Figure 1. Cryo-EM structure of the core TIM23 complex from S. cerevisiae.
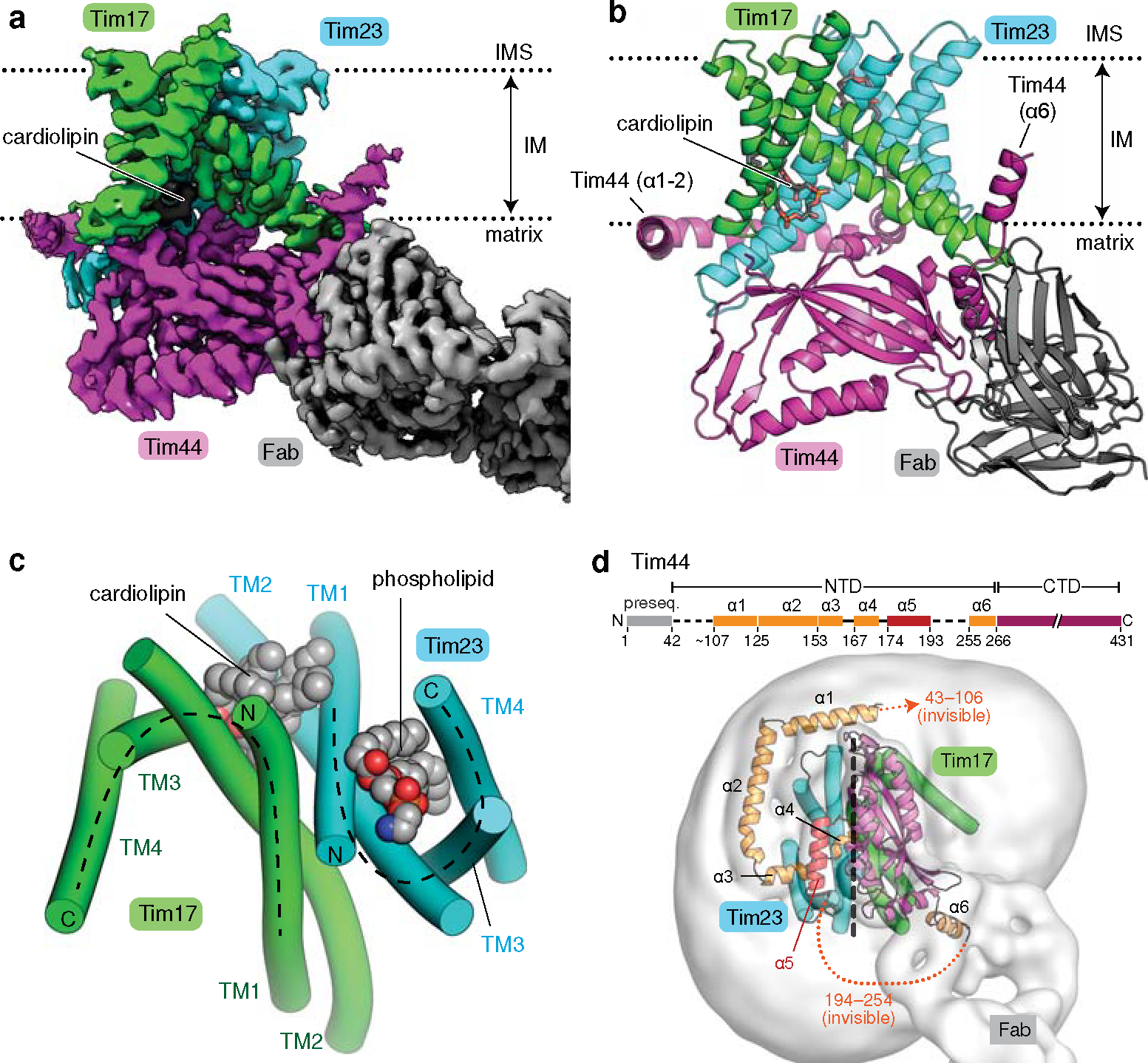
a, Cryo-EM reconstruction of Fab-bound core TIM23 complex. b, As in a, but showing the atomic model. We note that the amphipathic helix α6 of Tim44 is tilted upwards because it sits on a curved micelle edge (see d). c, Arrangement of the TMs of Tim17 and Tim23 in a view from the IMS. TMs are represented as cylinders (arranged in the N to C order), and lipids are shown as spheres. The topology of the cavities is highlighted with dashed lines. d, Organization of Tim44 domains. Upper panel, linear diagram; lower panel, view from the matrix. The detergent micelle is shown with a lowpass-filtered cryo-EM map (gray semitransparent surface). Tim44 is shown as a ribbon diagram (orange, amphipathic helices; red, α5; purple, CTD). Dashed line, plane of the Tim17-Tim23 interface.
