Abstract
The actin cytoskeleton plays fundamental roles in ciliogenesis and the actin depolymerizing factor destrin regulates actin dynamics by treadmilling actin filaments and increasing globular actin pools. However, the specific developmental roles of destrin in ciliogenesis have not been fully elucidated. Here, we investigated the function of destrin in ciliogenesis using Xenopus laevis and human retinal pigmented epithelial (hRPE1) cells. We discovered the loss of destrin increased the number of multiciliated cells in the Xenopus epithelium and impeded cilia motility. Additionally, destrin depletion remarkably reduced the length of primary cilia in the Xenopus neural tube and hRPE1 cells by affecting actin dynamics. Immunofluorescence using markers of ciliary components indicated that destrin controls the directionality and polarity of basal bodies and axonemal elongation by modulating actin dynamics, independent of basal body docking. In conclusion, destrin plays a significant role during vertebrate ciliogenesis regulating both primary and multicilia development. Our data suggest new insights for understanding the roles of actin dynamics in cilia development.
Keywords: Destrin, F-actin, Ciliogenesis, Primary cilia, Multiciliated cells, Xenopus laevis
INTRODUCTION
Actin is an abundant and highly conserved intracellular protein that exists either in a monomeric globular actin (G-actin) or as filamentous actin (F-actin) composed of G-actin monomers (Dominguez & Holmes 2011). F-actin, a major component of the actin cytoskeleton, assembles into bundles or branched networks and participates in various cellular processes, including cell motility, growth, polarity, and migration (Rajan et al., 2023). Despite its ubiquitous presence in cells, numerous studies have highlighted its localization in and around cilia, and the critical role of the actin cytoskeleton in ciliogenesis (Brücker et al., 2020). In primary cilia, mutations in ciliopathy genes that lead to increased actin stress fibers disrupt ciliogenesis, while treatment with actin polymerization inhibitor like cytochalasin D enhances ciliogenesis (Kim et al., 2010; Smith et al., 2020). Conversely, cytochalasin D treatment does not affect cilia morphology but disrupts cilia polarity and basal body spacing in multiciliated cells (MCCs), suggesting distinct roles for actin dynamics in primary cilia and MCCs.
The actin depolymerizing factor family, comprising destrin (Dstn) and coflins (Cfl1 and Cfl2), regulates actin dynamics by severing filaments and promoting depolymerization (Bamburg 1999; Bamburg & Wiggan 2002). Several studies have implicated Cfl1 in ciliogenesis; its depletion induces ciliogenesis in mammalian cells, and Cfl1 knockdown leads to ciliary defects in zebrafish (Kim et al., 2015; Zhang et al., 2016). Additionally, Cfl1 cooperates with Vangl2 to initiate planar cell polarity in mouse node cilia (Mahaffey et al., 2013). However, the role of Dstn in ciliogenesis remains unexplored. Understanding the regulatory mechanisms of ciliogenesis vi actin dynamics regulators is crucial for better insight into cilia biology in both monociliated cells and MCCs.
Therefore, this study aimed to elucidate the role of Dstn in ciliogenesis using Xenopus laevis and human retinal pigment epithelial cells (hRPE1). We performed loss-of-function experiments using morpholino oligonucleotides (MOs) in developing Xenopus embryos and siRNA in hRPE1 cells. Our results show that inhibition of dstn in Xenopus led to an increase in the number of MCCs and significantly disrupted the polarity of basal bodies in MCCs. In contrast, dstn knockdown in both hRPE1 cells and the Xenopus’ neural tube inhibited primary cilia elongation without influencing basal body docking. These findings suggest that Dstn regulates ciliogenesis by modulating actin dynamics in both primary and MCCs.
MATERIALS AND METHODS
1. Ethics statement
Experiments were conducted strictly following the guidelines of the Animal Care and Use Committee, consistent with international laws and policies (National Institute of Health) Guide for the Care and Use of Laboratory Animals, publication no. 85-23, 1985. The Institutional Review Board of Kyungpook National University in Korea approved the experimental use of amphibians (2021-0017). All members of the research group received training for the appropriate care and use of experimental organisms.
2. Xenopus laevis embryos microinjection
X. laevis eggs were obtained and in vitro fertilized using standard methods as previously described (Sive et al., 2007). For mRNA injection or in situ hybridization using RNA probe, dstn and alpha-tubulin primers were designed based on National Center for Biotechnology Information (NCBI) and Xenbase sequences. Plasmids were constructed for mRNA synthesis using the pCS107 vector with BamH1 and Xho1 restriction enzymes. The RNA probe sequence was labeled with digoxigenin (DIG, Roche, Basel, Switzerland) and inserted into a T-easy vector (pGEM, Promega, Madison, WI, USA). The pCS107 and T-easy vectors were linearized with the Apa1 restriction enzyme, and capped mRNAs were synthesized using the SP6 mMessage mMachine kit (Invitrogen, Carlsbad, CA, USA). DIG-labeled RNA probes were generated using the T7 mMessage mMachine kit (Invitrogen).
dstn MO with the nucleotide sequence (5′-TCCGAACACCTGATGCCATTGTTGA-3′) was purchased from Gene Tools. For the rescue experiments, mutant constructs (dstn*) not recognized by dstn MO were sub-cloned into the pCS107 vector. Embryos at the two-cell stage were microinjected with mRNAs (dstn, dstn*, centrin-RFP, clamp-GFP) and dstn MO.
3. Whole-mount in situ hybridization (WISH)
Embryos at stage 32 were fixed in MEMFA (4% paraformaldehyde, 0.1 M MOPS (pH 7.4), 1 mM MgSO4, and 2 mM EGTA) overnight at 4°C. Antisense digoxigenin (DIG)-labeled probes were generated from plasmids containing dstn or alpha-tubulin linearized by ApaI. The probes were synthesized using the mMESSAGE mMACHINE™ SP6 Transcription Kit (Invitrogen) and detected using an alkaline phosphatase-labeled anti-digoxigenin antibody (1:1,000, Roche) and NBT/BCIP staining solution (Roche).
4. Reverse transcriptase-PCR (RT-PCR)
Total RNA was extracted from hRPE1 cells, and cDNAs were synthesized using the PrimeScript™1st strand cDNA synthesis kit (Takara, Shiga, Japan). RT-PCRs were performed using Emerald Amp PCR Master Mix (Takara) and specific primers listed in Table 1.
Table 1. The primer sequences for RT-PCR.
| Gene | Forward primer | Reverse primer |
|---|---|---|
| GAPDH | 5′-CCACTCCTCCACCTTTGACG-3′ | 5′-CCACCACCCTGTTGCTGTAG-3′ |
| Human DSTN | 5′-GGCACCAGAACTAGCACCTC-3′ | 5′-AGCCCGATTGAGATCTTCTGG-3′ |
5. Measurement of fluid flow velocity by ciliary beating
Embryos were anesthetized using 0.1X benzocaine, fluorescent microbeads were added to the culture media, and videos were recorded. The velocity of fluorescent beads was measured using the fluorescent bead measurement program, Tracker 6.0.
6. Cell culture and siRNA transfection
Human telomerase-immortalized retinal pigmented epithelial cells (hRPE1) were cultured at 37 °C and 5% CO2 in DMEM/F12 media (Thermo Fisher Scientific, Waltham, MA, USA) containing 10% fetal bovine serum (Welgene, Gyeongsan, Korea) and 100 U/mL streptomycin/penicillin. dstn siRNA (Table 2) and control siRNA (Bioneer, Daejeon, Korea) were transfected into hRPE1 cells for 24 hrs using Lipofectamine RNA iMAX (Invitrogen). Next, the culture medium was replaced with a serum-free medium for cells with 90% confluency and cultured for an additional 48 hrs. Finally, DSTN-flag plasmids were transfected using Lipofectamine 2000 (Invitrogen).
Table 2. The sequences for DSTN siRNA.
| Target gene | Sequence 5’-3’ |
|---|---|
| DSTN siRNA #1 | CAGAACUAGCACCUCUGAA |
| UUCAGAGGUGCUAGUUCUG | |
| DSTN siRNA #2 | CUCCUUCAGAAGUUUGCUU |
| AAGCAAACUUCUGAAGGAG |
7. Immunofluorescence and confocal microscopy
For immunofluorescence, Xenopus embryos were fixed with MEMFA, and hRPE1 cells were fixed in 4% paraformaldehyde or methanol, depending on antibodies. Primary antibodies used were anti-acetylated tubulin (1:1,000, Sigma-Aldrich, St. Louis, MO, USA), CEP164 (1:1,000, Proteintech, Rosemont, IL, USA), IFT20 (1:200, Proteintech), and Rab11 (1:500, Invitrogen). The fluorescent-labeled secondary antibody was Alexa Fluor (1:2,000, Invitrogen). For F-actin staining, Xenopus embryos at stage 32 were fixed in MEMFA and stained with Alexa Fluor 488 phalloidin (1:1,000, Invitrogen). Images were obtained using an Olympus FV1200 confocal microscope.
8. Western blotting
Cell lysates were prepared using RIPA lysis buffer (150 mM NaCl, 10 mM Na2HPO4, 0.5% sodium deoxycholate, 1% NP40) supplemented with 1 mM PMSF (phenylmethylsulfonyl fluoride), 5 mM sodium vanadate, and 1 mM protease inhibitor cocktail (Roche). Lysates were loaded onto 12% SDS-polyacrylamide gels and transferred to a nitrocellulose membrane. The membrane was blocked in a 5% skim milk TBST buffer for 1 h at room temperature. Proteins were detected using polyclonal anti-DSTN (1:1,000, https://www.antibodies-online.com), and GADPH (1:1,000). The secondary antibody used was anti-rabbit IgG HRP-linked antibodies (1:2,000, Santa Cruz, Dallas, TX, USA).
9. Quantification and statistics
ImageJ software (National Institutes of Health) was used for the analysis of confocal imaging and optical microscope imaging data. Statistical analyses were conducted by GraphPad Prism version 9. The results are presented as the means±SE from the indicated number of independent experiments in graphs (n). The data were analyzed using the unpaired t-test and the level of significance was considered as * p<0.05.
RESULTS
1. Loss of dstn impairs cilia-induced fluid flow and increases the number of multiciliated cells (MCCs) in Xenopus embryos
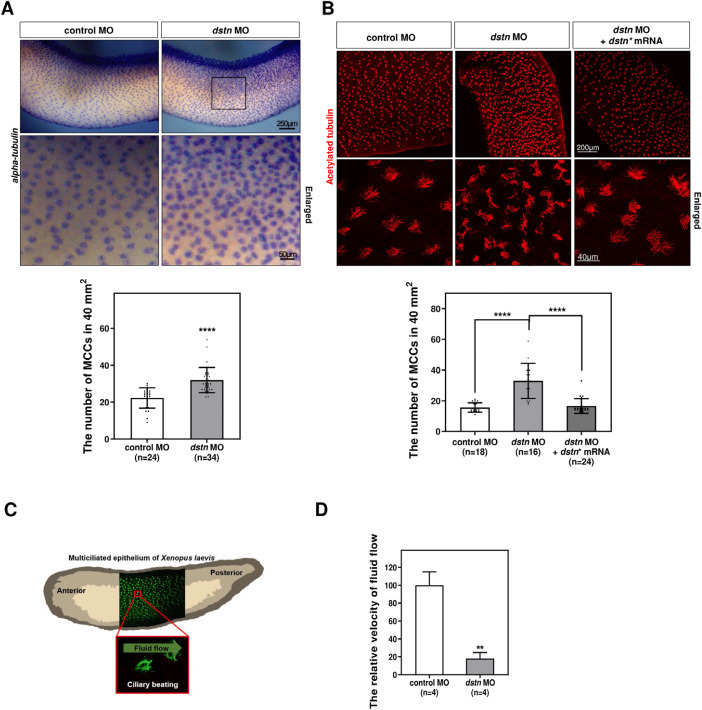
To investigate the role of Dstn in MCC formation during Xenopus embryogenesis, we conducted loss-of-function studies using dstn MO. We observed MCCs across larval skin through whole¬mount in situ hybridization (WISH) analysis, using an alpha-tubulin probe. The analysis revealed a significant increase in the number of MCCs in dstn knockdown embryos compared to control embryos (Fig. 1A). This increase in MCCs was effectively rescued in embryos injected with dstn* mRNAs (Fig. 1B). In addition to ciliogenesis, we also investigated whether dstn knockdown affects the function of multicilia, which are evolutionarily conserved structures that beat synchronously and coordinately to direct fluid flow across vertebrate epithelium (Brooks & Wallingford, 2014; Walentek et al., 2014) (Fig. 1C). Fluorescent microbeads were used to the velocity of cilia-driven fluid flow, using fluid flow tracker (Kim et al., 2023). The velocity of microbeads was significantly reduced in morphants compared to control embryos (Fig. 1D; Supplementary videos 1 & 2). These results suggest that Dstn plays a crucial role in MCC development and the synchronized beating of cilia during Xenopus embryogenesis.
Fig. 1. dstn depletion increases/ reduces the number of MCCs and ciliary fluid flow in Xenopus embryos.
(A) dstn MO was injected into the Xenopus embryos and the embryos were processed for WISH analysis. WISH analysis using α-tubulin was conducted to visualize MCCs. Statistical analysis of the data revealed a significant increase of MCCs induced by dstn knockdown per 40 mm2 as compared to control embryos (scale bar=250 μm; enlarged scale bar=50 μm). (B) The co-injection of dstn* mRNAs together with dstn MOs was carried out followed by immunofluorescence using acetylated tubulin to mark MCCs. The microinjection of dstn* along with dstn MO effectively rescued the number of MCCs in Xenopus epithelium (scale bar=200 μm; enlarged scale bar=40 μm). Statistical analysis of data exhibited a significant increase in the number of MCCs in three sets of dstn morphant embryos per 40 mm2 area compared with control embryos. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001. (C) Illustration showing Xenopus embryos’ epidermis is composed of MCCs labeled by acetylated tubulin (green). Polarized cilia beating directs fluid flow across Xenopus epidermis from the anterior toward the posterior end of embryos. Adapted from König & Hausen (1993) with permission of Elsevier; Hayes et al. (2007) with permission of Elsevier. (D) The cilia-driven epidermal flow was analyzed by fluorescent microbeads. The statistical quantification for the relative velocity of fluorescent microbeads clearly showed that relative velocity was considerably reduced in dstn morphant embryos compared with control embryos. MCC, multiciliated cells; MO, morpholino oligonucleotides; WISH, whole-mount in situ hybridization.
2. Dstn is required for multiciliated cells (MCC) development during Xenopus embryogenesis
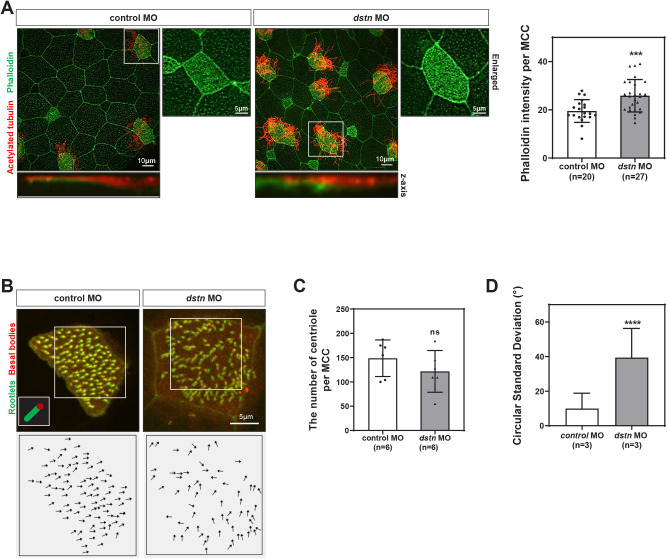
Given the close relationship between microtubule-based cilia and actin filaments, we hypothesized that both cytoskeletons are intrinsically linked during ciliogenesis (Werner et al., 2011). MCCs require an apical actin network for the docking of basal bodies at the apical cell surface during ciliogenesis (Smith et al., 2020). Compelling evidence supports the involvement of DSTN in actin filaments depolymerization (Hotulainen et al., 2005; Kumar et al., 2012; Zhang et al., 2016). To assess whether Dstn affects actin dynamics during MCC development, we visualized actin filaments on the larval epithelial surface using phalloidin staining. The phalloidin intensity (green) in MCCs labeled with acetylated tubulin (red) was significantly higher in dstn morphant embryos compared to control embryos (Fig. 2A). Additionally, confocal microscopic images revealed that actin fibers accumulated on the apical surface of dstn morphants along the z-axis (Fig. 2A).
Fig. 2. Dstn is required for ciliogenesis during Xenopus embryonic development.
(A) Xenopus epidermal MCCs marked by immunofluorescence using anti-acetylated tubulin (red) for cilia and phalloidin (green) for actin filament network (scale bar=10 μm; enlarged scale bar=5 μm). Statistical quantitation of phalloidin intensity revealed a significant increase in phalloidin intensity per MCC in dstn-depleted embryos as compared to control embryos. (B) Centrin-RFP and Clamp-GFP exhibited perturbation of basal body polarity in dstn-depleted embryos compared with coordinated polarity and alignment in control embryos. It is highlighted by black arrows (scale bar=5 μm). (C) We have quantified the number of centrioles per MCC in Xenopus epidermis and data showed that dstn knockdown did not significantly affect the number of centrioles in dstn morphant embryos. (D) The polarization was analyzed by angular measurements of Centrin/Clamp pairs in MCCs. Statistical analysis of data showed that dstn inhibition increased the circular standard deviation compared with control embryos (n=3). * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001, MO, morpholino oligonucleotides; MCC, multiciliated cells.
The docking of basal bodies at the cell membrane and the establishment of polarity are necessary for coordinated ciliary beating to generate fluid flow (Boisvieux-Ulrich et al., 1985; Mitchell et al., 2007). Basal body docking in MCCs requires an apical actin network (Sedzinski et al., 2017). Additionally, actin and microtubules regulate different aspects of planar cell polarity in MCCs by influencing ciliary beating (Werner et al., 2011). To better understand the roles of Dstn in basal body docking and polarity, we used fluorescent-tagged Centrin (red) and Clamp (green) to visualize basal bodies and rootlets respectively. Dstn knockdown led to basal body clumping and a loss of ciliary polarity (Fig. 2C and D). Quantification of basal body orientation, based on the basal body/rootlet angle relative to the body axis, showed a significant disruption in directionality following dstn knockdown (Fig. 2D). These results suggest that inhibition of Dstn causes an accumulation of F-actin beneath cilia in MCCs, disrupting basal body polarity and, consequently, the overall process of ciliogenesis.
3. Dstn regulates primary cilia formation in Xenopus and RPE1 cells
There is ample evidence supporting the role of actin destabilization in primary ciliogenesis, while MCC formation requires the apical actin network (Hotulainen et al., 2005; Werner et al., 2011). However, the roles of actin-depolymerizing in the regulation of primary and multicilia formation remain controversial, warranting further investigation (Zhang et al., 2016). To clarify the role of Dstn in multicilia and primary cilia formation, we studied Xenopus embryos and human retinal pigmented epithelial (hRPE1) cells.
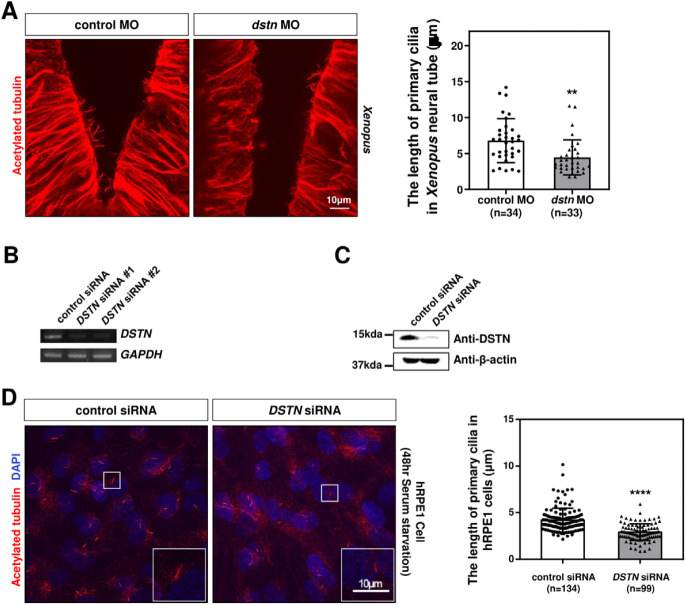
In the developing Xenopus neural tube, cells possess primary cilia that mediate signaling necessary for neural tube patterning (Walentek & Quigley, 2017). To test whether Dstn affects neural tube primary cilia, we used immunofluorescence on head sections. Immunostaining with acetylated tubulin revealed that the primary cilia of dstn-depleted embryos were significantly shorter than those of control embryos (Fig. 3A). To further assess the effects of DSTN inhibition on primary cilia, we transfected hRPE1 cells with DSTN siRNA and induced primary cilia formation through serum starvation. RT-PCR and western blot analyses confirmed the efficiency of DSTN siRNA, showing substantial reductions in RNA and protein levels (Fig. 3B and C). Similar to Xenopus, DSTN depletion in hRPE1 cells led to a significant reduction in primary cilia length (Fig. 3D). Altogether, these findings demonstrate that the Dstn regulates primary cilia formation in the same context in Xenopus and hRPE1 cells.
Fig. 3. Loss of Dstn results in length reduction of primary cilia in Xenopus and hRPE1 cells.
(A) Acetylated tubulin immunostaining of dstn-deficient embryos revealed a considerable reduction in the length of neural tube primary cilia of Xenopus compared with control embryos (scale bar=10 μm). Statistical analysis showed that the dstn morphants exhibited a length of primary cilia less than 5 μm compared with approximately 8 μm long cilia in control embryos. (B) RT-PCR analysis revealed the specificity of DSTN siRNAs as the expression of DSTN was significantly reduced in RPE1 cells transfected with DSTN siRNAs. GADPH was used as the internal control. (C) Western blotting showed that the protein expression of DSTN was considerably reduced in DSTN siRNAs transfected hRPE1 cells. β-actin was used as the loading control. (D) hRPE1 cells were transfected with DSTN siRNAs and serum-starved for 48 hrs to induce ciliogenesis. As analyzed by immunostaining with acetylated tubulin, depletion of DSTN induced shorter primary cilia in hRPE1 cells with less than 2.5 μm compared with 5 μm long primary cilia in control hRPE1 cells (scale bar=10 μm). Statistical analysis of data revealed that the length of primary cilia was significantly reduced in DSTN-depleted cells as compared to the control cells. * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
4. Dstn is required for axoneme elongation in primary cilia, but not for ciliary assembly
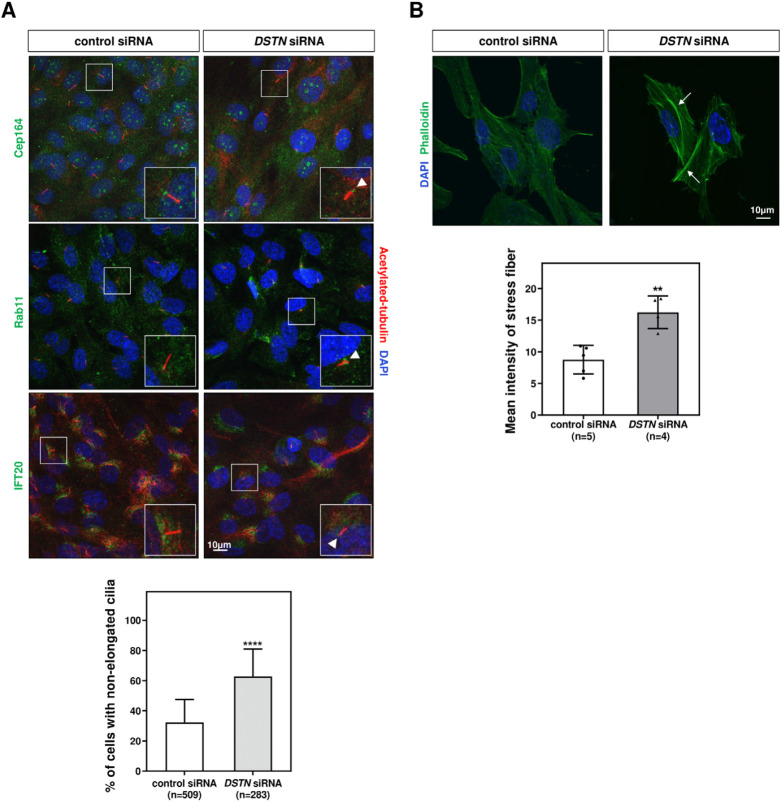
Ciliogenesis is a multistep process that includes the docking of ciliary vesicles, which is regulated by protein components such as Cep164 (distal centriolar protein) (Cao et al., 2012), Rab11 (ciliary vesicle trafficking protein) (Knödler et al., 2010), and IFT 20 (intraflagellar transport protein/ciliary membrane transport protein) (Zhu et al., 2017), followed by ciliary axoneme elongation (Nigg & Raff, 2009). To evaluate the role of Dstn in ciliary assembly, we transfected hRPE1 cells with DSTN siRNA, serum-starved the cells for 48 hrs, and performed immunostaining for Cep164, Rab11, and IFT20. Confocal images showed that DSTN depletion did not affect the localization or distribution of these basal body-associated proteins, but axoneme length was significantly reduced (Fig. 4A).
Fig. 4. Depletion of DSTN affects axonemal elongation but does not perturb the docking of ciliary vesicles to centrioles.
(A) hRPE1 cells were transfected with DSTN siRNAs and serum-starved for 48 hrs to induce ciliogenesis. The transfected cells were immunostained with antibodies against Cep164, Rab11, and IFT20 (green) along with acetylated tubulin (red). Loss of DSTN did not perturb the docking of ciliary vesicles to centrioles in hRPE1 cells. However, the axonemal length was considerably reduced in DSTN-depleted hRPE1 cells, as indicated by white arrowheads compared with control cells. A graph indicated the significant reduction in axonemes in hRPE1 cells devoid of DSTN, but no effect was observed for vesicle docking to centrioles after the loss of DSTN (scale bar=10 μm). (B) Immunofluorescence revealing stress fibers using phalloidin indicated that loss of DSTN in hRPE1 cells resulted in thicker stress fibers compared to thin fibers in control cells. White arrows indicated the thickness of stress fibers in control versus knockdown cells. Statistical analysis demonstrated that DSTN depletion resulted in increased mean phalloidin intensity (scale bar=10 μm). The level of significance is shown as * p<0.05, ** p<0.01, *** p<0.001, **** p<0.0001.
In earlier experiments, we demonstrated that Dstn influences actin filament accumulation during motile ciliogenesis (Fig. 2A). To determine whether Dstn similarly regulates actin dynamics in primary cilia, we performed immunofluorescence staining using phalloidin in DSTN-depleted hRPE1 cells. DSTN depletion resulted in thicker stress fibers compared to control siRNA-transfected cells (Fig. 4B). Given prior reports that increased stress fiber formation hinders primary cilia development, our results underscore the crucial role of Dstn in regulating primary ciliogenesis in hRPE1 cells.
DISCUSSION
Remodeling of the actin filament network provides insights into the regulation of ciliogenesis and cilium length (Pan et al., 2007). While CFL1, a member of the actin depolymerizing factors family, has been studied for its roles in ciliogenesis through the regulation of actin dynamics, the function of Dstn in ciliogenesis remains less understood. In this study, we used X. laevis as an in vivo model (Hayes et al., 2007) to demonstrate that Dstn governs cilia development and the polarized beating of cilia. Firstly, Dstn depletion increased the number of MCCs (Fig. 1A and B). Additionally, the loss of dstn resulted in distorted ciliary beating and a significant reduction in cilia-driven fluid flow (Fig. 1D). This indicates that Dstn is essential for organizing actin filaments in MCCs, as actin dynamics and ciliogenesis are intrinsically linked (Smith et al., 2020). As shown in Fig. 2A, the loss of dstn resulted in the accumulation of actin filaments at the apical surface of MCCs. However, this loss did not affect the docking and number of basal bodies but instead led to basal body clumping, significantly impacting their directionality and polarity (Fig. 2B–D). Our data suggest that Dstn governs the ciliogenesis of MCCs by affecting basal body polarity while not influencing basal body docking.
Further examination of Dstn’s role in primary cilia formation revealed that DSTN depletion leads to a reduction in primary cilia length in both Xenopus and hRPE1 cells (Fig. 3A and D). The shorter primary cilia after depletion of Dstn align with previous studies that highlighted the positive roles of CFL1 in ciliogenesis through the influence of LIMK2 and TESK1 modulation of YAP/TAZ activity (Kim et al., 2015). However, contrasting findings have also been reported, where CFL1 depletion increased the length of primary cilia in hRPE1 cells, influenced by NudC (Sedzinski et al., 2017). Thus, our results suggest that the regulation of ciliogenesis by Dstn involves the orchestration of a complex network and it can have both positive and negative roles in ciliogenesis. Understanding how cells integrate various signals to regulate ciliogenesis, particularly in reconciling these seemingly contradictory findings about actin depolymerizing factors’ roles in primary cilia length regulation, will be a key challenge for future studies.
Our investigation into the mechanistic details of Dstn in primary cilia regulation showed that while Dstn controls axoneme elongation, it does not affect the localization and distribution of basal bodies, mirroring its effects in MCCs (Figs. 2 and 4). Additionally, Dstn influences primary cilia formation by regulating actin stress fibers (Fig. 4B). Altogether, our data suggest that Dstn regulates ciliogenesis in both primary and motile cilia by modulating actin dynamics.
The paradoxical behavior of Dstn in primary cilia and multicilia can be explained by the distinct impacts of actin destabilization: promoting ciliogenesis in primary cilia, while having opposite effects in multicilia (Pan et al., 2007; Smith et al., 2020). In primary cilia, destabilized F-actin facilitates vesicle trafficking, which promotes ciliogenesis and increases ciliary length (Kim et al., 2010, 2015). Therefore, our findings that Dstn depletion reduces primary cilia length are consistent with previous reports (Wheway et al., 2015; Yeyati et al., 2017), indicating that Dstn positively regulates ciliogenesis by actin destabilization in primary cilia. The contradictory response observed in multicilia is again in agreement with reported data (Sedzinski et al., 2017) as multicilia formation requires an apical actin network for basal body docking and polarity establishment to allow synchronous ciliary beating (Mitchell et al., 2007; Pan et al., 2007; Kulkarni et al., 2018). F-actin stabilization and apical actin network support ciliogenesis by facilitating basal body docking in mouse tracheal epithelial cells, as the actin web expands the docking surface and spaces out the basal bodies across cell membrane (Werner et al., 2011; Sedzinski et al., 2017).
In conclusion, Dstn plays a significant role in ciliogenesis, regulating both primary and multicilia development. Our findings provide new insights into the roles of actin dynamics in ciliogenesis, contributing to a deeper understanding of this complex biological process.
Acknowledgements
This work was supported by the Korea Environment & Technology Institute (KEITI) through Core Technology Development Project for Environmental Diseases Prevention and Management funded by Korea Ministry of Environment (MOE) [grant number 2022003310001], and the National Research Foundation of Korea and the Ministry of Science & ICT [grant number 2021R1A2C1010408].
ORCID
Youni Kim https://orcid.org/0000-0002-3052-6754
Hyun-Kyung Lee https://orcid.org/0000-0003-4989-698X
Kyeong-Yeon Park https://orcid.org/0009-0009-2509-9047
Tayaba Ismail https://orcid.org/0000-0001-9970-4793
Hongchan Lee https://orcid.org/0000-0001-9009-209X
Hyun-Shik Lee https://orcid.org/0000-0002-3837-9867
Conflict of interests
The authors declare no potential conflict of interest.
Authors’ contributions
Conceptualization: Kim Y, Park KY, Lee HS.
Data curation: Kim Y, Park KY, Lee HK.
Methodology: Kim Y, Park KY.
Writing-original draft: Kim Y, Park KY.
Writing-review & editing: Kim Y, Lee HK, Park KY, Ismail T, Lee H, Lee HS.
Ethics approval
The Institutional Review Board of Kyungpook National University in Korea approved the experimental use of amphibians (2021-0017). All members of the research group received training for the appropriate care and use of experimental organisms.
Supplementary Materials
Supplementary Materials
REFERENCES
- Bamburg JR. Proteins of the ADF/cofilin family: Essential regulators of actin dynamics. Annu Rev Cell Dev Biol. 1999;15:185–230. doi: 10.1146/annurev.cellbio.15.1.185. [DOI] [PubMed] [Google Scholar]
- Bamburg JR, Wiggan OP. ADF/cofilin and actin dynamics in disease. Trends Cell Biol. 2002;12:598–605. doi: 10.1016/S0962-8924(02)02404-2. [DOI] [PubMed] [Google Scholar]
- Boisvieux-Ulrich E, Laine MC, Sandoz D. The orientation of ciliary basal bodies in quail oviduct is related to the ciliary beating cycle commencement. Biol Cell. 1985;55:147–150. doi: 10.1111/j.1768-322X.1985.tb00417.x. [DOI] [PubMed] [Google Scholar]
- Brooks ER, Wallingford JB. Multiciliated cells. Curr Biol. 2014;24:R973–R982. doi: 10.1016/j.cub.2014.08.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brücker L, Kretschmer V, May-Simera HL. The entangled relationship between cilia and actin. Int J Biochem Cell Biol. 2020;129:105877. doi: 10.1016/j.biocel.2020.105877. [DOI] [PubMed] [Google Scholar]
- Cao J, Shen Y, Zhu L, Xu Y, Zhou Y, Wu Z, Li Y, Yan X, Zhu X. miR-129-3p controls cilia assembly by regulating CP110 and actin dynamics. Nat Cell Biol. 2012;14:697–706. doi: 10.1038/ncb2512. [DOI] [PubMed] [Google Scholar]
- Dominguez R, Holmes KC. Actin structure and function. Annu Rev Biophys. 2011;40:169–186. doi: 10.1146/annurev-biophys-042910-155359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayes JM, Kim SK, Abitua PB, Park TJ, Herrington ER, Kitayama A, Grow MW, Ueno N, Wallingford JB. Identification of novel ciliogenesis factors using a new in vivo model for mucociliary epithelial development. Dev Biol. 2007;312:115–130. doi: 10.1016/j.ydbio.2007.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hotulainen P, Paunola E, Vartiainen MK, Lappalainen P. Actin-depolymerizing factor and cofilin-1 play overlapping roles in promoting rapid F-actin depolymerization in mammalian nonmuscle cells. Mol Biol Cell. 2005;16:649–664. doi: 10.1091/mbc.e04-07-0555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim CY, Lee HK, Lee H, Lee HS. Ruvbl1 is essential for ciliary beating during Xenopus laevis embryogenesis. Dev Reprod. 2023;27:159–165. doi: 10.12717/DR.2023.27.3.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Jo H, Hong H, Kim MH, Kim JM, Lee JK, Heo WD, Kim J. Actin remodelling factors control ciliogenesis by regulating YAP/TAZ activity and vesicle trafficking. Nat Commun. 2015;6:6781. doi: 10.1038/ncomms7781. [DOI] [PubMed] [Google Scholar]
- Kim J, Lee JE, Heynen-Genel S, Suyama E, Ono K, Lee KY, Ideker T, Aza-Blanc P, Gleeson JG. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464:1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knödler A, Feng S, Zhang J, Zhang X, Das A, Peränen J, Guo W. Coordination of Rab8 and Rab11 in primary ciliogenesis. Proc Natl Acad Sci USA. 2010;107:6346–6351. doi: 10.1073/pnas.1002401107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König G, Hausen P. Planar polarity in the ciliated epidermis of Xenopus embryos. Dev Biol. 1993;160:355–368. doi: 10.1006/dbio.1993.1312. [DOI] [PubMed] [Google Scholar]
- Kulkarni SS, Griffin JN, Date PP, Liem KF, Jr, Khokha MK. WDR5 stabilizes actin architecture to promote multiciliated cell formation. Dev Cell. 2018;46:595–610.E3. doi: 10.1016/j.devcel.2018.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar G, Srivastava R, Mitra K, Sahasrabuddhe AA, Gupta CM. Overexpression of S4D mutant of Leishmania donovani ADF/cofilin impairs flagellum assembly by affecting actin dynamics. Eukaryot Cell. 2012;11:752–760. doi: 10.1128/EC.00013-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahaffey JP, Grego-Bessa J, Liem KF, Jr, Anderson KV. Cofilin and Vangl2 cooperate in the initiation of planar cell polarity in the mouse embryo. Development. 2013;140:1262–1271. doi: 10.1242/dev.085316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell B, Jacobs R, Li J, Chien S, Kintner C. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 2007;447:97–101. doi: 10.1038/nature05771. [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- Pan J, You Y, Huang T, Brody SL. RhoA-mediated apical actin enrichment is required for ciliogenesis and promoted by Foxj1. J Cell Sci. 2007;120:1868–1876. doi: 10.1242/jcs.005306. [DOI] [PubMed] [Google Scholar]
- Rajan S, Kudryashov DS, Reisler E. Actin bundles dynamics and architecture. Biomolecules. 2023;13:450. doi: 10.3390/biom13030450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedzinski J, Hannezo E, Tu F, Biro M, Wallingford JB. RhoA regulates actin network dynamics during apical surface emergence in multiciliated epithelial cells. J Cell Sci. 2017;130:420–428. doi: 10.1242/jcs.194704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sive HL, Grainger RM, Harland RM. Xenopus laevis in vitro fertilization and natural mating methods. Cold Spring Harb Protoc. 2007;2007:pdb-prot4737. doi: 10.1101/pdb.prot4737. [DOI] [PubMed] [Google Scholar]
- Smith CEL, Lake AVR, Johnson CA. Primary cilia, ciliogenesis and the actin cytoskeleton: A little less resorption, a little more actin please. Front Cell Dev Biol. 2020;8:622822. doi: 10.3389/fcell.2020.622822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walentek P, Bogusch S, Thumberger T, Vick P, Dubaissi E, Beyer T, Blum M, Schweickert A. A novel serotonin-secreting cell type regulates ciliary motility in the mucociliary epidermis of Xenopus tadpoles. Development. 2014;141:1526–1533. doi: 10.1242/dev.102343. [DOI] [PubMed] [Google Scholar]
- Walentek P, Quigley IK. What we can learn from a tadpole about ciliopathies and airway diseases: Using systems biology in Xenopus to study cilia and mucociliary epithelia. Genesis. 2017;55:e23001. doi: 10.1002/dvg.23001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Hwang P, Huisman F, Taborek P, Yu CC, Mitchell BJ. Actin and microtubules drive differential aspects of planar cell polarity in multiciliated cells. J Cell Biol. 2011;195:19–26. doi: 10.1083/jcb.201106110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wheway G, Schmidts M, Mans DA, Szymanska K, Nguyen TMT, Racher H, Phelps IG, Toedt G, Kennedy J, Wunderlich KA et al. An siRNA-based functional genomics screen for the identification of regulators of ciliogenesis and ciliopathy genes. Nat Cell Biol. 2015;17:1074–1087. doi: 10.1038/ncb3201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeyati PL, Schiller R, Mali G, Kasioulis I, Kawamura A, Adams IR, Playfoot C, Gilbert N, van Heyningen V, Wills J, von Kriegsheim A, Finch A, Sakai J, Schofield CJ, Jackson IJ, Mill P. KDM3A coordinates actin dynamics with intraflagellar transport to regulate cilia stability. J Cell Biol. 2017;216:999–1013. doi: 10.1083/jcb.201607032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang C, Zhang W, Lu Y, Yan X, Yan X, Zhu X, Liu W, Yang Y, Zhou T. NudC regulates actin dynamics and ciliogenesis by stabilizing cofilin 1. Cell Res. 2016;26:239–253. doi: 10.1038/cr.2015.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu X, Liang Y, Gao F, Pan J. IFT54 regulates IFT20 stability but is not essential for tubulin transport during ciliogenesis. Cell Mol Life Sci. 2017;74:3425–3437. doi: 10.1007/s00018-017-2525-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Materials