Abstract
An RNA fragment of 75 nucleotides, which is located between the primer binding site and the 5′ major splice donor site in human immunodeficiency virus type 1, has been shown to participate in specific encapsidation of viral RNA. Compensation studies have identified two second-site mutations, namely, MP2 (a T12I substitution in p2) and MNC (a T24I substitution in the nucleocapsid [NC] protein) that were involved in the rescue of various deletions in the aforementioned RNA region (i.e., BH-D1, BH-D2, and BH-LD3). To study whether the MP2 and MNC point mutations exert their compensatory effects in a cis manner, production of Gag proteins was blocked by insertion of stop codons into LD3, LD3-MP2-MNC, and wild-type BH10 such that the constructs generated, i.e., LD3-DG, LD3-MP2-MNC-DG, and BH-DG, only provided RNA transcripts for packaging. The results of cotransfection experiments showed that the LD3-MP2-MNC-DG viral RNA was packaged as inefficiently as LD3-DG; in contrast, BH-DG was efficiently packaged. Therefore, nucleotide substitutions in MP2 and MNC did not act in a cis manner to correct the packaging deficits in LD3. Next, we deliberately changed the T12 in p2 or the T24 in the NC to each of 19 other amino acids. We found that amino acids with long hydrophobic side chains, i.e., V, L, I, and M, were favored at either position 12 in p2 or at position 24 in NC to compensate for the above-mentioned deletions. Further studies showed that only a few amino acids could not be used at these two sites by the wild-type virus due to decreased RNA levels in the virion or abnormal Gag protein processing. In this case, W, D, and E could not substitute for T12 in p2, and S, D, and N could not substitute for T24 in NC, without affecting viral infectivity. Therefore, the long hydrophobic side chains of V, L, I, and M are necessary for these amino acids to rescue the BH-D1, BH-D2, and BH-LD3 mutated viruses.
Human immunodeficiency virus type 1 (HIV-1) encapsidates two copies of full-length viral RNA that form a dimer through noncovalent linkage at the 5′ end (5). The cis-acting elements that are involved in the specific packaging and dimerization of viral RNA are located in the 5′ viral RNA leader sequence. Complex secondary structures have been proposed to exist in the leader region, among which SL1 and SL3 can bind to nucleocapsid (NC) protein with high affinity and are thought to be the major RNA elements responsible for viral RNA encapsidation (2, 7, 17, 19, 23, 27, 28). SL1 has also been shown to function as the dimerization initiation site (DIS) for viral RNA (1, 3, 9, 13, 19–22, 26, 30–34, 37). In addition, although the TAR and poly(A) hairpins, which are located at the 5′ end of the leader sequence, bind to NC protein with low affinity, they have also been shown to contribute to the packaging process (10, 11, 16, 17, 29).
We have been particularly interested in the roles of RNA sequences, located upstream of the 5′ major splice donor (SD) site, in encapsidation of viral RNA, since these RNA sequences are contained in both spliced and unspliced viral RNA, yet help the virus to selectively recruit unspliced RNA while excluding spliced RNA. To shed light on this issue, a number of deletion mutations, termed BH-D1, BH-D2, and BH-LD3, have been constructed to selectively remove RNA sequences in this region (23–25). These deletions had adverse impacts on both viral RNA encapsidation and viral replication. Interestingly, when the mutated viruses were cultured for a prolonged period, revertants with wild-type replication kinetics arose, and the results of sequencing analysis revealed two substitutional mutations, i.e., MP2 (T12I in p2) and MNC (T24I in NC), that can rescue such mutated viruses. We believe that exploration of the mechanism by which the MP2 and MNC mutations are able to compensate for the functions of the deleted RNA sequences will add new insights into the roles of RNA sequences upstream of the 5′ SD site in viral RNA encapsidation and viral replication.
The first question we sought to answer was whether changes of nucleotides by the MP2 and MNC point mutations had altered cis-acting RNA signals in the gag coding region and whether the compensatory effects obtained were controlled in a cis-acting manner. This subject was pursued by insertion of stop codons into the gag gene in LD3, LD3-MP2-MNC, and BH10 to block Gag production, in order to enable a comparison of packaging efficiency among these three viral RNA transcripts. The second issue was whether the I residue is the only amino acid at either position 12 in p2 or at position 24 in NC that can rescue the above deletions. Answers to this question will shed light on the strictness of the steric interactions at these two sites for the rescue process. This subject was approached by substituting either the T12 in p2 or the T24 in NC by each of 19 other amino acids and screening for those residues that are able to rescue the deletions described above.
MATERIALS AND METHODS
HIV-1 DNA mutagenesis.
The BH10 clone of infectious HIV-1 cDNA was employed as starting material to generate the following mutant constructs. The BH-D1, BH-D2, and BH-LD3 deletions eliminated sequences at nucleotides (nt) +200 to +226, +200 to +233, and +238 to +253, respectively, and were constructed as previously described (23, 25). To prevent translation of Gag proteins, stop codons were inserted at both the 6th and 8th amino acid positions in matrix (MA) protein and the 82nd and 85th amino acid positions in capsid (CA) protein (Fig. 1A). The former two stop codons in MA were engineered by PCR through use of primer pair pBssH-S (5′-CTGAAGCGCGCACGGCAAGAGG-3′ [positions 706 to 727])–p800 (5′-CTAATTCTCCCCCGCTTCATACTCACGCTCTCGCACCC-3′ [positions 829 to 792]); the latter two stop codons in CA were inserted by PCR through use of primer pair pBssH-S–p1430 (5′-GCCCTGCATGCACTTAATGCACTCAATCCCATTCTGC-3′ [positions 1453 to 1417]). The entire DIS sequences at nt +243 to +277 were deleted in construct ΔDIS that was generated by PCR using the primer pair pHpa-S (5′-CTGCAGTTAACTGGAAGGGCTAATTCACTCCC-3′ [positions 1 to 21])–pDIS (5′-TACTCACCAGTCGCCGCCCTCCTGCGTCGAGAGAGC-3′ [positions 750 to 680]).
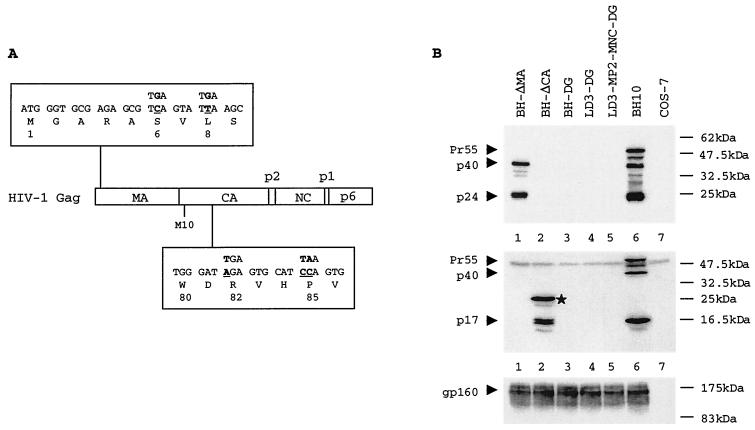
FIG. 1.
(A) Schematic illustration of the inserted stop codons in the gag gene. The mutated nucleotides are underlined. (B) Analysis of viral protein expression from various constructs in transfected COS-7 cells. The top, middle, and bottom panels represent Western blots performed with MAbs against HIV-1 CA (p24), MA (p17), and Env proteins, respectively. A fusion protein of ∼25 kDa that contains MA and truncated CA sequences is indicated by a star in the middle panel. Protein markers are shown on the right of the gels. BH-ΔMA contains stop codons in MA. BH-ΔCA contains stop codons in CA. BH-DG contains stop codons in both MA and CA. LD3-DG contains the LD3 deletion, as well as stop codons in MA and CA. LD3-MP2-MNC-DG contains the LD3 deletion and the MP2 and MNC point mutations, as well as stop codons in MA and CA.
An amino acid T at position 12 in the p2 protein was substituted by each of 19 other amino acids through use of primer p2-12X (5′-CCAAGTAACAAATTCAGCTNNNATAATGATGCAGAGAGGC-3′ [positions 1893 to 1932]), in which N represents A, C, G, or T; this permits each of the 20 amino acids to appear at this site (see Fig. 3). PCR was performed with the primer pair p2-12X–pNC-A (5′-TTAGCCTGTCTCTCAGTACAATC-3′ [positions 2084 to 2062]), and the initial PCR product was used as a primer in a second round of PCR along with primer pSph-S (5′-AGTGCATCCAGTGCATGCAGGGCC-3′ [positions 1431 to 1454]). The final PCR product was digested with restriction enzymes SphI and ApaI and inserted into BH-D1 or BH10 to generate clones D1-p2-12X or BH-p2-12X (X represents any of the 20 amino acids). A large number of bacterial colonies were screened to ensure representation of substitutions of all 20 amino acids at position 12 in p2.
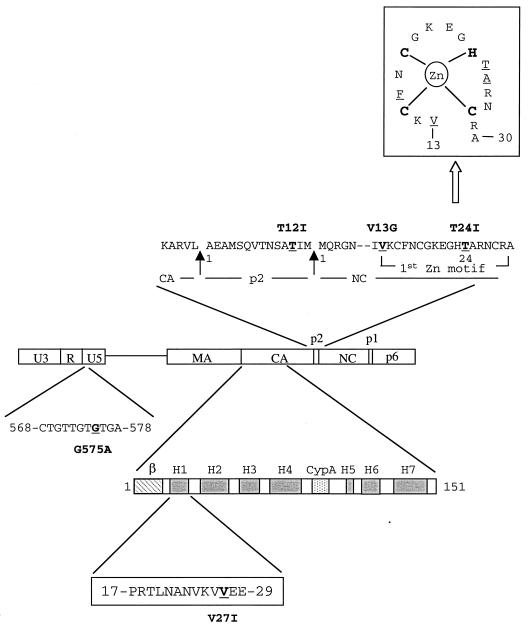
FIG. 3.
Schematic illustration of mutations in the U5 region, CA, p2, and NC. The amino acid sequence of p2, as well as parts of the CA and NC proteins, is shown. The arrows indicate viral protease cleavage sites that result in formation of p2. The substituted nucleotide in U5 or amino residues in CA, p2, and NC are underlined. Nucleotides in U5 are numbered from the beginning of the U3 region. The numbering of the amino acid positions starts from the first residue of each protein. Secondary structural domains in the amino terminus of CA protein (amino acids 1 to 151) are illustrated that include a β-hairpin (β), seven α-helixes (H1 to H7), and a cyclophilin A (Cyp A) binding site located between H4 and H5 (14). The V27I substitution in CA exists in H1. A sketch of the first Zn finger motif, in which some of the second-site mutations are located, is shown at the top of the figure. Interactions between the C, C, H, and C amino residues and the Zn ion are indicated; moreover, the underlined V13, F16, T24, and A25 amino acid residues are close in tertiary structure and form a hydrophobic cleft that can interact with the loop nucleotides of the region of viral genomic RNA SL3 (12).
The amino acid T residue at position 24 in NC was substituted by each of 19 other amino acids with the same cloning strategy using primer pair pNC-24X (5′-CCTAGGGGCCCTGCAATTTCTGGCNNNGTGCCCTTCTTTGC-3′ [positions 2007 to 1966])–pSph-S to generate clones D2-MP2-NC24X (containing the D2 deletion and MP2 substitution) or BH-NC24X. Primer positions refer to the beginning of the 5′ U3 region. All primers were synthesized by Gibco-BRL.
Cell culture, transfection, and infection.
COS-7 and MT-2 cells were grown in Dulbecco modified Eagle medium (DMEM) and RPMI 1640 medium, respectively, each supplemented with 10% fetal calf serum. COS-7 cells were transfected with HIV-1 DNA constructs in the presence of Lipofectamine (Gibco-BRL, Montreal, Quebec, Canada). Progeny virus was harvested 48 h after transfection and quantified by measuring levels of viral CA antigen (Ag) by enzyme-linked immunosorbent assay (Abbott Laboratories, Abbott Park, Ill.).
For infectivity assays, similar amounts of virus (i.e., 3 ng of CA Ag) were used to infect 5 × 105 MT-2 cells. After 2 h, cells were washed twice to remove unbound virus and grown in complete RPMI 1640 medium. Culture fluids were collected at various times, and reverse transcriptase (RT) activity was measured (23).
For mutated viruses with diminished infectiousness, the infected cells were split upon confluence and kept in culture until extensive formation of syncytia and high levels of RT activity were observed. At this stage, culture fluids were used to infect fresh MT-2 cells. Viruses were passaged until the infections observed were as virulent as those caused by wild-type virus. Cellular DNA was purified and amplified by PCR through the use of primer pair pHpa-S–pBcl-A (5′-CTATGAGTATCTGATCATACTG-3′ [positions 2445 to 2424]). The PCR product was cloned and sequenced to confirm the presence of the original mutations and to identify novel mutations that might have been present in noncoding leader RNA sequences and in the gag gene.
Radiolabeling and immunoprecipitation assays.
COS-7 cells that had been transfected with HIV-1 recombinant DNA constructs were starved in DMEM without l-Met and l-Cys at 37°C for 1 h, after which the cells were metabolically radiolabeled with [35S]-l-Met and [35S]-l-Cys (ICN) at a concentration of 100 μCi/ml for 30 min at 37°C. After an extensive washing with complete DMEM, supplemented with 30 μg of l-Met and 60 μg of l-Cys per ml, the cells were cultured for 1 h. Virus particles from culture fluids were pelleted by ultracentrifugation and analyzed on sodium dodecyl sulfate (SDS)–12% polyacrylamide gels. The cells were washed twice with cold phosphate-buffered saline and lysed with 1 ml of NP-40 lysis buffer (50 mM Tris-Cl [pH 8.0], 150 mM NaCl, 0.02% sodium azide, 100 μg of phenylmethysulfonyl fluoride per ml, 1 μg of aprotinin per ml, and 1% NP-40). The cell lysates were clarified in a bench-top Eppendorf centrifuge at 13,000 rpm for 10 min at 4°C and then incubated with anti-HIV-1 CA monoclonal antibody (MAb) for 1 h at 4°C, after which 5 μl of protein-A linked Sepharose 4B (Pharmacia, Montreal, Quebec, Canada) was added for a further 30 min of incubation. The Sepharose 4B was then centrifuged and washed in turn with NET-gel buffer and Tris–NP-40 buffer (35). Pellets were suspended in 20 μl of 1× SDS-containing gel-loading buffer, boiled, and analyzed by SDS–12% polyacrylamide gels. Viral proteins were visualized by exposure to X-ray films.
Encapsidation of viral RNA by RT-PCR.
Supernatants from COS-7 cells that had been transfected with various HIV-1 recombinant DNA constructs were clarified in a Beckman GS-6R centrifuge at 3,000 rpm for 30 min at 4°C and quantified on the basis of CA Ag levels. Viral RNA was purified from an amount of virus containing 2 ng of CA Ag using an RNA extraction kit (Qiagen). Viral RNA was dissolved in 50 μl of diethyl pyrocarbonate-treated water and treated with 20 U of RNase-free DNase (Gibco-BRL, Montreal, Quebec, Canada) at 37°C for 30 min to remove any DNA contamination. Five-microliter volumes of viral RNA were amplified for 20 cycles with a Titan One-Tube RT-PCR system (Boehringer Mannheim, Mannheim, Germany) using primer pair pGAG1-pST to analyze full-length viral RNA (23). DNA products were analyzed on 5% native polyacrylamide gels and visualized following exposure to X-ray films.
Transfected COS-7 cells were washed twice with cold phosphate-buffered saline and were lysed with NP-40 lysis buffer. A portion of the lysates was removed for p24 determination, and the rest were subjected to RNA extraction with Trizol Reagent (Gibco-BRL). RNA from 200 pg of cell lysates was analyzed by RT-PCR as described above.
RESULTS
The MP2 and MNC point mutations do not exert their compensatory effects via a cis-acting mechanism.
Since MP2 and MNC compensate for cis-acting viral RNA packaging signals, it may be speculated that these two mutations act by changing cis-acting RNA signals in the gag-coding region. It was shown that MP2 and MNC restored the diminished content of viral RNA in LD3 to wild-type levels (23). We now asked whether the MP2 and MNC point mutations might act in cis to enable the LD3-MP2-MNC RNA to be more efficiently incorporated by wild-type Gag proteins than LD3 viral RNA. To pursue this subject, Gag production was eliminated from LD3 and from LD3-MP2-MNC, such that the potential cis effects of MP2 and MNC on viral RNA packaging could be assessed. For this purpose, two stop codons were inserted at the sixth and eighth amino acid positions in the MA protein to yield construct BH-ΔMA (Fig. 1A). Surprisingly, substantial levels of the Gag-derived proteins, p40 and p24, but not Pr55Gag, were detected in transfected COS-7 cells by Western blots (Fig. 1B, lane 1). It was later realized that the continuous presence of p40 and p24 was due to reinitiation of translation from an “AUG” codon at the tenth amino acid position in the CA coding region, an event that results in the appearance of a p40 Gag product (6). To entirely terminate Gag production, two stop codons were inserted at the 82nd and 85th amino acid positions in the CA protein. As expected, no Gag expression was now detected with BH-ΔCA in the transfected COS-7 cells (Fig. 1B, lane 2). However, normal production of Env protein was seen (lane 2), indicating efficient transfection and normal viral gene transcription. The aforementioned four stop codons were then inserted into each of the BH10, LD3, and LD3-MP2-MNC constructs in order to engineer clones BH-DG, LD3-DG and LD3-MP2-MNC-DG, which were all defective in Gag production and yet active in the generation of Env proteins, as seen on Western blots (Fig. 1B, lanes 3, 4, and 5).
To further assess production of Gag protein from these recombinant constructs, Western blots were conducted with monoclonal antibodies (MAbs) against HIV-1 matrix (MA) antigen. We found that stop codons inserted at the sixth and eighth positions in MA completely arrested MA expression (Fig. 1B, lane 1). In contrast, stop codons inserted at the 82nd and 85th positions in CA still allowed efficient synthesis of a MA protein that was fused with the first 81 amino acids of CA (lane 2). When the two groups of stop codons were recombined in the same construct, no MA expression was detected (lanes 3, 4, and 5). Therefore, the inserted stop codons in MA and CA are both necessary to effectively eliminate Gag production. It should be pointed out that a truncated version of CA protein (i.e., 81 amino acids) should still be produced at low levels by each of the BH-DG, LD3-DG, and LD3-MP2-MNC-DG constructs because of weak translation initiation associated with CA. Since this low abundance peptide does not include the p2 and NC portions, wherein the compensatory mutations reside, they should not have significantly affected our cotransfection assays.
Next, we cotransfected the above DNA clones with ΔDIS, a construct that lacks the entire DIS sequence from nt +243 to +277 (Fig. 2A). Since BH-DG, LD3-DG, and LD3-MP2-MNC-DG did not produce Gag proteins, they only transcribed viral RNA available for packaging by the wild-type Gag proteins from ΔDIS. Primer pair pD (5′-CCAGAGGAGCTCTCTCGACGC-3′ [positions 672 to 692])–pA (5′-CCATCGATCTAATTCTCCC-3′ [positions 837 to 819]) was designed in such a way that RT-PCR yielded different lengths of DNA products from BH-DG, LD3-DG, and ΔDIS (Fig. 2A). To assess the possible preferential synthesis of the shorter ΔDIS fragment over either LD3 or BH10 in competitive PCR using the above-mentioned primer pair, similar amounts of three types of plasmid DNA were mixed as templates. Relative amounts of BH10, LD3, and ΔDIS plasmid DNA before mixing were first examined using primer pair pGAG1-pST that directs the amplification of a 119-bp fragment from each of the three constructs (23). We found that equivalent amounts of each plasmid DNA were synthesized (Fig. 2B, lanes 1 to 9). When the mixed plasmid DNA samples were examined in PCR using primer pair pD-pA (lanes 10 to 12), the relative amounts of different plasmid DNA samples observed in a single reaction were similar to those detected individually with primer pair pGAG1-pST. Therefore, no preferential amplification of ΔDIS product was seen in our competitive PCR system, possibly due to the limited difference in length between the PCR products.
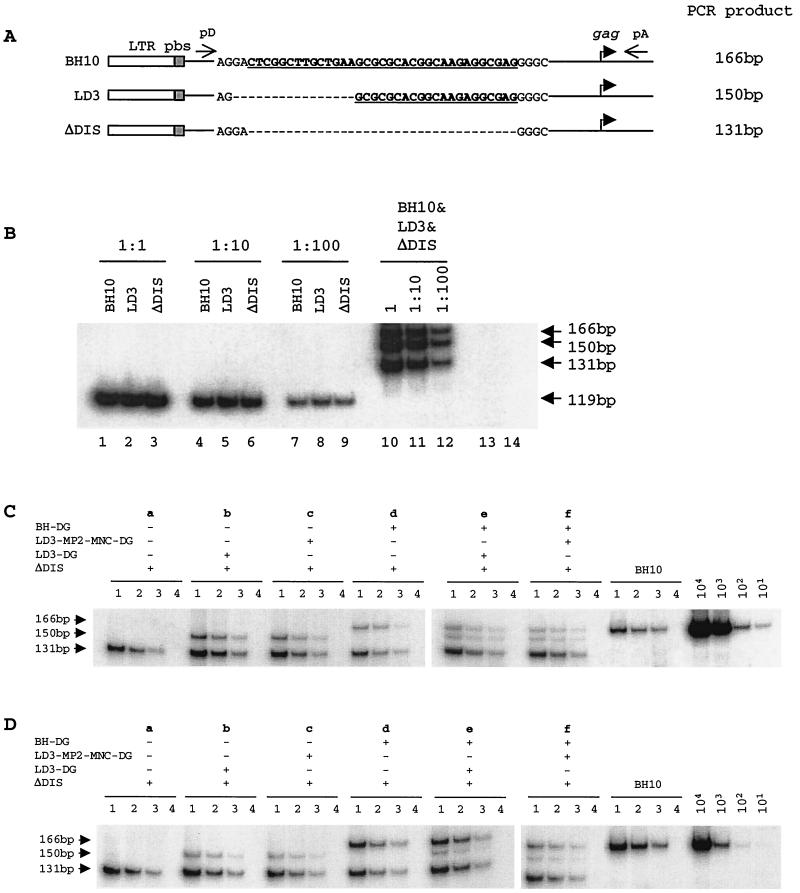
FIG. 2.
(A) Schematic illustration of the LD3 and ΔDIS deletions. The DIS sequences are underlined. Deleted nucleotides are indicated by dashed lines. RT-PCR using primer pair pD-pA yields DNA products of 166 bp from wild-type BH10 RNA, 150 bp from LD3 RNA, and 131 bp from ΔDIS RNA. (B) PCR using the BH10, LD3, and ΔDIS plasmid DNA. Each of the three types of plasmid DNA was first quantified individually in separate reactions using primer pair pGAG1-pST (lanes 1 to 9) (23). The same plasmid samples were then mixed and subjected to reactions using primer pair pD-pA (lanes 10 to 12). Three dilutions were included to ensure the linear range of reactions. Lanes 13 and 14 represent negative controls with primer pairs pGAG1-pST and pD-pA, respectively. (C) RT-PCR analysis of viral RNA in transfected COS-7 cells. Total cellular RNA that was extracted from cell lysates equivalent to 100 pg of HIV-1 p24 was subjected to RT-PCR. Three dilutions, i.e., 1:1 (lane 1), 1:3 (lane 2), and 1:9 (lane 3), of each RNA sample were measured. RNase A digestion was also performed as a negative control to rule out any possible plasmid DNA contamination (lane 4). Wild-type viral DNA standards of 101, 102, 103, and 104 copies were used in RT-PCR to determine the linear range of the reactions. Combinations of constructs in each transfection experiment are shown at the top of the gels. (D) RT-PCR analysis of RNA that was extracted from virus particles. Labeling of the lanes refers to that in panel C.
To minimally interfere with ΔDIS gene expression, 2 μg of ΔDIS DNA was used, compared with 0.5 μg each of BH-DG, LD3-DG, and LD3-MP2-MNC-DG, in cotransfection experiments involving two constructs, and 0.25 μg of each was used in cotransfection experiments involving three constructs. Cellular RNA from transfected COS-7 cells was first analyzed by RT-PCR. The results of Fig. 2C show that viral RNA levels from each of the constructs in cotransfection experiments reflect the relative amount of plasmid DNA used. For instance, in cotransfection experiment e, similar levels of viral RNA were detected from BH-DG and LD3-DG, both of which were much lower than that from ΔDIS, since 0.25 μg of BH-DG or LD3-DG, in comparison to 2 μg of ΔDIS, was used in transfection. When the RNA content in virus particles were examined, ΔDIS packaged moderately lower levels of viral RNA than BH10 (Fig. 2Da). When LD3-DG was cotransfected with ΔDIS, LD3-DG RNA was detected in virus particles, but at a level similar to that in cells (Fig. 2Db). Similar observations were made in cotransfection with ΔDIS and LD3-MP2-MNC-DG (Fig. 2Dc). However, when BH-DG was cotransfected with ΔDIS, similar levels of both viral RNAs were detected in virus particles despite the lower levels of BH-DG RNA in cells (Fig. 2Dd). Therefore, the BH-DG RNA was more efficiently packaged into virus particles than ΔDIS RNA; in contrast, the MP2 and MNC point mutations did not help LD3 RNA to be more efficiently incorporated into virus particles. To further test the above results, cotransfection experiments were performed with the three constructs. When both LD3-DG and BH-DG RNA were present in cells, BH-DG was much more efficiently packaged into virus particles than LD3-DG (Fig. 2De). Although the total levels of viral RNA in cotransfection f were slightly lower than those in cotransfection e, the BH-DG signal was significantly stronger than that of LD3-MP2-MNC-DG. Thus, LD3-MP2-MNC-DG was not able to successfully compete with BH-DG for packaging (Fig. 2Df). Therefore, the nucleotide substitutions in MP2 and MNC cannot help LD3 RNA to be more efficiently packaged via cis-acting mechanisms.
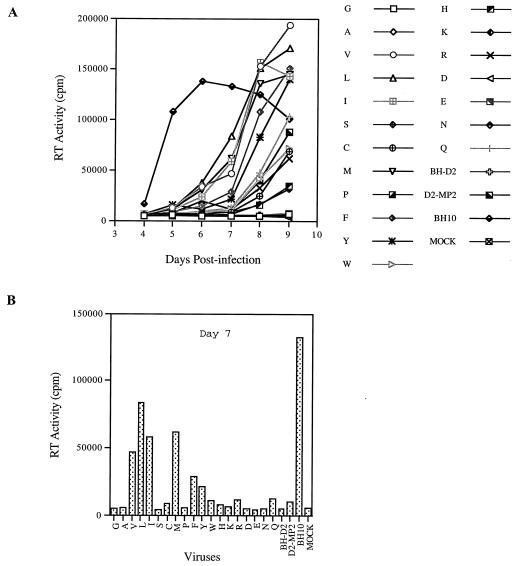
Substitutions of the T amino acid at position 12 in p2 with V, L, I, C, or M can dramatically increase the infectiousness of BH-D1 mutant viruses.
We have previously shown that the BH-D1 deletion mutation, eliminating HIV sequences at nt +200 to +226, attenuated viral replication. Long-term culture of the mutated virus in MT-2 cells led to a revertant containing a mutation at position 12 in p2, i.e., T12I (MP2), that helped to restore viral infectiousness to near wild-type levels (25). We now sought to answer whether other amino acids at position 12 in p2 were also able to rescue the BH-D1 deletion. Toward this end, the T12 in p2 in BH-D1 was substituted by each of 19 other amino acids (Fig. 3) (Table 1). Relevant recombinant DNA constructs were transfected into COS-7 cells, and the progeny viruses thus generated were quantified on the basis of CA Ag levels. Equivalent amounts of virus were used to infect MT-2 cells and the infectiousness of each preparation was determined by the formation of syncytia and RT activity in culture fluids. The results of Table 1 show that four amino acids, including V, L, C, and M, in addition to the previously detected I, can help to correct the adverse effects of BH-D1 on virus replication. This subject was further evaluated by replication kinetic studies (Fig. 4A). Viruses containing either a G, K, D, or E at position 12 in p2 were barely infectious. In contrast, when T12 was replaced with either V, L, I, C, or M, the recombinant viruses thus generated were able to generate high levels of RT activity (Fig. 4A). Among the five amino acids that could play this role, C was the least efficient.
TABLE 1.
Infectiousness of various D1-p2-12X viruses in MT-2 cells
| Amino acid substitutiona | Genetic codon(s) | Infectivityb |
|---|---|---|
| Gly (G) | GGC | − |
| Ala (A) | GCC | − |
| Val (V) | GUU, GUG | + |
| Leu (L) | UUA | + |
| Ile (I) | AUU, AUA | + |
| Ser (S) | UCC, UCU | − |
| Thr (T) | ACU | − |
| Cys (C) | UGU | + |
| Met (M) | AUG | + |
| Pro (P) | CCU | − |
| Phe (F) | UUU | − |
| Tyr (Y) | UAU | − |
| Trp (W) | UGG | − |
| His (H) | CAC | − |
| Lys (K) | AAG | − |
| Arg (R) | CGA | − |
| Asp (D) | GAC | − |
| Glu (E) | GAG | − |
| Asn (N) | AAU | − |
| Gln (Q) | CAG | − |
| Stop codon | UAA | − |
| BH10 (T) | ACC | + |
The amino acid T12 (ACC) in p2 of BH-D1 was randomly mutated to the others. The constructs thus generated were named after the mutated amino acids, e.g., D1-p2-12G and D1-p2-12A.
Evaluation of infectivity of each mutated virus was based on the formation of syncytia and levels of RT activity in culture fluids. A value of 5.0 × 105 cpm RT activity from 50 μl of culture fluids was considered positive (+). BH10 wild-type virus served as a positive control.
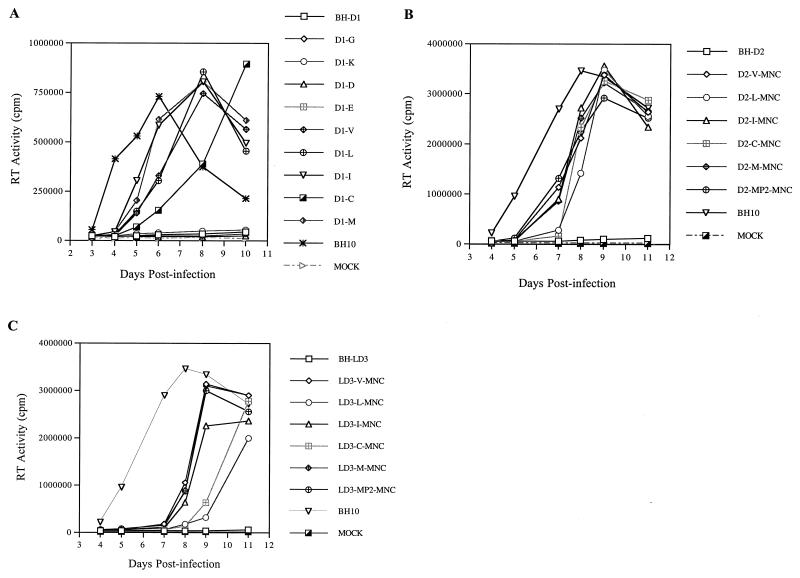
FIG. 4.
(A) Replication kinetics of wild-type BH10 and mutated viruses containing the BH-D1 deletion and substitutions of T12 in p2. (B) Infectiousness of mutated viruses containing the BH-D2 deletion, substitutions of T12 in p2, and a T24I point mutation in NC. (C) Growth curves of mutated viruses possessing the BH-LD3 deletion, substitutions of T12 in p2 and a T24I point mutation in NC. In these studies, 5 × 105 MT-2 cells were infected by viruses equivalent to 3 ng of p24 Ag. RT activity in culture fluids was determined at various times. The D2-MP2-MNC and LD3-MP2-MNC constructs, both containing a T12I substitution in p2 and a T24I mutation in NC, were generated previously (24, 25) and served as positive controls. Mock infections represent negative controls performed with heat-inactivated BH10 virus.
The T12I substitution in p2 was also identified in revertants of the BH-D2 and BH-LD3 mutated viruses (23, 25). We therefore wished to determine whether the amino acids shown to compensate for BH-D1 (see above), i.e., V, L, C, and M, were also able to rescue the BH-D2 and BH-LD3 defective viruses. Toward this end, the T12 in the p2 of BH-D2 or BH-LD3 was changed to either V, L, C, or M. In addition, a mutation at position 24 in the NC protein, termed MNC, was also included in these constructs, since it has been previously shown that this mutation works synergistically with the T12I mutation in p2 to rescue both BH-D2 or BH-LD3 (23, 25). The various constructs, i.e., D2-V-MNC, D2-L-MNC, D2-C-MNC, D2-M-MNC, LD3-V-MNC, LD3-L-MNC, LD3-C-MNC, and LD3-M-MNC, were transfected into COS-7 cells and the progeny viruses thus generated were used to infect MT-2 cells. The results of Fig. 4B and C show that all of the above-described constructs produced viable viruses in contrast to the noninfectiousness of BH-D2 and BH-LD3. Therefore, each of V, L, C, and M can help to rescue the BH-D2 and BH-LD3 deletions. The C and L substitutions were less efficient in this regard than either V, I, or M.
The W, D, and E amino acids at position 12 in p2 dramatically decreased infectiousness of wild-type virus.
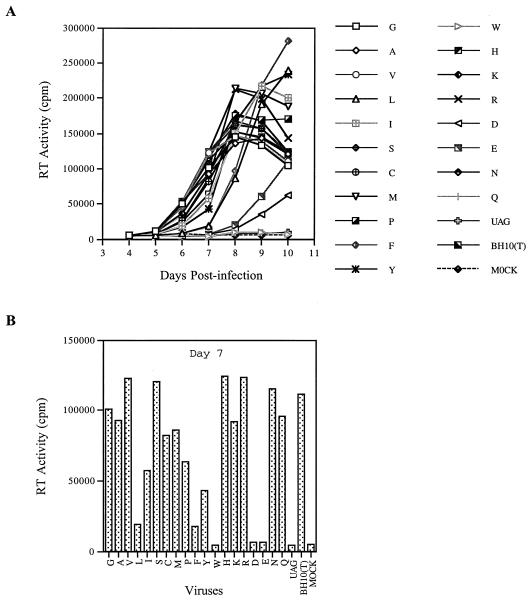
We next studied the effects of other substitutions at position 12 in p2 on the replication potential of wild-type BH10 virus. Accordingly, T12 in BH10 was changed to each of 19 other amino acids. These various constructs were transfected into COS-7 cells, and the progeny viruses thus generated were quantified according to the levels of CA Ag. The infectiousness of the viruses produced by these constructs was examined by infection of MT-2 cells. The data showed that the T12W substitution in p2 abolished viral replication and that T12D and T12E dramatically decreased viral infectiousness (Fig. 5). Substitutions to either L or F markedly diminished viral replication and yet were permissive for production of high levels of RT activity after 10 days, while I, P, and Y each only slightly delayed viral replication (Fig. 5). Therefore, in terms of impact on viral replication, the various substitutions can be ranked as follows: W>D, E>L, F>I, P, Y>G, A, V, S, C, M, H, K, R, N, Q.
FIG. 5.
Replication capacities of wild-type BH10 and mutated viruses containing substitutions of T12 in p2. Mutations are represented by single-letter abbreviations of amino acids. UAG is a stop codon that was inserted at position 12 in p2. (A) Growth curves of wild-type and mutated viruses after infection of MT-2 cells. (B) RT activity in culture supernatants at day 7 after infection. Mock represents a negative control performed with heat-inactivated wild-type virus.
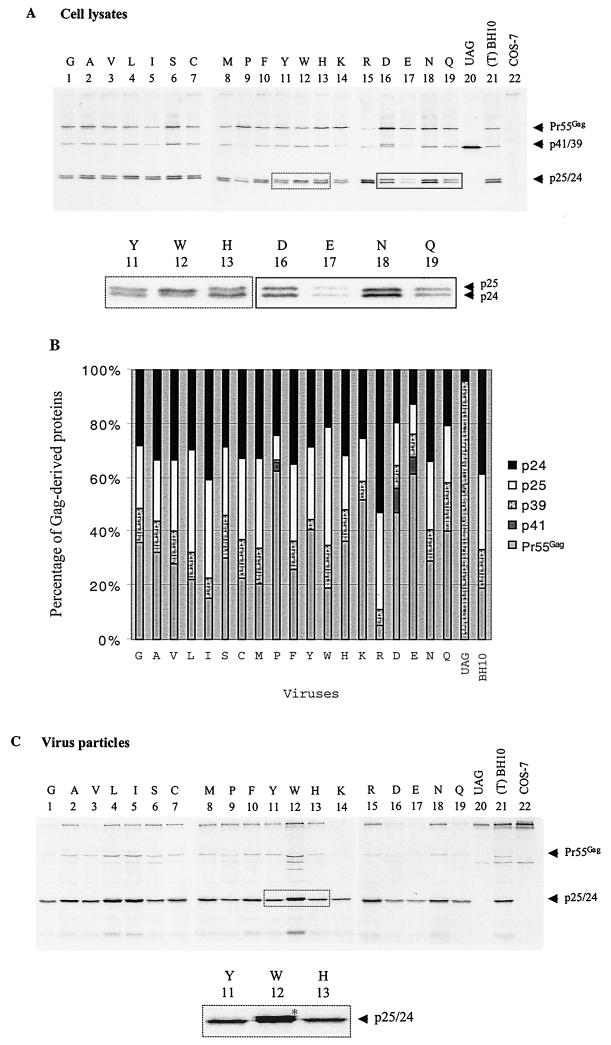
T12 is one of eight amino acids in the HIV-1 p2 and NC proteins that are recognized and cleaved by the viral protease. Hence, mutations of T12 may affect the efficiency of cleavage at this site and decrease viral replication. To test this hypothesis, the processing of Gag in our various mutated viruses was examined through short-term radiolabeling and immunoprecipitation of viral proteins. For this purpose, we employed anti-CA MAb to detect each of Pr55Gag, intermediate cleavage products, including p41/p39 and p25, as well as mature p24 in cell lysates (Fig. 6A). The intensities of the protein bands were quantified, and the percentage of each viral protein was plotted to assess the efficiency of Gag processing (Fig. 6B).
FIG. 6.
Processing of Gag precursor protein Pr55Gag in wild-type BH10 and mutated viruses containing substitutions of the T12 amino acid in p2. Mutations are represented by single-letter abbreviations of substituted amino acids. (A) Transfected COS-7 cells were labeled with l-[35S]Met and l-[35S]Cys for 30 min and cultured for 1 h. Cells were then lysed, and the lysates were subjected to immunoprecipitation through the use of MAb against CA (p24) Ag. The observed viral proteins, including Pr55Gag, p41, p39, p25, and p24, are labeled on the right of the gels. To clearly visualize the p25 and p24 proteins in certain mutants, enlarged portions of the gels are shown. Mock-transfected COS-7 cells were also labeled with l-[35S]Met and l-[35S]Cys and serve as a negative control. (B) The intensities of the viral protein bands were quantified through the use of the NIH Image Program, and the percentages of each viral protein in mutant or wild-type virus were plotted. (C) After 1 h in culture, labeled virus particles were pelleted from the supernatants and analyzed on SDS–12% polyacrylamide gels. A portion of the gel was enlarged to clearly show accumulation of p25 protein in the W mutant.
In the case of the BH-p2-12UAG construct, that produced a truncated Gag protein (i.e., MA-CA-p2 but missing three amino acids at the C terminus of p2 because of the UAG stop codon), a protein band of 39 kDa was observed. When a number of aliphatic amino acids, i.e., G, A, V, L, and I, were substituted for T at position 12 in p2, we found that the levels of Pr55Gag diminished as the length of the side chain of the amino acid increased (Fig. 6A and B). Yet, this did not directly correlate to viral infectiousness, since the G, A, V, and I mutants had similar replication capacity (Fig. 5). Of these, the L substitution resulted in a modest accumulation of p25 (CA-p2) fusion product in comparison with wild type (T) (Fig. 6A and B); this result correlates with the decreased infectiousness of the L mutant (Fig. 5). Substitution of T12 in p2 by S, C, or M, the side chains of which contain hydroxyl or sulfur groups, did not affect the processing of Gag proteins (Fig. 6A and B). Mutation of T to the cyclic amino acid P resulted in significant accumulation of Pr55Gag, diminished levels of the intermediate p39 and p25 products, as well as enhanced cleavage of p24 from p25 (Fig. 6A and B). Among the substitutions involving the aromatic amino acids F, Y, and W, W led to both enhanced accumulation of p25 and slightly different migration patterns for both the p25 and p24 proteins (Fig. 6A and B), deficits that may account for the noninfectiousness of the T12W mutant. In contrast, mutations to the basic amino acids H, K, or R yielded differential results, with both H and K causing an accumulation of Pr55Gag, while R facilitated cleavage between p2 and NC and led to the rapid appearance of the p25 and p24 products (Fig. 6A and B). The acidic amino acids D and E resulted in enhanced accumulation of Pr55Gag, the appearance of the p41 intermediate protein, and a slightly slower migration rate of p25. Both N and Q mutants showed moderate accumulation of Pr55Gag.
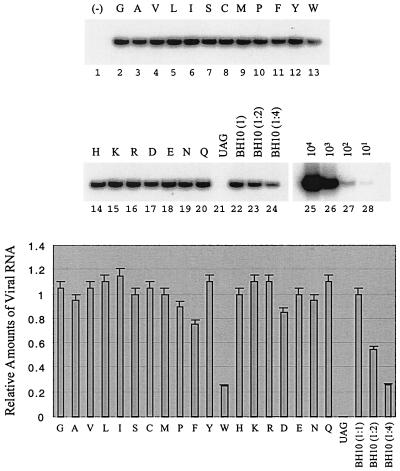
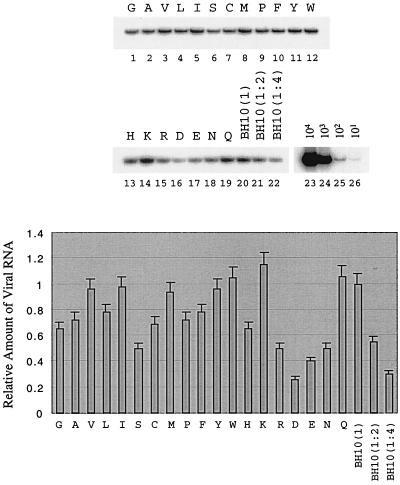
After the radiolabeling, COS-7 cells were cultured for 1 h in complete DMEM, after which culture supernatants were pelleted by ultracentrifugation and virus particles were lysed and analyzed on SDS–12% polyacrylamide gels. No viral protein signal was detected in the case of the BH-p2-12UAG construct; this confirms that this construct was unable to produce virus particles (Fig. 6C). Among all amino acids, W resulted in a great accumulation of p25 (Fig. 6C) and thus had the highest impact on processing of Pr55Gag, a result consistent with elimination of viral infectivity. The levels of pr55Gag in some viruses appear to be low (lanes 1, 14, 16, 17, and 19); yet, considering the low levels of p24/25, these viruses still exhibit similar ratios between Pr55Gag and p24/p25 as the wild-type BH10. We further examined levels of full-length viral RNA packaged by various recombinant virus particles. The majority of the 19 amino acids used to substitute for T at position 12 in p2 did not markedly affect RNA encapsidation (Fig. 7). However, W resulted in substantially decreased levels of viral RNA in virus particles (Fig. 7).
FIG. 7.
Levels of viral RNA in wild-type virus and in mutated viruses harboring substitutions at position 12 in p2. Levels of full-length viral RNA were assessed by RT-PCR through the use of primer pair pGAG1-pST (23). RNA samples were treated with DNase to remove any contaminating DNA. RNA samples were also digested with RNase A and then subjected to RT-PCR as a negative control to exclude the possibility of DNA contamination. A negative control of wild-type BH10 is shown in lane 1. RNA obtained from wild-type virus was diluted 1:2 and 1:4 before RT-PCR to ensure a linear range of these reactions (lanes 23 and 24). Additional controls were performed with wild-type DNA of 101, 102, 103, and 104 copies to determine the linear range of the reaction (lanes 25 to 28). The results represent three independent experiments. Band intensities were quantified with the NIH Image Program and plotted. The amount of BH10 RNA was arbitrarily set at 1.0.
A V27I mutation in CA, together with a G575A substitution in the U5 region, can partially correct the attenuated infectiousness caused by the T12D substitution in p2.
The T12W, T12D, and T12E mutations severely impaired infectiousness of wild-type virus. To further study these mutations, relevant viruses were cultured in MT-2 cells over prolonged periods to generate revertant viruses with increased growth potential. In the case of the construct containing the T12W substitution, no formation of syncytia was observed after 8 weeks in culture. In contrast, the T12D and T12E mutants showed rapid syncytium formation and high levels of RT activity after 6 weeks in culture. Conceivably, these latter substitutions may have served to stimulate the occurrence of other compensatory mutations that, in turn, enhanced viral replication.
Therefore, a fragment of viral DNA (nt 1 to 2430) was amplified from the genomes of the T12D and T12E revertant viruses. The results of cloning and sequencing of the T12E revertant virus revealed that a G-to-A nucleotide substitution had occurred, resulting in a change of codon GAG to AAG, i.e., amino acid E to K, at position 12 in p2. Thus, the mutated T12E virus was rescued by a further substitution of E to K. This finding is consistent with the result that a substitution of T by K at position 12 in p2 did not affect viral replication (Fig. 5).
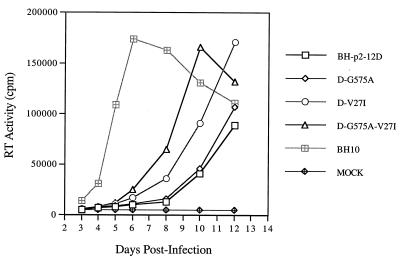
In the case of viruses that reverted from constructs containing the T12D mutation, the results of cloning and sequencing revealed that this mutation was retained in the viral genome. Interestingly, however, two additional mutations were discovered through this work, one in the U5 region (i.e., G575A) and the other in the CA protein (i.e., V27I) (Fig. 3). We next performed site-directed mutagenesis to generate constructs to directly examine the roles of these mutations in viral replication. The results of Fig. 8 show that the G575A substitution did not substantially improve the infectiousness of BH-p2-12D mutated virus, while V27I had a significant effect in this regard. Combining both G575A and V27I within BH-p2-12D had a further positive impact on infectivity (Fig. 8). Therefore, the V27I mutation in CA, together with the G575A substitution in the U5 region, appeared to be able to correct the defective infectiousness of the T12D mutant.
FIG. 8.
The V27I mutation in CA, together with the G575A substitution in the U5 region, stimulates growth of the BH-p2-12D mutant. For details, see Fig. 4.
The V, L, M, F, and Y amino acid residues can play the same role as I at position 24 in NC to rescue the BH-D2 mutated virus.
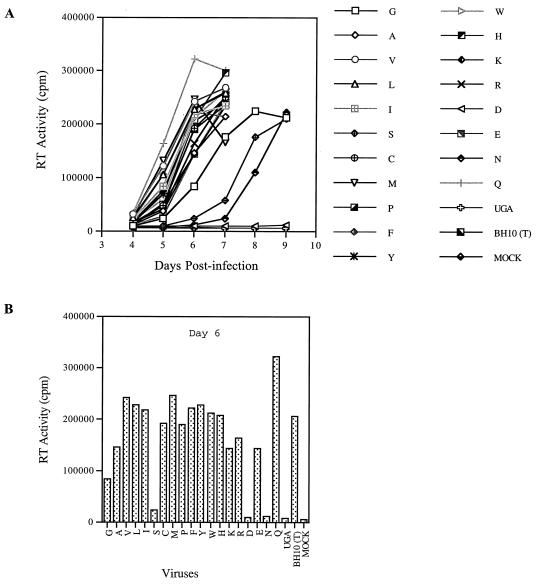
MNC was frequently identified together with MP2 in tissue culture able to rescue the BH-D2 and BH-LD3 deletion (25). It is intriguing to know whether similar findings to those obtained with MP2 would be observed in the case of MNC. Toward this end, T24 was changed to each of 19 other amino acids to generate constructs D2-MP2-NC24X (Fig. 3). The infectiousness of the viruses from these constructs was examined by infecting MT-2 cells. The results of Fig. 9 show that V, L, and M at position 24 in NC were able to rescue the BH-D2 mutated virus as well as did an I substitution. F and Y were less efficient in this regard. Therefore, amino residues with long hydrophobic side chains are favored at position 24 in NC to compensate for the BH-D2 deletion.
FIG. 9.
(A) Effects of various substitutions of the T24 residue in NC on the infectiousness of the BH-D2 mutated viruses. Each substitution is represented by the single-letter abbreviation of amino acids. (B) The RT activity of each virus at day 7 after infection is plotted to clearly show the difference of infectiousness between viruses. For details, refer to Fig. 4.
The S, D, and N amino residues at position 24 in NC dramatically decreased the infectiousness of wild-type virus.
Next, we inserted the above substitutions of T24 into the wild-type virus to examine their effects on viral replication. The results of Fig. 10 show that most of the substitutions did not affect or only moderately affected viral infectivity. The S and N mutations substantially decreased viral replication, and D eliminated viral infectiousness. Therefore, the wild-type virus can use most of the 20 amino acids at position 24 in NC without affecting viral replication. Of note, the presence of either C or H at position 24 in NC did not affect viral growth. This result implies that the presence of C or H at position 24 in NC does not interfere with the binding of Zn2+ ion by the CCHC motif in the first zinc finger.
FIG. 10.
(A) Growth curves of the BH10 viruses containing substitutions of T24 in NC in MT-2 cells. (B) RT activity of each virus at day 6 after infection is plotted as a bar graph to demonstrate the difference of infectivity between viruses. For details, refer to Fig. 4.
Since NC is the major viral structural protein that determines viral RNA packagin efficiency, the effects of the above NC substitutions on packaging was analyzed by RT-PCR. The results of Fig. 11 show that the acidic amino acid residue D displayed the most adverse impact in this regard, while G, A, S, H, R, E, and N moderately diminished viral RNA packaging.
FIG. 11.
Viral genomic RNA levels in the BH10 viruses containing various substitutions of the T24 residue in NC. Substitutions are represented by the single letters of amino acids. Viral RNA from viruses equivalent to 200 pg of CA antigen was subjected to RT-PCR using primer pair pGAG1-pST (23). The intensity of each band was quantified by the NIH Image program and plotted.
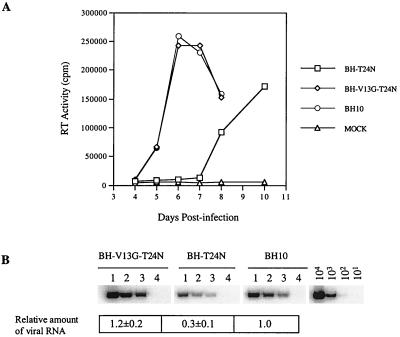
To gain further insight into the mechanisms whereby S, D, and N at position 24 in NC could affect HIV-1 replication, these mutant viruses were cultured in MT-2 cells for a prolonged period. Wild-type replication kinetics were observed for S and N, while neither cytopathology nor RT activity were detected in cultures in the case of the D mutant, even after 3 months. The results of cloning and sequencing of the S revertant virus revealed that the S amino acid had been changed to an L, which was shown to confer wild-type infectivity to the virus (Fig. 10). In the case of the N revertant, N was retained at position 24 in NC, while the V13 amino acid in NC was changed to a G (Fig. 3). When T24N and V13G were recombined through site-directed mutagenesis, the virus thus generated, i.e., BH-V13G-T24N, showed wild-type replication capacity in MT-2 cells (Fig. 12A). The results of RT-PCR indicated that this BH-V13G-T24N recombinant virus packaged wild-type levels of viral RNA (Fig. 12B).
FIG. 12.
(A) Infectiousness of the BH10 wild-type virus and the BH-T24N, as well as BH-V13G-T24N mutated viruses in MT-2 cells. (B) Viral genomic RNA levels in the wild-type and mutated viruses determined by RT-PCR using primer pair pGAG1-pST. RNA samples of each virus were diluted 1:2 and 1:4 to ensure the linear range of the reactions. Wild-type viral DNA standards of 101, 102, 103, and 104 copies were used to determine the linear range of reactions. Lane 1, viral RNA from viruses equivalent to 200 pg of CA Ag; lane 2, 100 pg of CA Ag; lane 3, 50 pg of CA Ag. Intensity of RNA signals was quantified using the NIH Image program with the levels of wild-type BH10 arbitrarily set at 1.0.
DISCUSSION
We have previously deleted various sequences between the primer-binding site and the 5′ SD site that impact on viral RNA packaging and dimerization. Interestingly, long-term culture of all the mutated viruses gave rise to the MP2 and MNC compensatory mutations in the Gag protein (23, 25). In this study, we further showed that, in addition to the MP2 (T12I in p2) substitution that was identified in culture, amino acids V, L, M, and C at position 12 in p2 can also rescue the aforementioned deletions. Among them, V, L, I, and M rescued the impaired infectivity of the mutated viruses to similar levels, while C acted in this regard to a lower extent. Similar findings were made with the MNC mutation (T24I in NC). Screening studies identified six amino acids, including V, L, I, M, F, and Y, at position 24 in NC that can help to rescue the deletions; among these, F and Y are the least efficient.
Our data argue that a nucleotide change from C to T in the MP2 and MNC mutations did not exert compensatory effects in a cis manner. The results of cotransfection experiments demonstrated that the nucleotide change from C to T in MP2 and MNC did not overcome the crippled packaging efficiency of the LD3 viral RNA (Fig. 2). Furthermore, the V, L, I, and M amino acids are coded for by different nucleotides, yet they all can rescue the LD3 deletions when present at position 12 in p2 and at position 24 in NC. Therefore, it is the hydrophobic nature of the substituted amino residues at position 12 in p2 and at position 24 in NC, rather than the altered nucleotides themselves, that rescued the deletion mutations.
One question concerning these findings is why only the I residue was identified in culture if the V, L, and M amino acids can also replace the T12 in p2 or T24 in NC to rescue the mutated viruses. This may simply be because only a single nucleotide change, i.e., ACC→AUC, is required to achieve a T-to-I substitution; however, each of the other amino acid changes require mutations at two or three base positions. It is also noted from the above results that the V, L, I, and M residues are identified both in p2 and in NC as the most efficient ones in the rescue process. Since V, L, I, and M are selected from 20 amino acids and all contain long hydrophobic side chains, we hypothize that specific interactions of a hydrophobic nature involving these residues in p2 and NC with other sequence of Gag or with viral RNA are essential to the rescue of the BH-D1, BH-D2, and BH-LD3 mutated viruses.
In addition to SL1, several other RNA elements are also involved in HIV-1 RNA packaging. Based on their affinity to the Gag protein, as assessed by in vitro binding experiments, the involved RNA elements can be divided into two groups. The TAR and poly(A) hairpin show low affinity for Gag, while SL1 and SL3 exhibit high affinity (5, 8). It is understood that SL1 and SL3 contribute to the packaging process through their tight binding to Gag, yet the mechanism remains elusive by which TAR and the poly(A) hairpin affect RNA packaging. Because of the multipartite nature of packaging signals, mutation of either one of these signals cannot totally exclude viral RNA from virus particles. Moreover, the mutated viruses may revert by modifying the Gag protein to increase its binding affinity to the unchanged packaging signals, so that the RNA packaging in the mutated viruses can be restored to wild-type levels. Our compensation analysis of the SL1 sequence may be an example of this kind.
The nuclear magnetic resonance (NMR) structure of the NL4-3 NC and the SL3 RNA complex indicated a direct contact between the I24 residue of NC and SL3 RNA, which implies that I24 participates in packaging (12). However, the 24th position of NC in BH10 is occupied by a T residue. Since multiple RNA elements are involved in the packaging process, although the NC of BH10 contains a T24 residue and may show a low affinity to SL3, interactions of NC with SL1 or other viral RNA elements may still allow the virus to recruit wild-type levels of viral RNA. However, when the SL1 RNA sequence is deleted, a T24I change is necessary to strengthen the interactions between NC and SL3 RNA, such that the mutant virus can package wild-type levels of viral RNA. Since V, L, and M contain similar hydrophobic side chains to I, they may establish similar interactions with SL3 RNA when present at position 24 in NC and correct the defective viral RNA packaging in the mutated viruses.
Another interesting finding is that a substitution of T24N in the first zinc finger motif of NC in BH10 can be compensated for by another point mutation, i.e., V13A in the same zinc finger. According to the NMR structure of NC and the SL3 RNA complex, the V13 and I24 amino acid residues, together with F16 and A25, form a hydrophobic cleft to which the G9 of SL3 can bind. Moreover, V13 and I24 contribute to the formation of a compact structure of NC by hydrophobic interactions with F6 (12). Therefore, V13 and I24 are important residues in NC. It is possible that substitution of T24 in BH10 with N may introduce steric interference with the V13 residue and further lead to distortion of NC conformation. The G amino acid contains an -H as its side chain and therefore can release the possible steric hindrance when substituted for V13. This issue needs to be tested by NMR analysis of the mutated NC protein binding to the SL3 RNA.
It is also interesting that the V27I substitution in CA can stimulate growth of the BH-p2-12D mutant. V27 is located in a region that forms “helix 1” in the CA protein (14, 15) (Fig. 3). Both crystallographic and mutagenesis studies suggest that proteolysis between MA and CA is followed by refolding and that the helices of two distinct CA molecules, including “helix 1,” then create a new CA-CA interface essential for the formation of a condensed conical core (36). The V27I mutation may adjust this process to correct deficits caused by the T12D mutation in p2. On the other hand, identification of the V27I mutation in CA also suggests possible interactions of p2 with certain regions of CA.
Proteolytic cleavage of Gag gives rise to four distinct proteins, i.e., MA, CA, NC, and p6. Although the structure of each of these proteins has been determined, their conformation within the context of the Gag protein is not fully resolved. The fact that wild-type virus can successfully accommodate 15 of 20 amino acids at position 12 in p2 suggests that this residue is not normally involved in crucial interactions with other regions of Gag. However, the presence of some amino acids at this position, e.g., W, D, or E, may conceivably exert negative effects on Gag conformation and may, therefore, result in abnormal Gag processing, RNA packaging, and viral replication. The V, L, I, and M substitutions may have established novel interactions that are crucial for the viability of the deleted virus but insignificant in regard to wild-type virus.
ACKNOWLEDGMENTS
This research was supported by grants from the Canadian Institutes of Health Research (CIHR).
We thank Mervi Detorio and Maureen Oliveira for technical assistance.
REFERENCES
- 1.Awang G, Sen D. Mode of dimerization of HIV-1 genomic RNA. Biochemistry. 1993;32:11453–11457. doi: 10.1021/bi00093a024. [DOI] [PubMed] [Google Scholar]
- 2.Baudin G, Marquet R, Isel C, Darlix J L, Ehresmann B, Ehresmann C. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229:382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 3.Berkhout B, van Wamel J L B. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Berkowitz R D, Fisher J, Goff S P. RNA packaging. Curr Top Microbiol Immunol. 1996;214:177–218. doi: 10.1007/978-3-642-80145-7_6. [DOI] [PubMed] [Google Scholar]
- 5.Berkowitz R D, Goff S P. Analysis of binding elements in the human immunodeficiency virus type 1 genomic RNA and nucleocapsid protein. Virology. 1994;202:233–246. doi: 10.1006/viro.1994.1339. [DOI] [PubMed] [Google Scholar]
- 6.Buck C B, Shen X, Egan M A, Pierson T C, Walker C M, Siliciano R F. The human immunodeficiency virus type 1 gag gene encodes an internal ribosome entry site. J Virol. 2001;75:181–191. doi: 10.1128/JVI.75.1.181-191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Clever J L, Parslow T G. Mutant human immunodeficiency virus type 1 genomes with defects in RNA dimerization or encapsidation. J Virol. 1997;71:3407–3414. doi: 10.1128/jvi.71.5.3407-3414.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Clever J, Sassetti C, Parslow T G. RNA secondary structure and binding sites for gag gene products in the 5′ packaging signal of human immunodeficiency virus type 1. J Virol. 1995;69:2101–2109. doi: 10.1128/jvi.69.4.2101-2109.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Clever J L, Wong M L, Parslow T G. Requirement for kissing-loop-mediated dimerization of human immunodeficiency virus RNA. J Virol. 1996;70:5902–5908. doi: 10.1128/jvi.70.9.5902-5908.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Das A T, Klaver B, Berkhout B. The 5′ and 3′ TAT elements of human immunodeficiency virus exert effects at several points in the virus life cycle. J Virol. 1998;72:9217–9223. doi: 10.1128/jvi.72.11.9217-9223.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das A T, Klaver B, Klasens B I F, van Wamel J L B, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 genome is essential for replication. J Virol. 1997;71:2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.De Guzman R N, Wu Z R, Stalling C C, Pappalardo L, Borer P N, Summers M F. Structure of the HIV-1 nucleocapsid protein bound to the SL3 Ψ-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 13.Fu W, Gorelick R J, Rein A. Characterization of human immunodeficiency virus type 1 dimeric RNA from wild-type and protease-defective virions. J Virol. 1994;68:5013–5018. doi: 10.1128/jvi.68.8.5013-5018.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gamble T R, Vajdos F F, Yoo S, Worthylake D K, Houseweart M, Sundquist W I, Hill C P. Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid. Cell. 1996;87:1285–1294. doi: 10.1016/s0092-8674(00)81823-1. [DOI] [PubMed] [Google Scholar]
- 15.Gitti R K, Lee B M, Walker J, Summers M F, Yoo S, Sundquist W I. Structure of the amino-terminal core domain of the HIV-1 capsid protein. Science. 1996;273:231–235. doi: 10.1126/science.273.5272.231. [DOI] [PubMed] [Google Scholar]
- 16.Harrich D, Hooker C W, Parry E. The human immunodeficiency virus type 1 TAR RNA upper stem-loop plays distinct roles in reverse transcription and RNA packaging. J Virol. 2000;74:5639–5646. doi: 10.1128/jvi.74.12.5639-5646.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harrison G P, Lever A M L. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66:4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Helga-Maria C, Hammarskjold M L, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficiency packaging of human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73:4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaddrick M, Lear A L, Cann A J, Heaphy S. Evidence that a kissing loop structure facilitates genomic RNA dimerization in HIV-1. J Mol Biol. 1996;259:58–68. doi: 10.1006/jmbi.1996.0301. [DOI] [PubMed] [Google Scholar]
- 20.Laughrea M, Jette L. A 19-nucleotide sequence upstream of the 5′ major splice donor is part of the dimerization domain of human immunodeficiency virus 1 genomic RNA. Biochemistry. 1994;33:13464–13474. doi: 10.1021/bi00249a035. [DOI] [PubMed] [Google Scholar]
- 21.Laughrea M, Jette L. Kissing-loop model of HIV-1 genome dimerization: HIV-1 RNAs can assume alternative dimeric forms, and all sequences upstream or downstream of hairpin 248–271 are dispensible for dimer formation. Biochemistry. 1996;35:1589–1598. doi: 10.1021/bi951838f. [DOI] [PubMed] [Google Scholar]
- 22.Laughrea M, Jette L. HIV-1 genomic dimerisation: formation kinetics and thermal stability of dimeric HIV-1 LAI RNAs are not improved by the 1–232 and 292–790 regions flanking the kissing loop domain. Biochemistry. 1996;35:9366–9374. doi: 10.1021/bi960395s. [DOI] [PubMed] [Google Scholar]
- 23.Liang C, Rong L, Laughrea M, Kleiman L, Wainberg M A. Compensatory point mutations in the human immunodeficiency virus type 1 Gag region that are distal from deletion mutations in the dimerization initiation site can restore viral replication. J Virol. 1998;72:6629–6636. doi: 10.1128/jvi.72.8.6629-6636.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Liang C, Rong L, Quan Y, Laughrea M, Kleiman L, Wainberg M A. Mutations within four distinct Gag proteins are required to restore replication of human immunodeficiency virus type 1 after deletion mutagenesis within the dimerization initiation site. J Virol. 1999;73:7014–7020. doi: 10.1128/jvi.73.8.7014-7020.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang C, Rong L, Russell R S, Wainberg M A. Deletion mutagenesis downstream of the 5′ long terminal repeat of human immunodeficiency virus type 1 is compensated for by point mutations in both the U5 region and gag gene. J Virol. 2000;74:6251–6261. doi: 10.1128/jvi.74.14.6251-6261.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marquet R, Paillart J C, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of human immunodeficiency virus type 1 RNA involves sequences located upstream of the splice donor site. Nucleic Acids Res. 1994;22:145–151. doi: 10.1093/nar/22.2.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McBride M S, Panganiban A T. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McBride M S, Panganiban A T. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71:2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McBride M S, Schwartz M D, Panganiban A T. Efficiency encapsidation of human immunodeficiency virus type 1 vectors and further characterization of cis elements required for encapsidation. J Virol. 1997;71:4544–4554. doi: 10.1128/jvi.71.6.4544-4554.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Muriaux D, Girard P M, Bonnet-Mathoniere B, Paoletti J. Dimerization of HIV-1 LAI at low ionic strength. An autocomplementary sequence in the 5′ leader region is evidenced by an antisense oligonucleotide. J Biol Chem. 1995;270:8209–8216. doi: 10.1074/jbc.270.14.8209. [DOI] [PubMed] [Google Scholar]
- 31.Paillart J C, Berthoux L, Ottmann M, Darlix J L, Marquet R, Ehresmann B, Ehresmann C. A dual role of the putative RNA dimerization initiation site of HIV-1 in genomic RNA packaging and proviral synthesis. J Virol. 1996;70:8348–8354. doi: 10.1128/jvi.70.12.8348-8354.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paillart J C, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269:27486–27493. [PubMed] [Google Scholar]
- 33.Paillart J C, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc Natl Acad Sci USA. 1996;93:5572–5577. doi: 10.1073/pnas.93.11.5572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Paillart J C, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Noncanonical interactions in a kissing loop complex: the dimerisation initiation site of HIV-1 genomic RNA. J Mol Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 35.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 36.Schwedler U K, Stemmler T L, Klishko V Y, Li S, Albertine K H, Davis D R, Sundquist W I. Proteolytic refolding of the HIV-1 capsid protein amino-terminus facilitates viral core assembly. EMBO J. 1998;17:1555–1568. doi: 10.1093/emboj/17.6.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Skripkin E, Paillart J C, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]