted cardiomyocyte proliferation, SVHD iPSC-CMs displayed lower percentages of TNNT2+Ki67+ cardiomyocytes compared with controls (HLHS, Padj=0.021; HRHS, Padj=0.018). Cell cycle defects in SVHD iPSC-CMs were also observed with EdU and pHH3 (phosphorylated histone H3) staining (data not shown). These results suggest SVHD iPSC-CMs exhibit cell proliferation abnormalities.
Figure.
Enriched cell-free RNA from single ventricle heart defect (SVHD) pregnancies dysregulates human cardiomyocyte (CM) proliferation. A, A cardiac differentiation protocol and immunofluorescence image of induced pluripotent stem cell–derived CMs (iPSC-CMs) with expression of α-actinin (green) and cardiac TNNT2 (cardiac troponin T) (red). A′, Cell proliferative ability of SVHD and healthy D15 iPSC-CMs is evaluated at baseline or with CHIR99021. Ki67 (green), TNNT2 (red), and nuclei (blue). A″ and A‴, Quantitative analysis of Ki67+TNNT2+ iPSC-CMs from healthy (n=3), HLHS (hypoplastic left heart syndrome) (n=3), and HRHS (hypoplastic right heart syndrome) (n=3) subjects at baseline and with CHIR99021. One-way ANOVA with Tukey post hoc comparison test. B, Heatmap of differentially expressed miRNA (DEmiRNA) in culture media of HRHS and healthy iPSC-CMs (Padj≤0.05; fold change [FC]≥2). B′, Demographics of maternal blood donors. Healthy (green), HLHS (yellow), and HRHS (blue). B″, Heatmap clustering shows candidate miRNAs elevated in SVHD pregnancies. B‴, quantitative PCR (qPCR) confirmation of upregulated miRNA candidates in maternal blood, iPSC-CM culture media, and intracellular compartments. Green circles (healthy), pink triangles (HRHS), and inverted blue triangles (HLHS). Student t test. C through C‴, Quantitative analysis of Ki67+TNNT2+ proliferating cardiomyocytes and cell population after perturbation with mock (Mo), scramble (S), miRNA mimic (M), and inhibitor (I) for hsa-miR-487b-3p (C and C′) and hsa-miR-134-5p (C″ and C‴). One-way ANOVA with Dunnett post hoc correction. Data are shown as mean±SD. D, Representative immunofluorescence images of miRNA perturbations in cardiomyocytes from a healthy induced pluripotent stem cell line. Ki67 (green), TNNT2 (red), and nuclei (blue). E and E′, Gene set enrichment analysis identifies significant biological processes affected by hsa-miR-487b-3p (E) and hsa-miR-134-5p (E′) mimic vs inhibitor perturbations. F, Schematic overview of maternal biomarker discovery using cell-free RNA during pregnancy. Scale bars: 50 µm (A) and 100 µm (A′ and D). BMI indicates body mass index; NES, normalized enrichment score; and Reg, regulation. *Padj<0.05, **Padj<0.01, ***Padj<0.001, ****Padj<0.0001.
Next, we asked whether SVHD iPSC-CMs secrete signaling molecules indicative of their hypoplastic-like behavior. We collected cell culture media of D15 iPSC-CMs to assess differentially expressed miRNA profiles. Overall, we identified 24 differentially expressed miRNAs (Padj<0.05; fold change [FC]>2) that were consistently enriched in all HRHS iPSC-CM cultures (Figure [B]). We sought to compare secreted differentially expressed miRNA profiles to those in maternal circulation from healthy and SVHD pregnancies.
We recruited pregnant patients (n=19) between 20 and 40 weeks of pregnancy, with no significant difference in the gestational age, maternal age, or maternal body mass index (Figure [B′]). Seven carried a healthy fetus and 12 carried an SVHD fetus (n=8 for HLHS and n=4 for HRHS). Echocardiograms were used to diagnose HLHS or HRHS fetal phenotypes. All pregnant individuals were healthy and did not have congenital or acquired cardiovascular abnormalities. We first sequenced cell-free miRNA profiles in maternal blood from pregnant patients carrying a SVHD (n=5) or healthy (n=3) fetus and selectively assessed the upregulated 24 miRNA profiles from in vitro cultures. After a second batch of maternal plasma samples (n=4 healthy and n=4 SVHD), we found hsa-miR-134-5p, hsa-miR-433-3p, hsa-miR-487b-3p, and hsa-miR-889-3p were consistently enriched (Padj<0.05; FC>2) in both maternal blood and iPSC-CM culture media (Figure [B″]). To prioritize targets for downstream analysis, we selected the top 3 DEmiRs based on their statistical significance and fold change. We performed real-time quantitative PCR (RT-qPCR) on maternal plasma, D15 iPSC-CMs, and culture media and verified consistent elevation of these miRNAs (Figure [B‴]).
To interrogate biological function of upregulated cell-free miRNAs in maternal blood, we manipulated their activity through transfecting miRNA mimic, inhibitor, scramble, or buffer (mock) to D15 human iPSC-CMs. We quantified the percentage of TNNT2+Ki67+ cardiomyocytes and cell number and examined transcriptional profiles. The miR-487b mimic significantly improved cardiomyocyte proliferation compared with controls (Padj<1.0×10−4), while inhibition had no effect (Padj=0.17). The population of iPSC-CMs showed similar trends (Figure [C and C′]). miR-433 mimic and inhibitor had no significant effect on cardiomyocyte proliferation. miR-134 mimic significantly suppressed cardiomyocyte proliferation (Padj=2.8×10−3) by reducing the percentage of Ki67+TNNT2+ iPSC-CMs, whereas the miR-134 inhibitor displayed increased Ki67+TNNT2+ iPSC-CMs (Padj=1.1×10−3). The populations of iPSC-CMs mimicked these trends with a decrease in the cell number between scramble and mimic and an increase between scramble and inhibitor (Figure [C″, C‴, and D]). RNA-sequencing analysis indicates pathways related to cardiac muscle cell proliferation and cardiac chamber morphogenesis were upregulated in iPSC-CMs transfected with the miR-487b mimic (Figure [E]), whereas cell cycle and heart development were downregulated in those with the miR-134 mimic (Figure [E′]). These results suggest an inhibitory role of hsa-miR-134-5p but a compensatory role of hsa-miR-487b on cardiac proliferation.
In conclusion, we identified elevated cell-free miRNAs in maternal blood of pregnant subjects carrying an SVHD fetus and leveraged iPSCs to investigate their functions in human cardiomyocyte proliferation. These cell-free miRNAs in maternal blood could be developed as noninvasive biomarkers for prenatal detection of fetal SVHDs, pending further animal studies and clinical validation (Figure [F]).
DATA AVAILABILITY
The data that support findings of this study are available from the corresponding author upon reasonable request. Cell-free miRNA-sequencing data (GSE233362) and bulk RNA-sequencing data (GSE266333) are deposited in the NCBI Gene Expression Omnibus (GEO) database.
ARTICLE INFORMATION
Acknowledgments
The authors acknowledge Dr Dennis Lewandowski for his assistance in editing the manuscript. The authors thank clinical research nurses Brian Beckman and Jade Hayden for recruiting pregnant subjects and patients with HLHS (hypoplastic left heart syndrome) and HRHS (hypoplastic right heart syndrome) at Nationwide Children’s Hospital.
Sources of Funding
This study was partially supported by the National Institutes of Health (R01HL155282, R21HL165406, and F32HL170581), Additional Ventures (Additional Ventures Innovation Fund [AVIF], Single Ventricle Research Fund [SVRF], TTE [Expansion Award]), and American Heart Association (23IPA1046350, 24TPA1288584, 24AVCSASV1277994, and 24POST1194909).
Disclosures
Nationwide Children’s Hospital has filed a US patent application regarding maternal biomarker discovery for detecting single ventricle heart defect in fetuses during pregnancy, with M. Alonzo, K. Texter, V. Garg, and M.-T. Zhao named as inventors. The other authors report no conflicts.
Nonstandard Abbreviations and Acronyms
- EdU
- 5-ethynyl-2′-deoxyuridine
- HLHS
- hypoplastic left heart syndrome
- HRHS
- hypoplastic right heart syndrome
- iPSC
- induced pluripotent stem cell
- iPSC-CM
- induced pluripotent stem cell–derived cardiomyocyte
- SVHD
- single ventricle heart defect
For Sources of Funding and Disclosures, see page 1022.
REFERENCES
- 1.Oster ME, Lee KA, Honein MA, Riehle-Colarusso T, Shin M, Correa A. Temporal trends in survival among infants with critical congenital heart defects. Pediatrics. 2013;131:e1502–e1508. doi: 10.1542/peds.2012-3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rasmussen M, Reddy M, Nolan R, Camunas-Soler J, Khodursky A, Scheller NM, Cantonwine DE, Engelbrechtsen L, Mi JD, Dutta A, et al. RNA profiles reveal signatures of future health and disease in pregnancy. Nature. 2022;601:422–427. doi: 10.1038/s41586-021-04249-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moufarrej MN, Wong RJ, Shaw GM, Stevenson DK, Quake SR. Investigating pregnancy and its complications using circulating cell-free RNA in women’s blood during gestation. Front Pediatr. 2020;8:605219. doi: 10.3389/fped.2020.605219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ye S, Wan X, Su J, Patel A, Justis B, Deschenes I, Zhao MT. Generation and expansion of human cardiomyocytes from patient peripheral blood mononuclear cells. J Vis Exp. 2021;168:e62206. doi: 10.3791/62206 [DOI] [PubMed] [Google Scholar]
- 5.Buikema JW, Lee S, Goodyer WR, Maas RG, Chirikian O, Li G, Miao Y, Paige SL, Lee D, Wu H, et al. Wnt activation and reduced cell-cell contact synergistically induce massive expansion of functional human iPSC-derived cardiomyocytes. Cell Stem Cell. 2020;27:50–63.e5. doi: 10.1016/j.stem.2020.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support findings of this study are available from the corresponding author upon reasonable request. Cell-free miRNA-sequencing data (GSE233362) and bulk RNA-sequencing data (GSE266333) are deposited in the NCBI Gene Expression Omnibus (GEO) database.