Figure 2. The cavity of Tim17 forms the path for protein translocation.
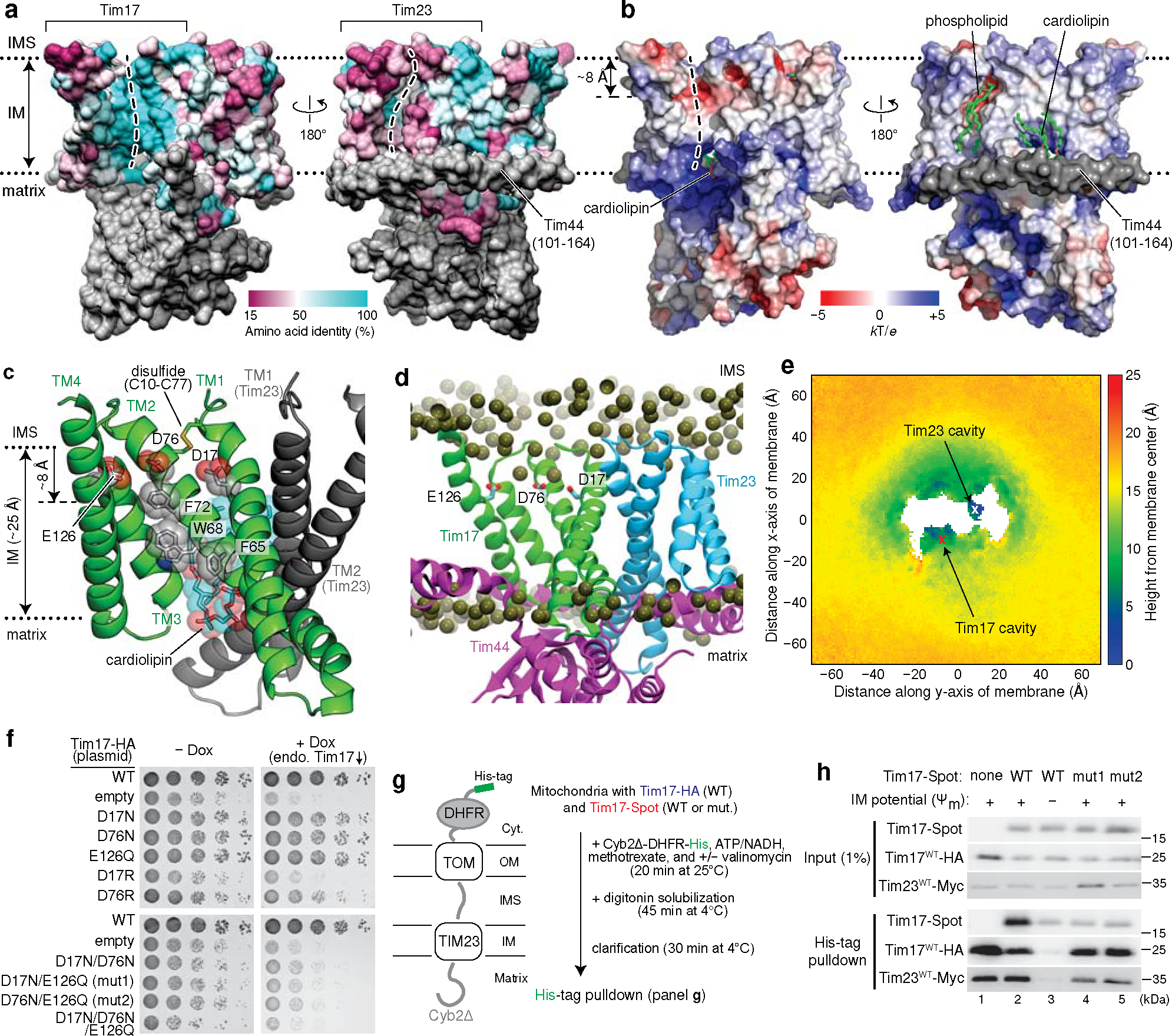
a, Mapping of amino acid sequence conservation of Tim17 and Tim23 onto the structure. Left, view into the Tim17 cavity; right, view into the Tim23 cavity. Dashed line, path along the cavity. b, Surface electrostatics of the TIM23 complex. The views are equivalent to those in a. α1–3 (residues ~107–164) of Tim44 (modelled as polyalanine) were not included in the calculation. c, Conserved acidic and aromatic amino acids lining the Tim17 cavity. Note that Tim17 contains a disulfide bond between Cys10 and Cys77, as shown previously47. d and e, All-atom MD simulation of Tim17–Tim23–Tim44 in a model mitochondrial membrane. In d, positions of lipid headgroup phosphorous atoms are shown as spheres. Panel e shows time-averaged thickness of the lipid hydrophobic layer from the membrane center to the IMS along the membrane plane measured by positions of lipid glycerol atoms (a view from the IMS; also see Extended Data Fig. 5 e–i). We note that a low value at the Tim23 cavity is due to the coordinated phosphatidylethanolamine (Fig. 1c). f, Indicated acidic amino acids lining the Tim17 cavity were mutated, and their functionality was tested by a cell growth assay. The addition of doxycycline (Dox) represses expression of chromosomal WT Tim17. Note that slower growth with the ‘empty/−Dox’ condition is likely due to hypomorphic expression of chromosomal TIM17 under a tetracycline promoter. g, Schematic for pulldown experiments of stalled Cyb2Δ-DHFR. h, Cyb2Δ-DHFR was pulled down with nickel resin after forming a stalled translocation intermediate, and samples were analyzed by immunoblotting. In lane 3, ΔΨm was dissipated by addition of valinomycin. Data in f and h are representative of three independent experiments.
