Extended Data Fig. 1 |. Purification of the TIM23 complex from S. cerevisiae and initial cryo-EM analysis.
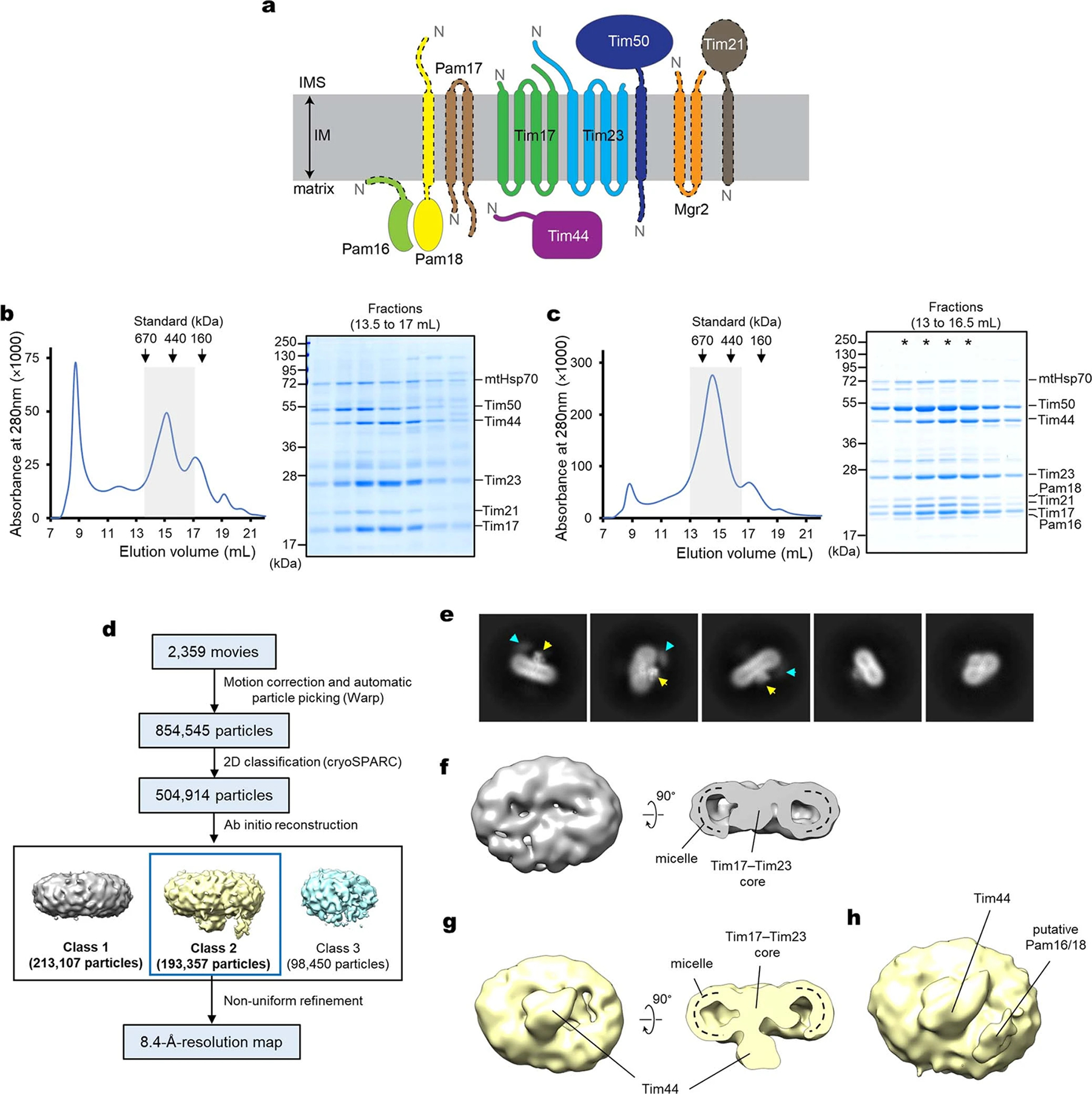
a, Schematic diagram of the membrane-associated components of the TIM23 complex. The two soluble matrix components mtHsp70 and Mge1 were not shown. The N-terminus of each subunit is indicated by an “N”. Note that Tim21, Pam17, and Mgr2 are not essential, and individual deletions of the portions of essential Pam16, Pam18, and Tim50 proteins outlined by a dashed line are viable13,17,19. b, Purification of the endogenous TIM23 complex. After affinity purification via Spot-tagged Tim17, the sample was subjected to Superose-6 size-exclusion chromatography (SEC). The gray box indicates fractions further analyzed by non-reducing SDS-PAGE and Coomassie staining (right panel). We note that copurified Tim50 often did not fully co-migrate with the other subunits in SEC, indicating its dissociation over time. c, As in b, but purification of the TIM23 complex from mitochondria co-overexpressing the nine subunits shown in a, omitting mtHsp70 and Mge1. Fractions marked with asterisks are pooled and used for cryo-EM analysis shown in d–h. d, Summary of single particle analysis of the sample shown in c. e, Example two-dimensional (2D) classes of the TIM23 complex. Putative matrix domains of Tim44-CTD and Pam16/18 are indicated by yellow and cyan arrowheads, respectively. f, Class 1 from ab-initio refinement of particles shown in d. g, As in f, but showing Class 2. h, As in the left panel of g, but shown at a lower isosurface threshold value. Data shown in b and c are representative of two experiments.
