Abstract
It is generally believed that cellular chaperones facilitate the folding of virus capsid proteins, or that capsid proteins fold spontaneously. Here we show that p73, the major capsid protein of African swine fever virus (ASFV) failed to fold and aggregated when expressed alone in cells. This demonstrated that cellular chaperones were unable to aid the folding of p73 and suggested that ASFV may encode a chaperone. An 80-kDa protein encoded by ASFV, termed the capsid-associated protein (CAP) 80, bound to the newly synthesized capsid protein in infected cells. The 80-kDa protein was released following conformational maturation of p73 and dissociated before capsid assembly. Coexpression of the 80-kDa protein with p73 prevented aggregation and allowed the capsid protein to fold with kinetics identical to those seen in infected cells. CAP80 is, therefore, a virally encoded chaperone that facilitates capsid protein folding by masking domains exposed by the newly synthesized capsid protein, which are susceptible to aggregation, but cannot be accommodated by host chaperones. It is likely that these domains are ultimately buried when newly synthesized capsid proteins are added to the growing capsid shell.
Chaperones prevent the irreversible aggregation of proteins in cells (15, 16). The Hsp70/DnaK chaperones bind short stretches of hydrophobic amino acids exposed in nascent chains emerging from ribosomes. Proteins released from Hsp70 attempt to fold and bury hydrophobic domains, but if unsuccessful they rebind or are transferred to ring chaperonins such as GroEL and GroES or the TCP1 (TriC/CCT) chaperonin. A broad spectrum of newly synthesized polypeptides associate with these chaperones “in vivo,” and the chaperone pathway is viewed as one of broad specificity and high capacity (15, 16, 19, 37). The folding of some proteins, however, requires specialized chaperones, and these may be needed to coordinate protein folding with subunit assembly (18). The PapD chaperones of Escherichia coli, for example, reduce nonproductive interactions between pilin subunits before they are assembled into the base of the growing pilus (3).
The careful coordination of protein folding and subunit assembly are important during the assembly of icosahedral viruses. Icosahedral capsids contain an exact number of protein subunits assembled into an ordered lattice; the simplest ones contain 60 identical subunits, while the largest ones contain several thousand. Capsid subunits are synthesized as monomers in the cytosol and expose domains that are ultimately buried during capsid assembly. In order to prevent nonproductive capsid aggregation, it is important that inappropriate interactions between these domains are minimized before delivery of the capsid subunit onto the growing capsid shell. For some viruses aggregation may be prevented by host chaperones (14, 17, 24, 25) or through assembly with scaffold proteins (22, 29).
The Iridoviridae and African swine fever virus (ASFV) virus are a group of cytoplasmic DNA viruses with very large capsids. ASFV shares the genomic organization of the Poxviridae and the striking icosahedral symmetry of the Iridoviridae (1, 7, 21) and has been described as a missing evolutionary link between poxviruses and iridoviruses (31). A possible evolutionary link between ASFV and the Iridoviridae is supported by the sequence homology between the major capsid proteins of the viruses (33) and close similarities in morphology and morphogenesis (2, 21, 26, 27, 30, 34, 35, 43). Early studies on negatively stained and freeze-dried ASF virions have shown capsid layers 190 nm in diameter containing as many as 2,000 capsomeres organized into a hexagonal lattice, suggesting icosahedral symmetry (1, 7, 21). More recent cryoreconstructions of mature capsids of the iridovirus, Paramecium bursari chlorella virus, reveal 1,680 hexavalent capsomers containing as many as 5,000 copies of individual capsid proteins (41). At present it is not known how cells ensure the correct assembly of these large structures. For ASFV, assembly is initiated by the recruitment of the major capsid protein, p73, from the cytosol onto the cytosolic face of the endoplasmic reticulum (ER) (11, 12). The capsid protein is then assembled progressively into a large complex on both sides of ER cisternae (2, 12, 30). This is an energy-dependent process that requires a continuous supply of newly synthesized capsid protein (12, 13). The localized and vectorial assembly of several thousand capsid subunits into virions on ER cisternae suggests that some mechanism prevents premature aggregation of the newly synthesized capsid protein in the cytosol. Given the documented ability of molecular chaperones to prevent protein aggregation, we have investigated the role played by chaperones during the early stages of ASFV assembly. Surprisingly, the major capsid protein of ASFV could not be folded by host chaperones. Instead, the virus encodes a specialized chaperone which prevents aggregation of the capsid protein before delivery onto the growing capsid shell.
MATERIALS AND METHODS
Reagents, cells, viruses, and antibodies.
Vero cells were infected with the BA71v isolate of ASFV as described previously (11). The monoclonal antibody 4H3 has been described (11), 17LD3 was purchased from Ingensa (Madrid, Spain), and polyclonal antisera recognizing p73 were produced by immunizing rabbits with recombinant p73 produced as an inclusion body in E. coli. Antibodies recognizing the N terminus of B602Lp were generated by immunizing rabbits with a peptide (CEETLKQLYQRTNPYKQFKNDSR) coupled to keyhole limpet hemocyanin.
Metabolic labeling, immunoprecipitation, and sucrose density sedimentation.
Metabolic labeling, immunoprecipitation, and sucrose density sedimentation were carried out as described previously (11–13). Proteins were resolved by sodium dodecyl sulfate-polyacrylamide gel electrophoreses (SDS-PAGE) and detected by autoradiography.
Expression of p73 and B602Lp in BSC40 cells.
The reading frames for p73 and B602Lp were isolated by PCR from genomic viral DNA and subcloned into the pT7 vector (Invitrogen BV, Leek, The Netherlands). BSC40 cells were infected for 1 h with VTF7.3 strain of vaccinia virus encoding T7 polymerase. Cells were washed with serum-free medium and transfected with T7 vectors using Lipofectin (Gibco-BRL/Life Technologies, Ltd., Paisley, United Kingdom). Cells were analyzed for the expression of protein 24 h later.
RESULTS
Rapid conformational maturation of the capsid protein occurs in the cytoplasm before binding to the ER.
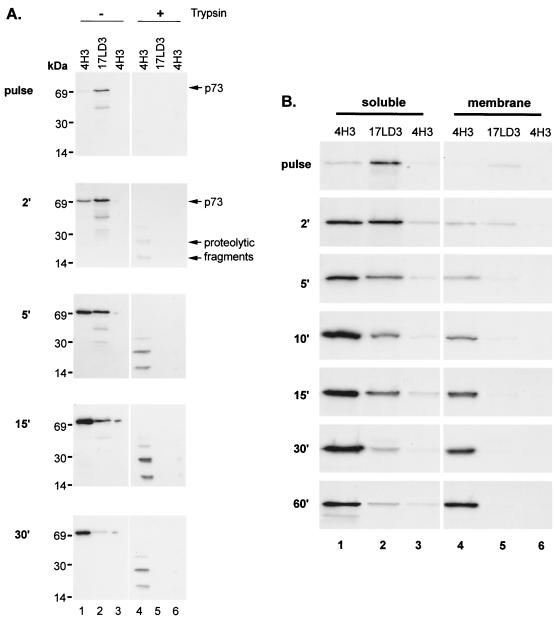
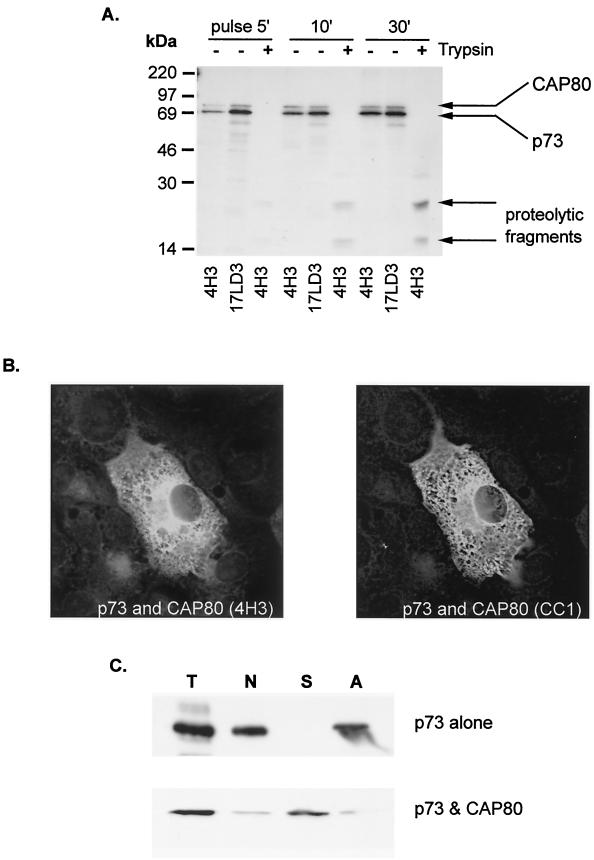
The folding of p73, the major capsid protein of ASFV, was studied by following the appearance of epitopes recognized by 4H3, a conformation-dependent antibody (Fig. 1A, left lanes). Infected cells were pulse-labeled for 2 min to label the nascent capsid protein and then chased for 30 min. The epitope recognized by 4H3 appeared at low levels after 2 min and increased to a peak at between 5 and 15 min. The increase in signal obtained using 4H3 was mirrored by a loss of signal when lysates were reprecipitated with a conformation-independent antibody, 17LD3. Taken together the immunoprecipitations indicated a rapid conformational maturation of p73 immediately after synthesis. The total quantity of p73 precipitated at each time point increased between 2 and 15 min; even so, the relative levels of folded versus unfolded p73 increased with time. The increase in signal during this period may be due to the slow elongation of nascent chains since the effect was reduced if the chase was repeated in the presence of cycloheximide (results not shown). The conformation of p73 was also tested by adding trypsin to cell lysates (Fig. 1A, right lanes). Significantly, protease-resistant fragments were absent from pulse-labeled cells but appeared by between 2 and 5 min into the chase period. Both experiments indicated a rapid conformational maturation of the capsid protein in infected cells.
FIG. 1.
Rapid conformational maturation of p73 occurs in the cytoplasm before binding to the ER. (A) Conformational maturation of p73 in infected cells. Infected Vero cells were pulse-labeled for 2 min and chased for the indicated times. Lysates were incubated in the absence (−) or presence (+) of trypsin and immunoprecipitated sequentially with the conformation-dependent antibody 4H3 and the conformation-independent antibody 17LD3. Any remaining capsid protein was detected by reprecipitation with 4H3. Proteins were resolved by SDS-PAGE and autoradiography. (B) Conformational maturation of p73 occurs before translocation onto the ER. Vero cells infected with ASFV were pulse-labeled for 2 min and chased for the indicated times. Crude membrane and cytosol preparations were lysed and immunoprecipitated sequentially with the conformation-dependent antibody 4H3 and the conformation-independent antibody 17LD3. Any remaining capsid protein was detected by reprecipitation with 4H3. Proteins were resolved by SDS-PAGE and autoradiography.
The capsid of ASFV is assembled on the cytoplasmic face of the ER (2, 11, 12, 30). The next experiment determined whether the capsid protein folded before binding to the ER membrane. If this was the case, it could be concluded that the early conformational maturation observed above took place before the addition of the capsid protein onto the growing capsid shell. Vero cells infected with ASFV were again pulse-labeled for 2 min and chased for increasing times. Crude membrane and cytosol fractions were prepared as indicated, and the conformation of p73 was probed by immunoprecipitation (Fig. 1B). As anticipated from the above experiments, 4H3 detected very little folded p73 during the pulse in either the cytosolic or membrane fraction (lanes 1 and 4, respectively). At this time point unfolded p73, as detected by 17LD3, was primarily located in the cytosolic fraction (lane 2). However, a very small proportion was present in the membrane fraction. This level fell during the experiment and was undetectable by 30 min into the chase (lane 5). After a 2-min chase approximately 40% of the p73 had folded into a conformation recognized by 4H3; the remainder was detected by 17LD3 and was therefore unfolded protein. Both pools were still confined to the cytosol at this time point. As the chase times were extended, the proportion of folded p73 increased, and this was paralleled by an increase in the levels of capsid detected on the membrane fraction (lane 4). We have shown previously that these membranes cosediment with ER marker proteins (11). Significantly, all of the membrane-associated capsid protein could be removed from lysates using the conformation-dependent antibody, indicating that only the conformationally mature form of p73 bound the ER. The results showed that the conformational maturation of p73 occurred before association with the ER membrane and therefore took place before the onset of assembly of the viral capsid.
Cellular chaperones are unable to assist capsid folding.
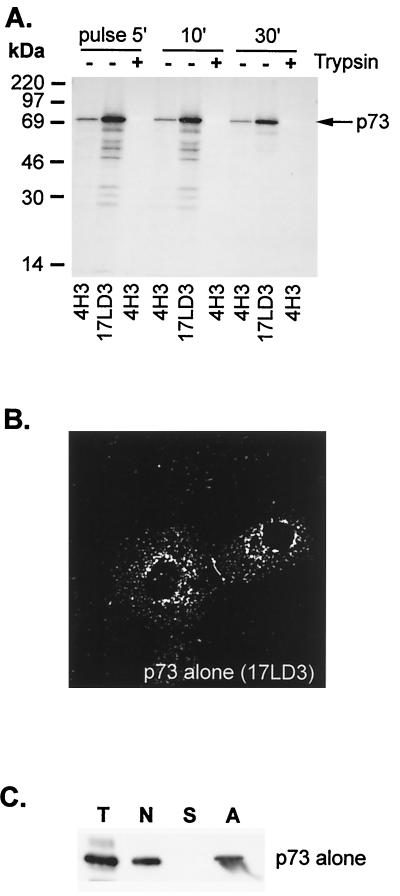
The ability of host chaperones to assist in the conformational maturation of p73 was tested by expressing the protein in the absence of other ASFV proteins (Fig. 2A). Low levels of p73 were recovered from lysates by 4H3, either during the pulse or during a 30-min chase, and p73 failed to mature into a conformation resistant to trypsin. Furthermore, ladders of proteins were immunoprecipitated by 17LD3, suggesting that misfolding resulted in degradation of the capsid. When the distribution of p73 expressed alone in cells was analyzed by immunofluorecence microscopy (Fig. 2B), the capsid protein was localized to clumps in the cytoplasm. These structures failed to colocalize with markers for cellular membrane compartments such as ERGIC, Golgi apparatus, endosomes, or lysosomes (not shown), suggesting aggregation of p73. The possible aggregation of p73 was tested further by extracting homogenized cells with mild detergent (Fig. 2C); under these conditions, more than half of the total p73 expressed in BSC40 cells pelleted with a crude membrane and nuclear fraction (N). Significantly, all the p73 contained within the pellet resedimented following extraction with immunoprecipitation buffer containing 1% Brij 35(A) Taken together, the lack of colocalization of p73 positive intracellular structures with membrane markers of the secretory pathway and the failure to solubilize p73 with detergent strongly suggested that p73 aggregated when expressed alone in cells.
FIG. 2.
The major capsid protein of ASF virus fails to fold when expressed alone in cells. (A) Conformational maturation of p73 expressed alone in cells. BSC40 cells were transfected with a plasmid encoding p73 under control of T7 polymerase promoter and infected with vaccinia virus encoding T7 polymerase (VTF7.3). Cells were pulse-labeled for 5 min with [35S]methionine and [35S]cysteine and chased as indicated. Matched lysates were precipitated with 4H3 or 17LD3 or incubated with trypsin (+) and then precipitated with 4H3 as indicated. (B) Subcellular distribution of p73 expressed alone in cells. Cells prepared as described in panel A above were fixed in methanol and blocked, and the location of p73 determined by immunofluorescence microscopy using 17LD3. (C) Subcellular fractionation of cells expressing p73. The capsid protein was expressed in BSC40 cells as described in the previous panel. Cells were homogenized by repeated passage through a 25-gauge needle and separated by centrifugation into a crude membrane and nuclear fraction (N) and a supernatant containing soluble protein (S). The nuclear-membrane fraction was extracted with mild detergent and recentrifuged to sediment aggregated protein (A). Representative samples of each fraction were separated by SDS-PAGE and probed by Western blot using 17LD3 (T, total; N, postnuclear membrane pellet; S, soluble protein; A, aggregate).
Association of p73 with viral protein, CAP80.
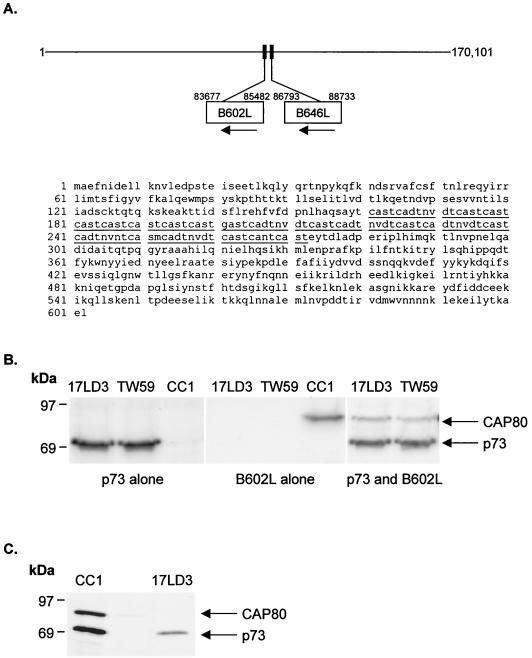
The aggregation of p73 expressed alone in cells showed that cellular chaperones could not assist the folding of the capsid protein and raised the interesting possibility that the virus may encode a chaperone. Newly synthesized p73 in infected cells migrates at between 150 and 200 kDa on sucrose gradients (12), suggesting that p73 may associate with a protein of similar size. Interestingly, analysis of the complete genome of ASFV (42) showed that the B602L reading frame, next to the gene encoding p73 (Fig. 3A), was one of the few viral genes encoding a protein of 70 kDa. A rabbit polyclonal antibody (CC1) was raised against a peptide representing a hydrophilic stretch of amino acids (residues 23 to 44) toward the N terminus of the protein encoded by the B602L gene. Immunoprecipitations of cells expressing p73 or the B602L protein alone are shown in panel B of Fig. 3. The capsid protein migrating at 70 kDa was precipitated by the conformation-independent monoclonal antibody 17LD3 and a rabbit polyclonal antibody raised against recombinant p73 expressed in bacteria (TW59). The B602L gene product migrated at 80 kDa. The 10-kDa increase in expected size of the B602L gene product was shown by deletion analysis to be caused by the central cysteine-rich domain, producing abnormal migration of the protein in SDS-PAGE (data not shown). Significantly, in cells expressing both proteins, complexes of p73 and the 80-kDa protein were recovered by both antibodies specific for p73, showing that the B602L gene product bound to p73. The presence of these complexes in cells infected with ASFV was confirmed by immunoprecipitation of metabolically labeled cells infected with ASFV (Fig. 3C). The antibody raised against the 80-kDa protein coprecipitated a 70-kDa protein from infected cells. When the complex was denatured, the 70-kDa protein could be reprecipitated by antibodies specific for p73. Given the properties of the B602L gene product, the protein was called capsid-associated protein 80 (CAP80).
FIG. 3.
AFSV encodes a capsid associated protein of 80 kDa. (A) Genome map of ASFV. The gene encoding p73 (B646L) lies next to the B602L reading frame. The amino acid sequence (20, 42) of the B602L reading frame is shown, and the central cysteine-rich domain is underlined. (B) The protein encoded by the B602L gene binds the major capsid protein of ASFV. The proteins were expressed in BSC40 cells either alone or together as described in the legend to Fig. 2. Cells were pulse-labeled for 30 min and immunoprecipitated using antibodies specific for p73 (17LD3 and TW59) or B602L (CC1). The migration of p73 and the B602L gene product (CAP80) following SDS-PAGE are indicated. (C) CAP80 binds the major capsid protein of ASFV in infected cells. Vero cells infected with ASFV were pulse-labeled for 30 min, lysed, and immunoprecipitated using antibody specific for CAP80 (CC1). Half of the precipitate was denatured in 1% SDS, diluted in lysis buffer, and reprecipitated using an antibody specific for p73 (17LD3). Proteins were resolved by reduced SDS-PAGE, and CAP80 and p73 are indicated.
Transient association of p73 with CAP80 in cells infected with ASFV.
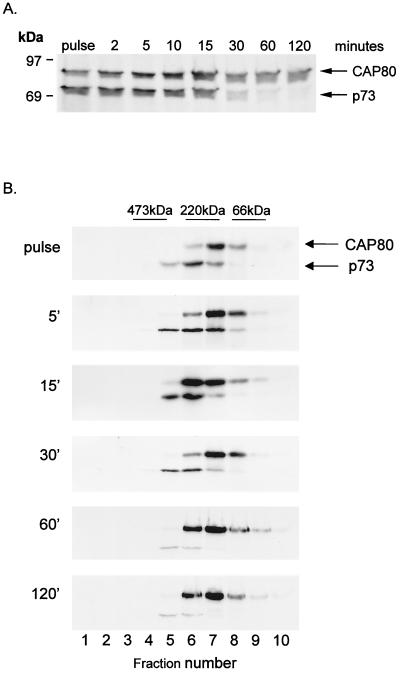
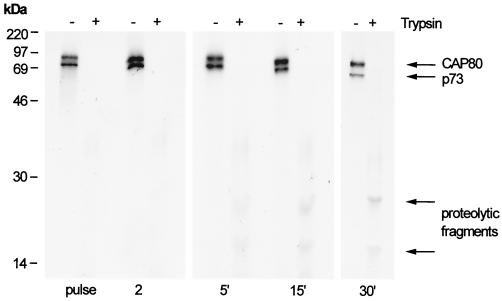
The time course of association of CAP80 with p73 in cells infected with ASFV was analyzed by pulse-chase immunoprecipitation (Fig. 4A). CAP80-p73 complexes were recovered from infected cells labelled for just 2 min, a time when the capsid was conformationally immature (see Fig. 1). Significantly, the capsid protein dissociated from CAP80 30 min into the chase, suggesting the release of CAP80 from the conformationally mature capsid protein. For many viruses, capsid assembly is assisted by scaffold proteins that form large procapsid complexes but dissociate during capsid maturation. To see if CAP80 were functioning as a scaffold during assembly, the sizes of complexes containing CAP80 were analyzed. Figure 4B shows that CAP80 migrated at 150 to 200 kDa on sucrose gradients, both during a short pulse and during the 30-min chase period coincident with the onset of dissociation of p73. We have shown previously (12) that assembly of p73 into a large protein complex indicative of a capsid begins 30 min after synthesis and that p73 migrates at the bottom of sucrose gradients at these time points. The lack of movement of CAP80 to the bottom of the sucrose gradient suggested that dissociation of CAP80 occurred before addition of p73 to the growing capsid shell. The results suggested that CAP80 is primarily involved in folding p73 subunits rather than acting as a scaffold protein to produce procapsids.
FIG. 4.
CAP80 associates transiently with the capsid protein of ASFV and does not form a procapsid. (A) Vero cells infected with ASFV were pulse-labeled for 2 min and chased for the indicated times. Cells were lysed and immunoprecipitated using antibodies specific for CAP80 (CC1). (B) Cell lysates were centrifuged for 20 h on 10 to 40% sucrose gradients. Fractions were immunoprecipitated using antibodies specific for CAP80 (CC1). Proteins were resolved by reduced SDS-PAGE, followed by autoradiography. The migrations of p73, CAP80, and molecular size standards (66 kDa, bovine serum albumin; 220 kDa, β-amylase; 473 kDa, apoferritin) are indicated.
CAP80 facilitates folding of virus capsid protein.
The ability of CAP80 to affect the folding of p73 was tested directly by expressing both proteins in fibroblasts (Fig. 5A). Importantly, when p73 was expressed with CAP80, the capsid protein folded rapidly after synthesis. The levels recovered by the conformation-dependent antibody 4H3 and the conformation-independent antibody 17LD3 from matched lysates taken from cells pulse-labeled for 5 min were the same (panel A), suggesting that most of the p73 was folded. Moreover, proteolytic fragments indicative of folding were recovered when lysates were incubated with trypsin, and the levels of these increased over 30 min. Immunofluorescence analysis of these cells (Fig. 5B) showed that both proteins were localized to a diffuse reticular stain in the cytoplasm rather than to aggregates, as observed in Fig. 2B. The reticular stain was provocative since it suggested association of a p73-CAP80 complex with the ER, the site of assembly of ASFV. Unfortunately, we were unable to confirm this since the reticular stain failed to colocalize with calnexin or the luminal ER protein ERP60 (not shown). The ability of CAP80 to increase the solubility of p73 was tested using the membrane extraction assay described above for Fig. 2C. Coexpression with CAP80 markedly increased the recovery of p73 from the soluble fraction, showing that the solubility of the capsid was substantially increased by CAP80 (Fig. 5C). This ability to promote folding and prevent the aggregation of p73 was highly indicative of a chaperone function for CAP80.
FIG. 5.
CAP80 facilitates the folding of p73. (A) Conformational maturation of AFSV capsid protein coexpressed with CAP80. Monkey BSC40 cells were transfected with plasmids encoding p73 and CAP80 as described in Fig. 2. Cells were pulse-labeled for 5 min and chased as indicated. Matched lysates were precipitated with 4H3 or 17LD3 or incubated with trypsin (+) and then precipitated using 4H3. The migrations of CAP80, p73, and proteolytic fragments are indicated. (B) Subcellular distribution of capsid protein coexpressed with CAP80. P73 and CAP80 were coexpressed in BSC40 cells as described above. Cells were fixed in methanol and blocked, and the locations of p73 and CAP80 were determined by immunofluorescence microscopy using 17LD3 and CC1, respectively. Primary antibodies were visualized using appropriate secondary antibodies coupled to coupled to Alexa Fluor 488 or 594. (C) Subcellular factionation of cells coexpressing p73. P73 protein and CAP80 were expressed in BSC40 cells as described above and homogenized and fractionated as described in the legend to Fig. 2. The distribution of p73 in representative samples was probed by Western blot using 17LD3 (T, total; N, postnuclear membrane pellet; S, soluble fraction; A, aggregate). The panel compares the distribution of p73 expressed alone (top, from Fig. 2) with the distribution when the protein was coexpressed with CAP80 (bottom).
The capsid protein dissociates from CAP80 after folding.
A crucial feature of molecular chaperones is the ability to bind unfolded or conformationally immature proteins and then be released once folding is completed. The above data showed that CAP80 bound unfolded p73 in infected cells and in cells expressing just the two ASFV proteins, but the change in conformation of p73 associated with CAP80 had not been tested directly. The conformation of p73 associated with CAP80 was probed by adding trypsin to washed immunoprecipitates of CAP80 obtained from cells infected with ASFV (Fig. 6). The production of tryptic fragments would indicate that p73 bound to CAP80 was folded. When immunoprecipitates isolated from pulse-labeled cells were probed with trypsin, we were unable to detect proteolytic fragments, suggesting that the associated p73 was conformationally immature. Importantly, after a 5-min chase, proteolytic fragments were observed, indicating that the p73 associated with CAP80 was now folded. At 30 min the levels of p73 decreased, indicating dissociation of folded capsid. These data show directly that in infected cells the capsid protein folds while associated with CAP80 and is ultimately released and so provide further convincing evidence for the chaperone function of CAP80.
FIG. 6.
The conformation of p73 matures while the capsid protein is bound to CAP80. Infected cells were pulse-labeled for 2 min and chased as indicated. Cell lysates were immunoprecipitated with CC1 to capture CAP80-p73 complexes. The conformation of p73 bound to CAP80 was tested by adding trypsin to one half of the immunoprecipitate (+), and the presence of protease-resistant fragments was detected by SDS-PAGE and autoradiography.
DISCUSSION
Since viruses are obligate intracellular parasites, it is generally believed that the folding of viral capsid proteins is accommodated by host chaperones (14, 17, 24, 25) or that capsid proteins fold spontaneously (28). This is based on the premise that the basic structures of virus capsids, and the cellular pathways that fold them, were put in place very early in viral evolution (4, 5). This study has shown that the major capsid protein of ASFV undergoes a rapid conformational change very soon after synthesis. Remarkably, host chaperones were unable to facilitate this conformational maturation, and the capsid protein aggregated and precipitated when expressed alone in cells. The conformational maturation of the viral capsid protein was, instead, dependent on association with a virally encoded protein, termed CAP80. Several properties of CAP80 suggested that the protein functioned as a chaperone to aid the conformational maturation of the capsid. First, CAP80 prevented the aggregation of p73 and increased the solubility of the protein, suggesting an ability to mask hydrophobic domains exposed on the newly synthesized capsid protein. Second, coexpression of CAP80 and p73 facilitated the folding of p73 and allowed the capsid protein to fold with kinetics similar to that seen in infected cells. Third, in common with molecular chaperones, CAP80 associated transiently with its substrate. Sucrose density gradient sedimentation and membrane fractionation experiments showed that, in infected cells, CAP80 bound to the newly synthesized “unfolded” capsid protein but dissociated before the conformationally mature capsid protein was assembled into virions on the ER membrane.
The Hsp70 and Hsp40 proteins and ring chaperonins prevent protein aggregation by burying hydrophobic domains exposed on nascent chains emerging from the ribosome until sufficient structural information is available for the protein to fold productively. It is interesting to consider why host chaperones were unable to prevent aggregation of p73. The primary sequence of p73 contains several short stretches of hydrophobic residues flanked by basic residues able to bind Hsp70 (6). It is likely, therefore, that p73 binds Hsp70. The observed aggregation of p73 indicates that the capsid protein exposes aggregation prone domains that cannot be masked by Hsp70 or does not have sufficient structural information to fold productively. Since aggregation was prevented by CAP80, the viral chaperone must mask aggregation prone sites that are missed by host chaperones and/or provide the structural information necessary for productive folding. Immunoprecipitation analysis failed to detect proteins other than p73 associated with CAP80. CAP80 does not therefore appear to bind other viral proteins and appears to be a specialized chaperone dealing with specific protein folding problems posed by p73. We cannot at this point exclude the possibility that CAP80 associates with host chaperones since the short metabolic labeling times used in the immunoprecipitation experiments may not have detected host chaperones with low turnover rates.
It is not unusual for chaperones to show specificity for individual proteins (18). In most cases, specialized chaperones bind proteins which, in common with capsid proteins, ultimately self-associate. The PapD chaperone of E coli, for example, prevents aggregation of individual pillus subunits and facilitates the assembly of pili (3). In vertebrates, Hsp47 binds to procollagen specifically (18, 36), and the chaperone auxillin regulates clathrin coat assembly and disassembly (38). It is thought that specialized chaperones bind specific domains in nascent protein chains that are prone to cause aggregation and dissociate when these domains are masked during protein self-assembly. This model would explain the specificity of CAP80 for p73. One function for CAP80 could be to mask such aggregation-prone sites on p73 until they can be used as binding sites during transfer to the growing capsid shell. Cycles of binding and release of CAP80 would prevent premature assembly or aggregation in the cytosol and enable vectorial assembly of the virus on the ER membrane. For several viruses, the delivery of capsid subunits onto growing capsid shells induces a conformational change that exposes a site for the binding of the next subunit. This process, called conformational switching, allows the capsid to increase in size in a stepwise manner, eventually establishing icosahedral symmetry (8). If further conformational changes in p73 took place following assembly into the capsid shell, these could trigger release of CAP80.
Our results show that the conformation of p73 changed while the protein was bound to CAP80 and raise the interesting possibility that CAP80 can actively induce the conformational maturation of the capsid protein. If so, CAP80 appears to function differently from Hsp70 and the ring chaperonins that prevent aggregation but do not actively change the conformation of the associated proteins (15, 16). The functions of CAP80 are more similar to PapD that induces a conformational change in pillin to ensure vectorial assembly at the base of the pilus (3). PapD prevents aggregation of the newly synthesized pillin subunit by donating a β strand to an exposed hydrophobic pocket. During the assembly of the pilus, PapD dissociates, and the pocket is filled by a β strand from the neighboring pilin subunit (10, 23, 32). This process of donor strand complementation is similar in principal to conformational switching employed by icosahedral viruses and allows protein folding to be coordinated with particle assembly. Whether CAP80 provides similar transient structural information to p73 prior to assembly of the ASFV capsid will have to await analysis of the crystal structure of CAP80-p73 complexes.
Since the discovery of cellular chaperones more than 20 years ago (17), there are few detailed studies of virally encoded chaperones required for capsid assembly. The Gp31 protein of the T4 bacteriophage, for example, is a functional homolog of GroES (39), and the adenovirus p100 protein mediates the assembly of hexon trimers (9). Although not strictly involved in capsid folding, vaccinia virus protein A33R acts as a chaperone to recruit viral protein A36R into virion envelopes and, in the absence of A33R, the A36R protein is incorrectly localized to the Golgi apparatus (40). A requirement for a virally encoded chaperone during capsid folding may be rare because of the evolutionary risk imposed on the virus. Database searches revealed little homology between CAP80 and known chaperones or host proteins. The evolutionary origins of CAP80 therefore remain obscure. The reading frame for CAP80 lies next to the gene encoding p73 in the center of the ASFV genome (20, 42). Interestingly, both reading frames read from the same direction, raising the possibility that they may originally have been joined and encoded a single structural protein. If the reading frames were separated during the evolution of ASFV, this would explain why the two proteins now have to be expressed together to provide sufficient structural information for productive protein folding.
ACKNOWLEDGMENTS
This work supported by the Biology and Biotechnology Research Council.
We are grateful to Saski Van der Vies, John Ellis, and Martin Carden for helpful discussions about viral chaperones and to Steve Archibald for graphics.
REFERENCES
- 1.Almeida J D, Waterson A P, Plowright W. The morphological characteristics of African Swine fever virus and its resemblance to Tipula iridescent virus. Archiv Gesamte Virusforsch. 1967;20:392–396. doi: 10.1007/BF01241958. [DOI] [PubMed] [Google Scholar]
- 2.Andres G, Garcia-Escuderu R, Simon-Mateo C, Vinuela E. African swine fever virus is enveloped by a two-membraned collapsed cisternae derived from the endoplasmic reticulum. J Virol. 1998;72:8988–9001. doi: 10.1128/jvi.72.11.8988-9001.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnhart M M. PapD-like chaperones provide the missing information for folding of pilin proteins. Proc Natl Acad Sci USA. 2000;97:7709–7714. doi: 10.1073/pnas.130183897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Belnap D M, Steven A C. ‘Déjà vu all over again’: the similar structures of bacteriophage PRD1 and adenovirus. Trends Microbiol. 2000;8:91–93. doi: 10.1016/s0966-842x(00)01704-2. [DOI] [PubMed] [Google Scholar]
- 5.Benson S D, Bamford J K H, Bamford D H, Burnett R M. Viral evolution revealed by bacteriophage PRD1 and human adenovirus coat protein structures. Cell. 1999;98:825–833. doi: 10.1016/s0092-8674(00)81516-0. [DOI] [PubMed] [Google Scholar]
- 6.Blond-Elguindi S, Cwirla S E, Dower W J, Lipshutz R L, Sprang S R, Sambrook J F, Gething M-J H. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of Bip. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 7.Carrascosa J L, Carazo J M, Carrascosa A L, Garcia N, Santisteban A, Vinuela E. General morphology and capsid fine structure of African swine fever virus. Virology. 1984;132:160–172. doi: 10.1016/0042-6822(84)90100-4. [DOI] [PubMed] [Google Scholar]
- 8.Casjens S. Principles of virus structure, function, and assembly. In: Chiu W, Burnett R M, Garcea R L, editors. Structural biology of viruses. Oxford, England: Oxford University Press; 1996. pp. 4–37. [Google Scholar]
- 9.Cepko C L, Sharp P A. Assembly of adenovirus major capsid protein is mediated by a non viral protein. Cell. 1982;31:407–415. doi: 10.1016/0092-8674(82)90134-9. [DOI] [PubMed] [Google Scholar]
- 10.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinker J, Hultgren S J, Knight S D. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 11.Cobbold C, Whittle J T, Wileman T. Involvement of the endoplasmic reticulum in the assembly and envelopment of African swine fever virus. J Virol. 1996;70:8382–8390. doi: 10.1128/jvi.70.12.8382-8390.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cobbold C, Wileman T. The major structural protein of African swine fever virus, p73, is packaged into large structures, indicative of viral capsid or matrix precursors, on the endoplasmic reticulum. J Virol. 1998;72:5215–5223. doi: 10.1128/jvi.72.6.5215-5223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cobbold C, Brookes S M, Wileman T. Biochemical requirements of virus wrapping by the endoplasmic reticulum: involvement of ATP and endoplasmic reticulum calcium store. J Virol. 2000;74:2151–2160. doi: 10.1128/jvi.74.5.2151-2160.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cripe T P, Delos S E, Estes P A, Garcea R L. In vivo and in vitro association of Hsc70 with polyomavirus capsid proteins. J Virol. 1995;69:7807–7813. doi: 10.1128/jvi.69.12.7807-7813.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ellis R J, Hartl F U. Principles of protein folding in the cellular environment. Curr Opin Struct Biol. 1999;270:102–110. doi: 10.1016/s0959-440x(99)80013-x. [DOI] [PubMed] [Google Scholar]
- 16.Feldman D E, Frydman J. Protein folding in vivo: importance of molecular chaperones. Curr Opin Struct Biol. 2000;10:26–33. doi: 10.1016/s0959-440x(99)00044-5. [DOI] [PubMed] [Google Scholar]
- 17.Georgopoulus C, Hendrix R, Casjens S, Kaiser A. Host participation in bacteriophage lambda head assembly. J Mol Biol. 1973;76:45–60. doi: 10.1016/0022-2836(73)90080-6. [DOI] [PubMed] [Google Scholar]
- 18.Hendershot L M, Bulleid N J. Protein-specific chaperones: the role of Hsp47 begins to gel. Curr Biol. 2000;10:R912–R915. doi: 10.1016/s0960-9822(00)00850-2. [DOI] [PubMed] [Google Scholar]
- 19.Houry W A, Frishman D, Eckerskorn C, Lottspelch F, Hartl F U. Identification of in vivo substrates of the chapronin groEL. Nature. 1999;402:147–154. doi: 10.1038/45977. [DOI] [PubMed] [Google Scholar]
- 20.Irusta P M, Borca M V, Kutish G F, Lu Z, Caler E, Carrillo C, Rock D L. Amino acid tandem repeats within a late viral gene define the central variable region on African swine fever virus. Virology. 1996;220:20–27. doi: 10.1006/viro.1996.0281. [DOI] [PubMed] [Google Scholar]
- 21.Kelly D C, Robertson J S. Icosahedral cytoplasmic deoxyriboviruses. J Gen Virol. 1973;20:17–41. doi: 10.1099/0022-1317-20-Supplement-17. [DOI] [PubMed] [Google Scholar]
- 22.King J, Casjens S. Catalytic head assembly protein in virus morphogenesis. Nature. 1974;251:112–117. doi: 10.1038/251112a0. [DOI] [PubMed] [Google Scholar]
- 23.Kuehn M J, Ogg D J, Kihlberg J, Slonim L N, Flemmer K, Bergfors T, Hultgren S J. Structural basis of pilus subunit recognition by the PapD chaperone. Science. 1993;262:1234–1243. doi: 10.1126/science.7901913. [DOI] [PubMed] [Google Scholar]
- 24.Lingappa J R, Martin R L, Wong M L, Ganem D, Welch W J, Lingappa V R. A eukaryotic cytosolic chaperonin is associated with a high molecular weight intermediate in the assembly of hepatitis B capsid, a multimeric particle. J Cell Biol. 1994;125:99–111. doi: 10.1083/jcb.125.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Macejak D G, Sarnow P. Association of Hsp70 with enterovirus capsid precursor P1 in infected human cells. J Virol. 1992;66:1520–1527. doi: 10.1128/jvi.66.3.1520-1527.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Manyakov V F. Fine structure of the iridescent virus type 1 capsid. J Gen Virol. 1977;36:73–79. [Google Scholar]
- 27.Moura Nunes J F, Vigario J D, Terrinha A M. Ultrastructural study of African swine fever virus replication in cultures of swine bone marrow cells. Arch Virol. 1975;49:59–66. doi: 10.1007/BF02175596. [DOI] [PubMed] [Google Scholar]
- 28.Nicola A V, Chen W, Helenius A. Cotranslational folding of an alphavirus capsid protein in the cytosol of living cells. Nat Cell Biol. 1999;1:341–345. doi: 10.1038/14032. [DOI] [PubMed] [Google Scholar]
- 29.Prevelidge P E, Thomas D, King J. Scaffold proteins regulate the polymerization of P22 coat proteins into icosahedral shells in vitro. J Mol Biol. 1988;202:743–757. doi: 10.1016/0022-2836(88)90555-4. [DOI] [PubMed] [Google Scholar]
- 30.Rouiller I, Brookes S M, Hyatt A D, Windsor M, Wileman T. African swine fever virus is wrapped by the endoplasmic reticulum. J Virol. 1998;72:2327–2387. doi: 10.1128/jvi.72.3.2373-2387.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Salas J, Salas M L, Vinuela E. African swine fever virus: a missing link between poxviruses and iridoviruses. In: Domingo E, Webster R, Holland J, editors. Origin and evolution of viruses. New York, N.Y: Academic Press, Inc.; 1999. pp. 467–480. [Google Scholar]
- 32.Sauer F G, Futterer K, Pinker J S, Dodson K W, Hultgren S J, Waksman G. Structural basis of chaperone function and pilus biogenesis. Science. 1999;285:1058–1061. doi: 10.1126/science.285.5430.1058. [DOI] [PubMed] [Google Scholar]
- 33.Schnitzler P, Darai G. Identification of the gene encoding the major capsid protein of fish lymphocystis virus. J Gen Virol. 1993;74:2143–2150. doi: 10.1099/0022-1317-74-10-2143. [DOI] [PubMed] [Google Scholar]
- 34.Stoltz D B. The structure of icocahedral cytoplasmic deoxyriboviruses. J Ultrastruct Res. 1971;37:219–239. doi: 10.1016/s0022-5320(71)80052-7. [DOI] [PubMed] [Google Scholar]
- 35.Stoltz D B. The structure of icocahedral cytoplasmic deoxyriboviruses. II. An alternative model. J Ultrastruct Res. 1973;43:58–74. doi: 10.1016/s0022-5320(73)90070-1. [DOI] [PubMed] [Google Scholar]
- 36.Tasab M, Batten M R, Bulleid N J. Hsp47: a molecular chaperone that interacts with and stabilizes correctly folded procollagen. EMBO J. 2000;19:2204–2211. doi: 10.1093/emboj/19.10.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Thulasiraman V, Yang C-F, Frydman J. In vivo newly translated polypeptides are sequestered in a protected folding environment. EMBO J J. 1999;18:85–95. doi: 10.1093/emboj/18.1.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ungewickell E, Ungewickell H, Holstein S E H, Linder R, Prasad K, Barouch W, Martin B, Greene L, Eisenberg E. Role of auxilin in uncoating clathrin-coated vesicles. Nature. 1995;378:632–635. doi: 10.1038/378632a0. [DOI] [PubMed] [Google Scholar]
- 39.Van der Vies S M, Gatenby A A, Georgopoulus C. Bacteriophage T4 encodes a co-chaperonin that can substitute for Escherichia coli GroES in protein folding. Nature. 1994;368:654–656. doi: 10.1038/368654a0. [DOI] [PubMed] [Google Scholar]
- 40.Wolffe E J, Weisberg A S, Moss B. The vaccinia virus protein A33R provides a chaperone function for viral membrane localization and tyrosine phosphorylation of the A36R protein. J Virol. 2001;75:303–310. doi: 10.1128/JVI.75.1.303-310.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yan X, Olson N H, Van Etten J L, Bergoin M, Rossmann M S, Baker T S. Structure and assembly of a large lipid containing dsDNA virus. Nat Struct Biol. 2000;7:101–103. doi: 10.1038/72360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yanez R J, Rodriguez J M, Nogal M L, Yuste L, Enriquez C, Rodriguez J F, Vinuela E. Analysis of the complete sequence of African swine fever virus. Virology. 1995;208:249–278. doi: 10.1006/viro.1995.1149. [DOI] [PubMed] [Google Scholar]
- 43.Yule B G, Lee P E. A cytological and immunological study of Tipula iridescent virus-infected Galleria mellonella larval hemocytes. Virology. 1973;51:409–423. doi: 10.1016/0042-6822(73)90440-6. [DOI] [PubMed] [Google Scholar]