Abstract
4-Carboxymethyl-4-methylbut-2-en-4-olide (4-methyl-2-enelactone) isomerase, transforming 4-methyl-2-enelactone to 3-methyl-2-enelactone, was purified from a derivative strain of Pseudomonas sp. B13, named B13 FR1, carrying the plasmid pFRC2OP. This plasmid contained the isomerase gene cloned from Alcaligenes eutrophus JMP 134, which uses 4-methyl-2-enelactone as a carbon source. The enzyme consists of a single peptide chain of Mr 40,000 as judged by SDS/PAGE. In addition to 4-methyl-2-enelactone, the putative reaction intermediate, 1-methyl-3,7-dioxo-2,6-dioxy-bicyclo[3.3.0]octane (1-methylbislactone), was a substrate for the enzyme, but kinetic data presented did not favour its role as a reaction intermediate. Isomeric methyl-substituted 4-carboxymethylbut-2-en-4-olides were neither substrates nor inhibitors. Possible reaction mechanisms are discussed.
Full text
PDF


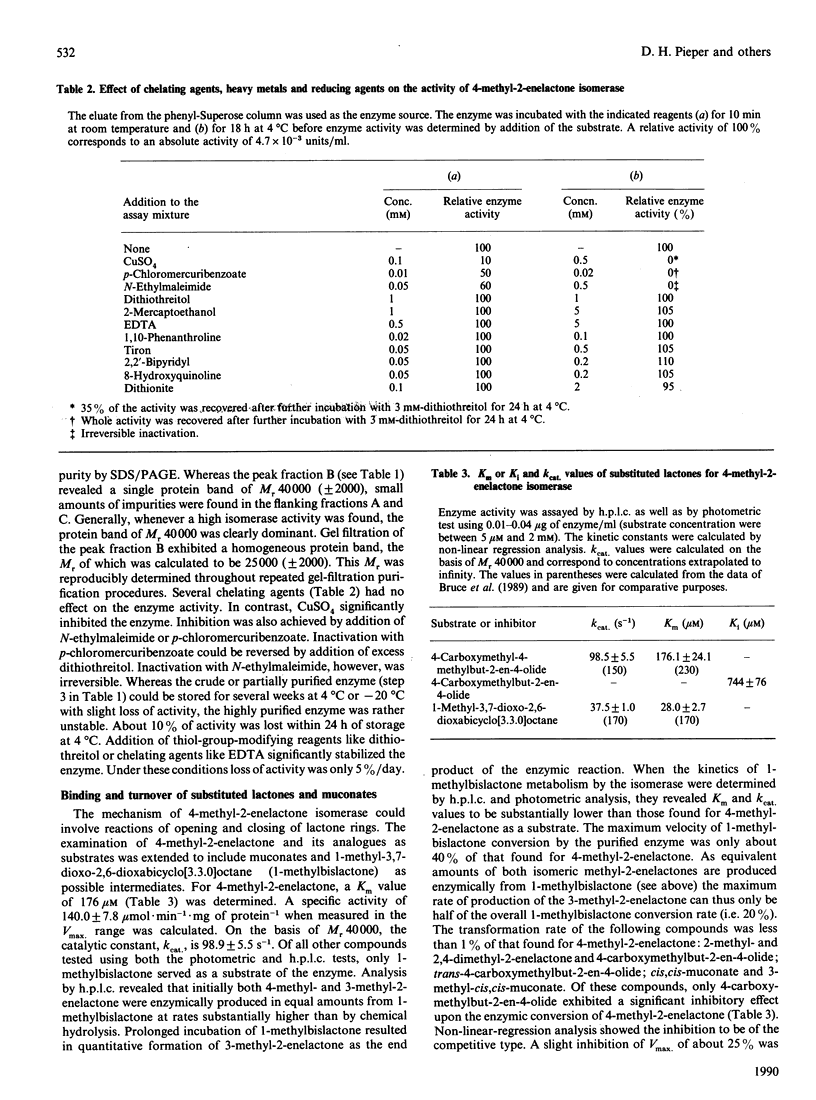
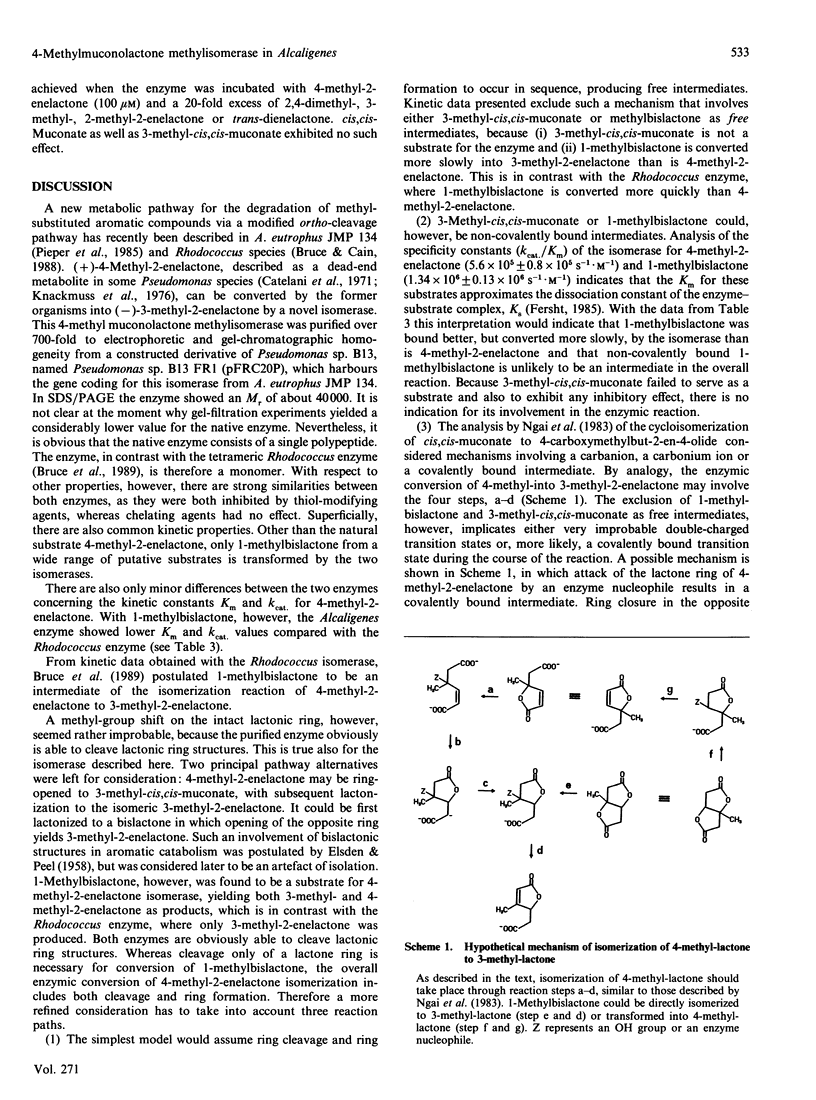

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Bruce N. C., Cain R. B., Pieper D. H., Engesser K. H. Purification and characterization of 4-methylmuconolactone methyl-isomerase, a novel enzyme of the modified 3-oxoadipate pathway in nocardioform actinomycetes. Biochem J. 1989 Aug 15;262(1):303–312. doi: 10.1042/bj2620303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catelani D., Fiecchi A., Galli E. Dextro-gamma-carboxymethyl-gamma-methyl-delta-alpha-butenolide. A 1,2-ring-fission product of 4-methylcatechol by Pseudomonas desmolyticum. Biochem J. 1971 Jan;121(1):89–92. doi: 10.1042/bj1210089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Don R. H., Pemberton J. M. Properties of six pesticide degradation plasmids isolated from Alcaligenes paradoxus and Alcaligenes eutrophus. J Bacteriol. 1981 Feb;145(2):681–686. doi: 10.1128/jb.145.2.681-686.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorn E., Hellwig M., Reineke W., Knackmuss H. J. Isolation and characterization of a 3-chlorobenzoate degrading pseudomonad. Arch Microbiol. 1974;99(1):61–70. doi: 10.1007/BF00696222. [DOI] [PubMed] [Google Scholar]
- ELSDEN S. R., PEEL J. L. Metabolism of carbohydrates and related compounds. Annu Rev Microbiol. 1958;12:145–202. doi: 10.1146/annurev.mi.12.100158.001045. [DOI] [PubMed] [Google Scholar]
- Harayama S., Rekik M., Timmis K. N. Genetic analysis of a relaxed substrate specificity aromatic ring dioxygenase, toluate 1,2-dioxygenase, encoded by TOL plasmid pWW0 of Pseudomonas putida. Mol Gen Genet. 1986 Feb;202(2):226–234. doi: 10.1007/BF00331641. [DOI] [PubMed] [Google Scholar]
- Hegeman G. D., Rosenberg S. L. The evolution of bacterial enzyme systems. Annu Rev Microbiol. 1970;24:429–462. doi: 10.1146/annurev.mi.24.100170.002241. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Merril C. R., Goldman D., Sedman S. A., Ebert M. H. Ultrasensitive stain for proteins in polyacrylamide gels shows regional variation in cerebrospinal fluid proteins. Science. 1981 Mar 27;211(4489):1437–1438. doi: 10.1126/science.6162199. [DOI] [PubMed] [Google Scholar]
- Murray K., Duggleby C. J., Sala-Trepat J. M., Williams P. A. The metabolism of benzoate and methylbenzoates via the meta-cleavage pathway by Pseudomonas arvilla mt-2. Eur J Biochem. 1972 Jul 24;28(3):301–310. doi: 10.1111/j.1432-1033.1972.tb01914.x. [DOI] [PubMed] [Google Scholar]
- Ngai K. L., Ornston L. N., Kallen R. G. Enzymes of the beta-ketoadipate pathway in Pseudomonas putida: kinetic and magnetic resonance studies of the cis,cis-muconate cycloisomerase catalyzed reaction. Biochemistry. 1983 Oct 25;22(22):5223–5230. doi: 10.1021/bi00291a024. [DOI] [PubMed] [Google Scholar]
- Powlowski J. B., Dagley S. beta-Ketoadipate pathway in Trichosporon cutaneum modified for methyl-substituted metabolites. J Bacteriol. 1985 Sep;163(3):1126–1135. doi: 10.1128/jb.163.3.1126-1135.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reineke W., Knackmuss H. J. Microbial metabolism of haloaromatics: isolation and properties of a chlorobenzene-degrading bacterium. Appl Environ Microbiol. 1984 Feb;47(2):395–402. doi: 10.1128/aem.47.2.395-402.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojo F., Pieper D. H., Engesser K. H., Knackmuss H. J., Timmis K. N. Assemblage of ortho cleavage route for simultaneous degradation of chloro- and methylaromatics. Science. 1987 Dec 4;238(4832):1395–1398. doi: 10.1126/science.3479842. [DOI] [PubMed] [Google Scholar]
- Sala-Trepat J. M., Murray K., Williams P. A. The metabolic divergence in the meta cleavage of catechols by Pseudomonas putida NCIB 10015. Physiological significance and evolutionary implications. Eur J Biochem. 1972 Jul 24;28(3):347–356. doi: 10.1111/j.1432-1033.1972.tb01920.x. [DOI] [PubMed] [Google Scholar]
- Schmidt E., Remberg G., Knackmuss H. J. Chemical structure and biodegradability of halogenated aromatic compounds. Halogenated muconic acids as intermediates. Biochem J. 1980 Oct 15;192(1):331–337. doi: 10.1042/bj1920331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scopes R. K. Measurement of protein by spectrophotometry at 205 nm. Anal Biochem. 1974 May;59(1):277–282. doi: 10.1016/0003-2697(74)90034-7. [DOI] [PubMed] [Google Scholar]


