Figure 4. Model for the polypeptide transport mechanism of the TIM23 complex.
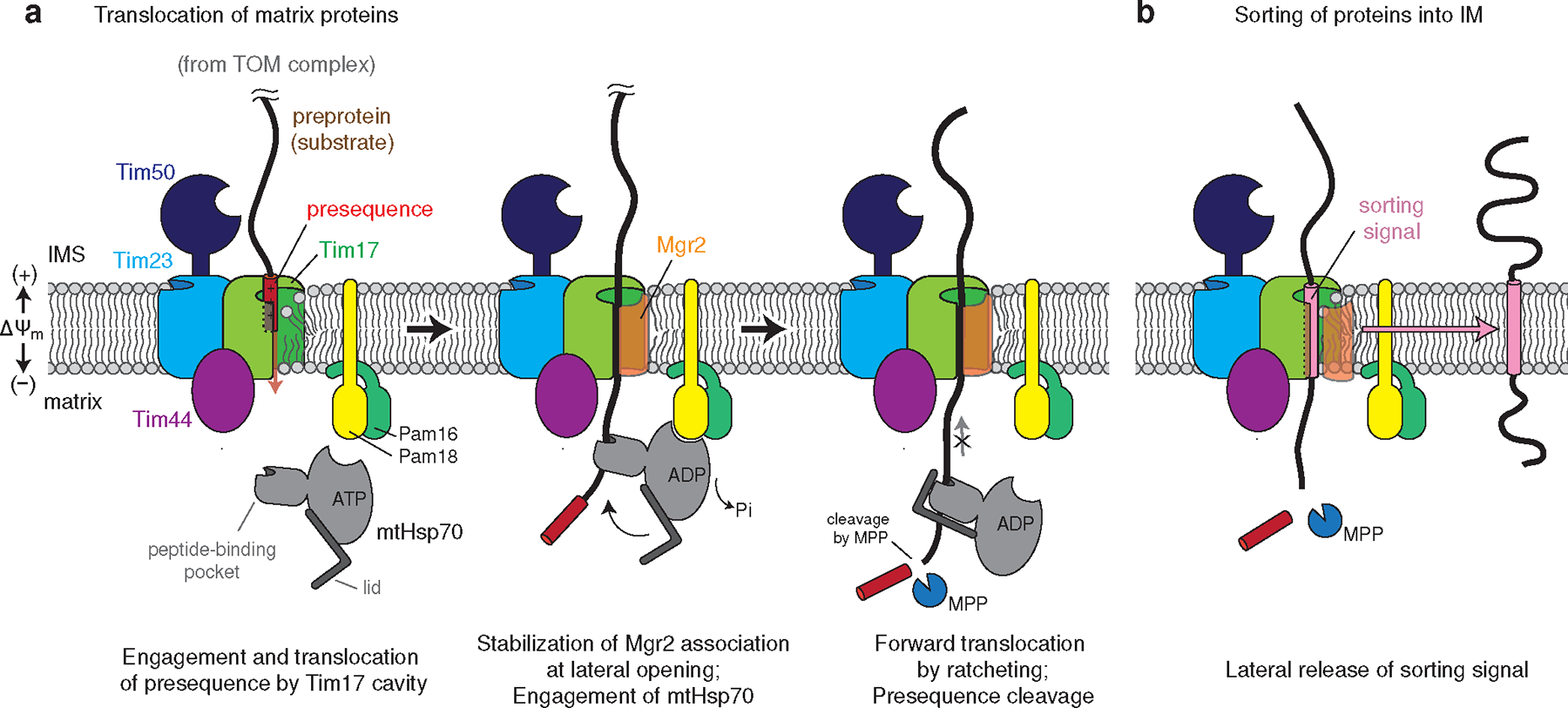
a, The N-terminal presequence first inserts into the cavity of Tim17. This step may be facilitated by Tim50, which may recruit the presequence to the TIM23 complex, and the negatively charged surface of the Tim17 cavity. Initial translocation of the presequence is likely powered by membrane potential (ΔΨm) across the inner membrane48,49. During translocation, the nonessential subunit Mgr2 stably associates with Tim17, sealing the lateral opening, and the preprotein is grasped by mtHsp70 as it emerges into the matrix. The J-domain of Pam18 activates ATP hydrolysis, leading to tight binding of mtHsp70 onto the polypeptide. The tight association of mtHsp70 will promote forward translocation of the polypeptide into the matrix while preventing backsliding. The presequence will be cleaved by the mitochondrial processing peptidase (MPP; not shown). For clarity, the tethers between the core TIM23 complex and Pam16–Pam18 and between Tim44 and mtHsp70 (ref. 26) are not shown. b, In the case of preproteins containing a TM sorting signal, dissociation of Mgr2 allows the signal to be released into the lipid phase through the open lateral gate of the Tim17 cavity.
